Abstract
Cardiac troponin I (cTnI) is the only sarcomeric protein identified to date that is expressed exclusively in cardiac muscle. Its expression in cancer tissues has not been reported. Herein, we examined cTnI expression in non-small cell lung cancer (NSCLC) tissues, human adenocarcinoma cells SPCA-1 (lung) and BGC 823 (gastric) by immunohistochemistry, western blot analysis and real-time PCR. Immunopositivity for cTnI was demonstrated in 69.4% (34/49) NSCLC tissues evaluated, and was strong intensity in 35.3% (6/17) lung squamous cell carcinoma cases. The non-cancer-bearing lung tissues except tuberculosis (9/9, 100%) showed negative staining for cTnI. Seven monoclonal antibodies (mAbs) against human cTnI were applied in immunofluorescence. The result showed that the staining pattern within SPCA-1 and BGC 823 was dependent on the epitope of the cTnI mAbs. The membrane and nucleus of cancer cells were stained by mAbs against N-terminal peptides of cTnI, and cytoplasm was stained by mAbs against the middle and C-terminal peptides of cTnI. A ~25 kD band was identified by anti-cTnI mAb in SPCA-1 and BGC 823 extracts by western blot, as well as in cardiomyocyte extracts. The cTnI mRNA expressions in SPCA-1 and BGC 823 cells were about ten thousand times less than that in cardiomyocytes. Our study shows for the first time that cTnI protein and mRNA were abnormally expressed in NSCLC tissues, SPCA-1 and BGC 823 cells. These findings challenge the conventional view of cTnI as a cardiac-specific protein, enabling the potential use of cTnI as a diagnostic marker or targeted therapy for cancer.
Keywords: Cardiac troponin I, non-small cell lung cancer, human lung adenocarcinoma cell line SPCA-1, human gastric adenocarcinoma cell line BGC 823
Introduction
Troponin I (TnI) is a critical regulatory thin filament protein which is expressed in striated muscle. Along with the other subunits of troponin and tropomyosin, it participates in the control of muscle contraction by Ca2+. Mammals have three genes expressing TnI known as slow twitch (TNNI1), fast twitch (TNNI2) and cardiac (TNNI3). Human TNNI3 gene is located at 19q13.4, and codes for approximately 210 amino acids (24-kD) protein. This cardiac TnI isoform (cTnI) protein contains a unique N-terminal extension about 30 amino acids, compared with the fast and slow skeletal isoforms of TnI [1-3].
The cTnI is the only sarcomeric protein identified to date that is expressed exclusively in cardiac muscle, and has been found in nuclei of cardiomyocytes by experimental evidences recently [4-6]. The expression of cTnI in tumor cells has not been reported. In the present study, we sought to demonstrate cTnI expression in lung tumor tissues and two cancer cell lines using immunohistochemistry, immunofluorescence, real-time PCR, and western blot analysis.
Material and methods
Tissues
Forty-nine formalin-fixed, paraffin-embedded blocks were provided by Departments of Cardiothoracic Surgery and Respiratory Diseases of Jinling Hospital. The specimens were obtained from patients diagnosed with lung adenocarcinoma (AC, n = 21), lung squamous cell carcinoma (SCC, n = 17), lung adenosquamous carcinoma (ASC, n = 3), lung large cell carcinoma (LC, n = 2), lung metastatic sarcoma (n = 1), lung tuberculosis (TB, n = 3), pulmonary sclerosing hemangioma (PSH, n = 1), and pulmonary inflammatory pseudotumor (n = 1) through pathological evidence from June 2007 to June 2008. Meanwhile, seven non-cancer-bearing lung tissues obtained away from the tumor area were submitted for histology. Patients with non-small cell lung cancer (NSCLC) were staged according to the tumor-node-metastasis (TNM) system. Patients did not receive chemo-, radio- or immuno-therapy prior to specimen collection. This research has been authorized by the principle committee of Jinling Hospital.
Cell culture
Human lung adenocarcinoma cell line SPCA-1 and human gastric adenocarcinoma cell line BGC 823 were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and characterized by mycoplasma detection, DNA-fingerprinting, isoenzymes analysis and cell viability detection. In addition, cultured neonatal Sprague-Dawley (SD) rat cardiomyocytes and endothelial cells were harvested as cTnI positive and negative control, respectively. These cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Wisent Inc., Montreal, QC, Canada) supplemented with 20% (v/v) fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), 100 U/ml of penicillin and 100 μg/ml of streptomycin. Cells were incubated at 37°C in a humidified air atmosphere supplemented with 5% CO2. The culture mediums were changed every two days. The study protocol was approved by Medical Ethical Committee of the First Affiliated Hospital of Nanjing Medical University.
Mouse anti-human cTnI monoclonal antibodies
The mouse anti-cTnI monoclonal antibody (mAb) 2F6.6 was a generous gift from Dr. Jack H. Ladenson (Division of Laboratory Medicine, Department of Pathology, Washington University School of Medicine, St. Louis, USA). The mouse anti-cTnI mAbs 19C7, 16A11 and 84 were purchased from Hytest (Turku, Finland). In addition, we obtained three mouse anti-cTnI mAbs XY15, XY13, and XY10 by conventional hybridoma method in our lab. The mAbs 19C7 and XY15 belong to IgG2b, and XY13, XY10, 16A11 and 84 belong to IgG1. The epitope specificities of 2F6.6, 19C7, XY15, 16A11, XY13, 84 and XY10 are 13-36, 41-49, 56-80, 86-90, 76-110, 117-126 and 181-210 of human cTnI, respectively.
Immunohistochemical (IHC) staining of cTnI on the tissues of patients with lung diseases
Sections of 4 μm were deparaffinized by xylene washes, and tissues were rehydrated by descending concentrations of ethanol (100%, 95%, 70%, 50%, 30%), finishing with H2O. Tissues were heated to boiling in citrate-based buffer (Vector Laboratories, Burlingame, CA, USA) 3 × 5 minutes for antigen retrieval. Endogenous peroxidase activity was inhibited using 1% hydrogen peroxide for 20 minutes. Then, tissues were blocked with 2.5% normal horse serum for 30 minutes (R.T.U. Vectorstain universal Elite ABC kit). After that, the blocking buffer were removed by aspiration, and tissues were incubated with the primary antibody 2F6.6 or 19C7 at concentration of 5 μg/ml diluted in TBST containing 1% BSA at 4°C overnight. After washing, horse anti-mouse/anti-rabbit biotinylated secondary antibody was applied for 30 minutes. Unbound secondary antibody was removed by washing with TBST. Tissues were then incubated with the avidin-peroxidase complex for 30 minutes at room temperature (R.T.U. Vectorstain universal Elite ABC kit), washed with TBST, and visualization of bound complex was achieved by adding a solution of 3,3’ diaminobenzidine (DAB kit; Vector Laboratories, Burlingame, CA, USA) for 8-10 minutes. Tissues were counterstained with hematoxylin (Sigma-Aldrich, St Louis, MO, USA) and finally dehydrated in ascending concentrations of ethanol, washed in xylene, and coverslipped using Permount mounting medium. The cTnI immunopositivity was scored according to staining intensity (0, negative; 1, weak staining; 2, moderate staining; and 3, strong staining).
Immunofluorescence (IF) staining of cTnI on cultured cancer cells
Cultured cells seeded on dishes (NEST, Wuxi, Jiangsu, China) were washed in phosphate-buffered saline (PBS) for three times. The cells were fixed with 4% paraformaldehyde (PFA; Amresco, Solon, OH, USA) for 20 minutes, incubated with 0.5% Triton-X 100 (Amresco, Solon, OH, USA) for 20 minutes to increase permeability, and blocked with 2.5% normal horse serum (R.T.U. Vectorstain universal Elite ABC kit, Vector Laboratories, Burlingame, CA, USA) for another 30 minutes at room temperature. After that, the blocking buffer was removed by aspiration and the cells were incubated with the primary antibody at concentration of 2 μg/ml diluted in PBS with 1% bovine serum albumin (BSA) at 4°C overnight. After washing, the cells were incubated with horse anti-mouse/anti-rabbit biotinylated secondary antibody for 30 minutes, and incubated for another 10 minutes with Fluorescein Avidin DCS (Vector Laboratories, Burlingame, CA, USA) at 10 μg/ml diluted in PBS. Then, the cells were mounted with VECTASHIELD Mounting medium with DAPI (Vector Laboratories, Burlingame, CA, USA), and captured images under LSM 5 Live DuoScan Laser Scanning Microscope (Zeiss, Oberkochen, Germany).
Western blot
Protein was isolated from the SPCA-1, BGC 823 cells and cultured neonatal rat cardiomyocytes by using RIPA lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail, Beyotime, Shanghai, China) supplemented with 1 mM phenylmethanesulfonyl fluoride (PMSF, Sigma-Aldrich, St. Louis, MO, USA), and was quantified using BCA Protein Assay (Pierce, Rockford, IL, USA). Twenty micrograms of protein per sample was run by 12% SDS-PAGE gel and transferred to the polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). Non-specific binding were blocked by 5% (w/v) skimmed milk in Tris-Buffered Saline containing 0.1% Tween 20 (TBST) for 2 hours at room temperature. Then, the membranes were incubated with anti-cTnI mAb XY15 at concentration of 4 μg/ml diluted in blocking solution at 4°C overnight. After extensively washed with TBST, the blots were then incubated with horseradish-peroxidase-conjugated anti-mouse antibody (1:10000, Sigma-Aldrich, St. Louis, MO, USA) for one hour at room temperature. Finally, the signals were developed by autoradiography using the ECL-Western blotting system (Bio-Rad, Hercules, CA, USA).
Real-time PCR analysis of cTnI gene expression
RNA was extracted from SPCA-1, BGC 823 and cultured neonatal rat cardiomyocytes using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. RNA (0.3 μg) was reverse transcribed using PrimeScriptTM RT Master Mix Perfect Real Time Kit (Takara, Dalian, China), and then the cDNA was amplified using SYBR Premix Ex TaqTM (Takara, Dalian, China) under the following cycle conditions: 30 s at 95°C for 1 cycle, 5 s at 95°C and 30 s at 60°C for 40 cycles with Mastercycler EP realplex (Eppendorf, Hamburg, Germany) in the ABI® 7500 Sequence Detection System (Applied Biosystems, Foster city, CA, USA). Each sample was measured in triplicate. No template controls and no reverse transcriptase controls for each sample were included. The GAPDH gene was used as an endogenous control. The fold change of TNNI3 gene expressions relative to that of cardiomyocytes was analyzed by the 2-ΔΔCt method. Primers sequences were for GAPDH sense 5’-GCACCGTCAAGGCTGAGAAC-3’ and antisense 5’-TGGTGAAGACGCCAGTGGA-3’ (NM_002046, 347-484, 138 bp); for TNNI3 sense 5’-CGTGTGGACAAGGTGGATGA-3’ and antisense 5’-TTCCTTCTCGGTGTCCTCCT-3’ (NM_000363, 450-695, 246 bp).
Results
Immunohistochemistry
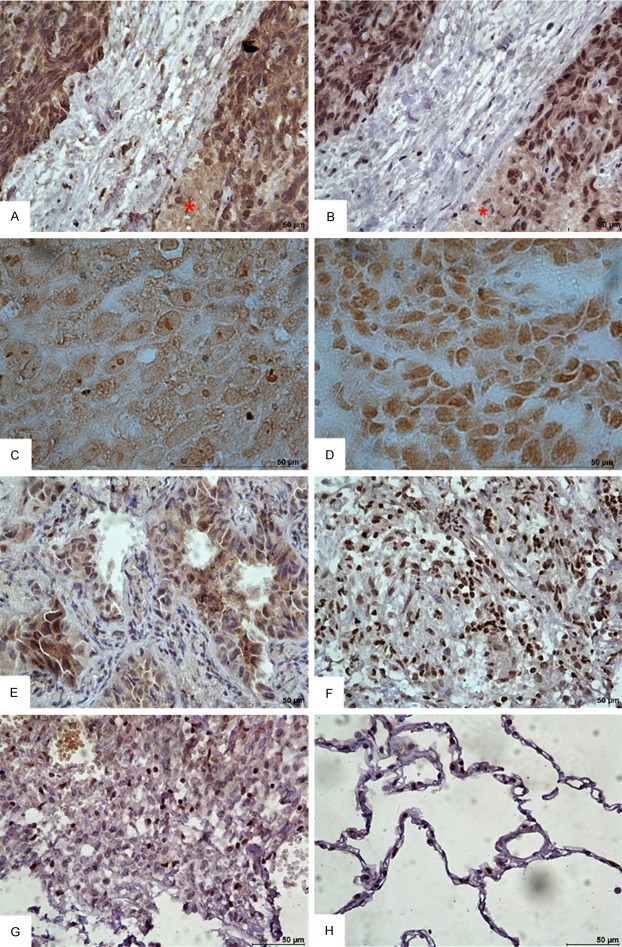
The results of cTnI immunohistochemical staining in lung tissues are summarized in Table 1. Of the 49 cases of NSCLC, 34 (69.4%) demonstrated positive staining with anti-cTnI mAb 2F6.6 or 19C7. The positive staining could be accumulated, diffused or scattered (Figure 1). However, non-cancer-bearing tissues (9/9, 100%) except tuberculosis were negative for cTnI. There was no significant difference in the overall ratios of positive staining (intensity 1-3) between lung adenocarcinoma and lung squamous cell carcinoma (14/21, 67% vs. 11/17, 65%, Pearson chi-square test, P = 0.899). Notably, all 6 cases of the strong staining were lung squamous cell carcinoma. The mAbs 2F6.6 and 19C7 staining overlapped in the tissues from the same patient (Figure 1A and 1B). But with higher magnification and without counterstaining, it could be distinguished that the cell membrane and the inner local area were stained by 2F6.6 (Figure 1C), and nucleus was stained by 19C7 (Figure 1D). The immunopositivity of cTnI had no connection with TNM stage (P = 0.839), lymph node status (P = 0.281), tumor size and invasiveness (P = 0.721) in these NSCLC patients.
Table 1.
The cTnI immunohistochemistry results in non-small cell lung cancer tissues
| Staining intensity (percentage of total) | ||||
|---|---|---|---|---|
|
|
||||
| 0 | 1 (weak) | 2 (moderate) | 3 (strong) | |
| Lung diseases tissues | ||||
| Adenocarcinoma | 7/21 (33%) | 10/21 (48%) | 4/21 (19%) | 0/21 (0%) |
| Squamous cell carcinoma | 6/17 (35%) | 3/17 (18%) | 2/17 (12%) | 6/17 (35%) |
| Adenosquamous carcinoma | 0/3 (0%) | 1/3 (33%) | 2/3 (67%) | 0/3 (0%) |
| Lung large cell carcinoma | 0/2 (0%) | 1/2 (50%) | 1/2 (50%) | 0/2 (0%) |
| Metastatic sarcoma | 0/1 (0%) | 1/1 (100%) | 0/1 (0%) | 0/1 (0%) |
| Tuberculosis | 1/3 (33%) | 1/3 (33%) | 1/3 (33%) | 0/3 (0%) |
| Non-cancer lung diseases | 2/2 (100%) | 0/2 (0%) | 0/2 (0%) | 0/2 (0%) |
| Non-cancer-bearing lung tissues | 7/7 (100%) | 0/7 (0%) | 0/7 (0%) | 0/7 (0%) |
| NSCLC TNM stage | ||||
| I + II | 8/27 (30%) | 9/27 (33%) | 7/27 (26%) | 3/27 (11%) |
| III + IV | 4/15 (27%) | 6/15 (40%) | 2/15 (13%) | 3/15 (20%) |
| NSCLC Lymph node status | ||||
| + | 7/19 (37%) | 8/19 (42%) | 1/19 (5%) | 3/19 (16%) |
| - | 5/23 (22%) | 7/23 (30%) | 8/23 (35%) | 3/23 (13%) |
| NSCLC Tumor size and invasiveness | ||||
| T1 + T2 | 9/33 (27%) | 11/33 (33%) | 8/33 (24%) | 5/33 (15%) |
| T3 + T4 | 3/9 (33%) | 4/9 (44%) | 1/9 (11%) | 1/9 (11%) |
Figure 1.
Representative immunohistochemical staining images for cTnI in human NSCLC tissues (DAB, bar = 50 μm). (A, B) Immunohistochemical staining (with hematoxylin counterstaining) in the lung squamous cell carcinoma tissue from the same patient. (A) Stained with 2F6.6; (B) Stained with 19C7. All showed strong staining. Asterisk showed cellular necrosis area. (C, D) Immunohistochemical staining without hematoxylin counterstaining and with higher magnification. (C) Stained with 2F6.6; (D) Stained with 19C7. (E, F) The tissues from adenocarcinoma and tuberculosis showed positive staining. (E) Adenocarcinoma, (F) Tuberculosis. (G, H) The pulmonary sclerosing hemangioma and non-cancer-bearing lung tissue showed negative staining. (G) Pulmonary sclerosing hemangioma, (H) Non-cancer-bearing lung tissue collected away from tumor area.
Immunofluorescence
Then, we performed the immunofluorescence staining in SPCA-1 and BGC 823 cells (Figures 2 and 3) by using different mAbs against human cTnI. The staining pattern within SPCA-1 and BGC 823 was dependent on the epitope of the cTnI antibodies. The intensive staining was occurred at cell membrane and nucleolus by using 2F6.6 (13-36) and nucleus except nucleolus by using 19C7 (41-49). The cytoplasmic staining by using XY15 (56-80), 16A11 (86-90), XY13 (76-110), 84 (117-126) and XY10 (181-210) showed differential intensity, in which XY15 was the strongest one in BGC 823 cells (Figure 3) and 16A11, XY13, 84 were weak in SPCA-1 (Figure 2). Furthermore, we confirmed the cTnI immunofluorescence staining in human hepatoma cell lines such as Huh-7,MHCC-97L and MHCC-97H cells (Figure 6) by using 2F6.6 and XY15. All the cells showed intensive staining in cytoplasm by XY15 and nucleolus by 2F6.6. The positive staining of cell membrane by using 2F6.6 was reappeared in MHCC-97L and MHCC-97H cells except Huh-7 cells. The anti-cTnI mAbs 2F6.6, XY15 and XY10 could stain in human embryonic kidney 293 cell (HEK 293) and human embryonic liver cell (L02) (Figure 7).
Figure 2.
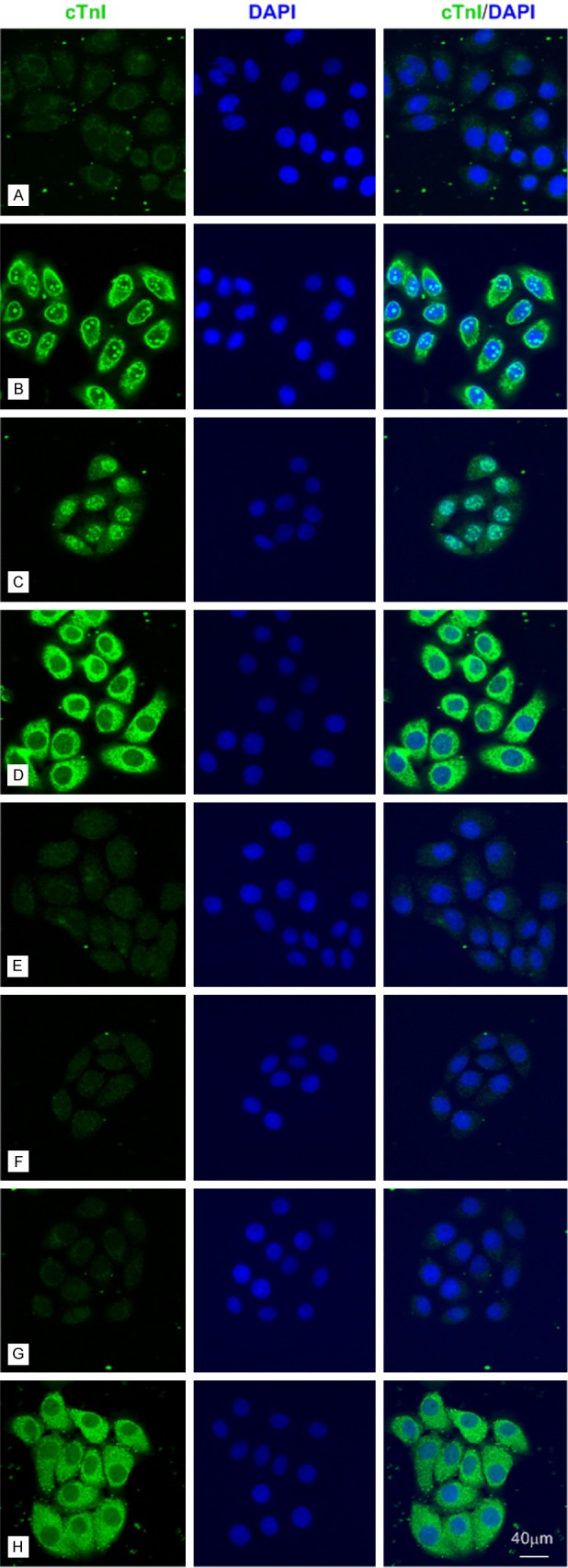
The anti-cTnI mAbs could stain different parts of SPCA-1 cells by immunofluorescence with an epitope-dependent pattern. Representative confocal images of SPCA-1 cells (bar = 40 μm). A: Negative control. B-H: SPCA-1 cells were stained for cTnI (Fluorescein Avidin DCS, green) by using 2F6.6, 19C7, XY15, 16A11, XY13, 84 and XY10 mAbs, which recognized cTnI epitopes embodied at 13-36, 41-49, 56-80, 86-90, 76-110, 117-126 and 181-210, respectively.
Figure 3.
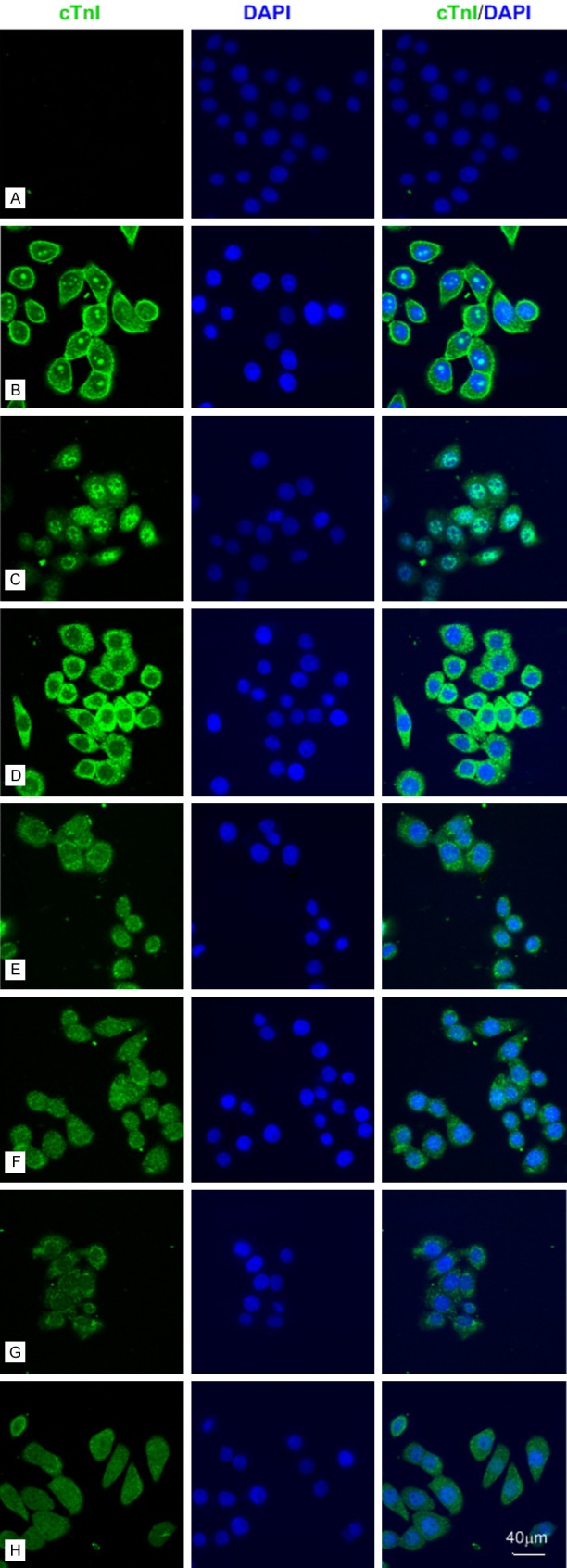
The anti-cTnI mAbs could stain different parts of BGC 823 cells by immunofluorescence with an epitope-dependent pattern. Representative confocal images of BGC 823 cells (bar = 40 μm). A: Negative control. B-H: BGC 823 cells were stained for cTnI (Fluorescein Avidin DCS, green) by using 2F6.6, 19C7, XY15, 16A11, XY13, 84 and XY10 mAbs, which recognized cTnI epitopes embodied at 13-36, 41-49, 56-80, 86-90, 76-110, 117-126 and 181-210, respectively.
Figure 6.
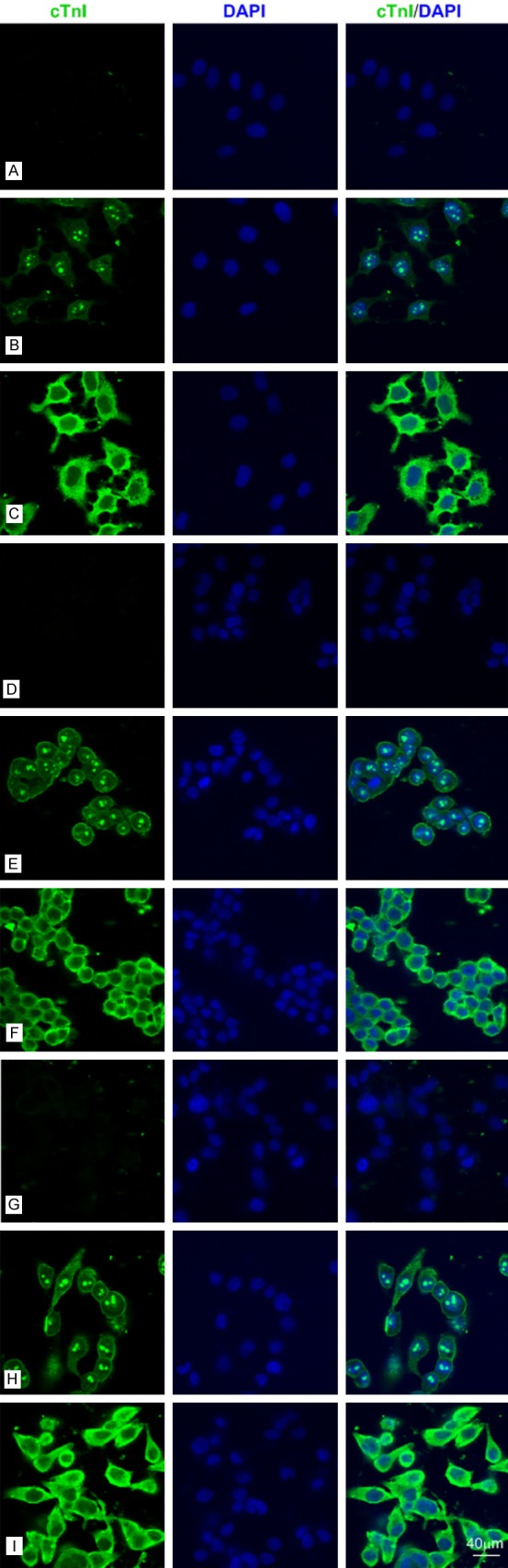
The anti-cTnI mAbs could stain Huh-7 cells, MHCC-97L cells and MHCC-97H cells by IF. A-C: Representative confocal images of Huh-7 cells (bar = 40 μm). A: Negative control. B, C: Huh-7 cells were stained for cTnI (Fluorescein Avidin DCS, green) using 2F6.6 and XY15 mAbs, which recognized epitopes embodied at 13-36 and 56-80, respectively. D-F: Representative confocal images of MHCC-97L cells (bar = 40 μm). D: Negative control. E, F: MHCC-97L cells were stained for cTnI (Fluorescein Avidin DCS, green) using 2F6.6 and XY15 mAbs, which recognized epitopes embodied at 13-36 and 56-80, respectively. G-I: Representative confocal images of MHCC-97H cells (bar = 40 μm). G: Negative control. H, I: MHCC-97H cells were stained for cTnI (Fluorescein Avidin DCS, green) using 2F6.6 and XY15 mAbs, which recognized epitopes embodied at 13-36 and 56-80, respectively.
Figure 7.
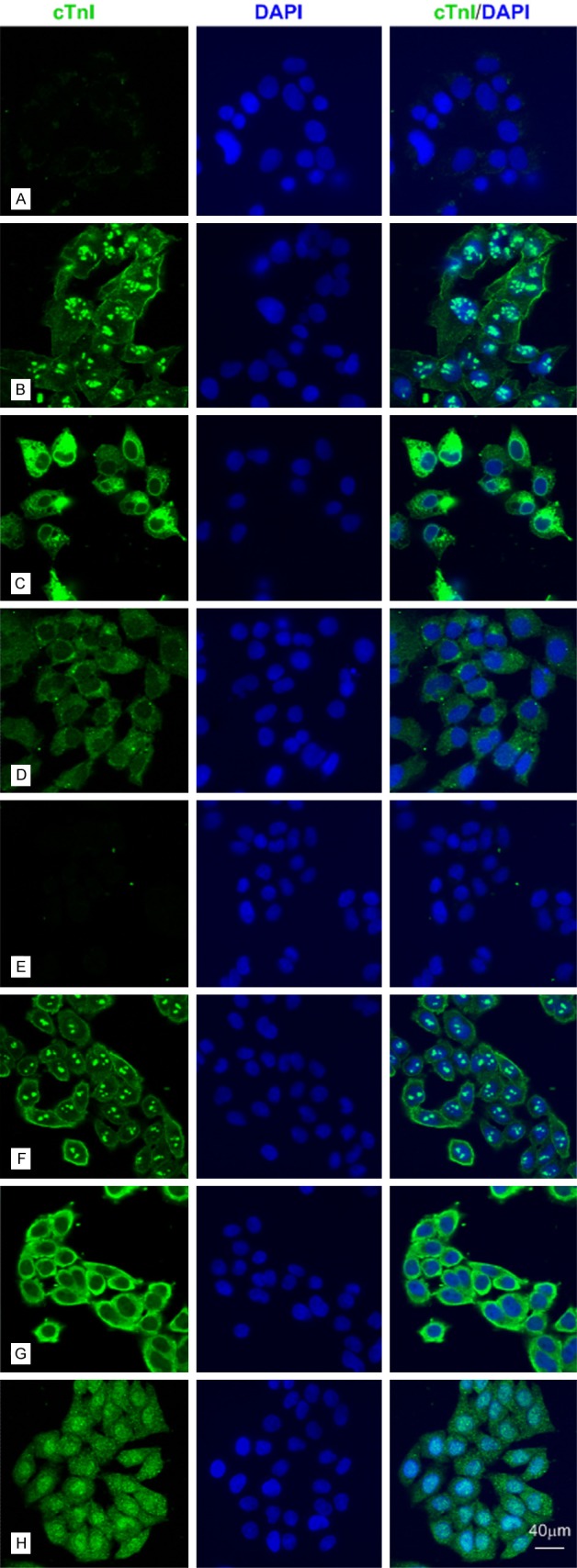
The anti-cTnI mAbs could stain HEK 293 cells and LO2 cells by IF. A-D: Representative confocal images of HEK 293 cells (bar = 40 μm). A: Negative control. B-D: HEK 293 cells were stained for cTnI (green) using 2F6.6, XY15 and XY10 mAbs, which recognized epitopes embodied at 13-36, 56-80 and 181-210, respectively. E-H: Representative confocal images of L02 cells (bar = 40 μm). E: Negative control. F-H: L02 cells were stained for cTnI (green) using 2F6.6, XY15 and XY10 mAbs, which recognized epitopes embodied at 13-36, 56-80 and 181-210, respectively.
The troponin complex is inherently stable. However, TnI is inherently of poor structural stability, and is subject to proteolytic cleavage by proteases. TnI must be complexed with TnC in order to help maintain its conformational structure and stability. We performed the double immunofluorescence staining with a rabbit anti-cTnI polyclonal antibody (Abcam, UK) and the mouse anti-cTnC mAb (H86274M, Biodesign, Memphis, TN, USA) in the cultured neonatal rat cardiomyocytes and endothelial cells, in which the two antibodies co-stained positively in cytoplasm of cardiomyocytes, but negatively in endothelial cells (Figure 4). In this study, the anti-cTnC mAb could stain nucleus of SPCA-1 and BGC 823 cells (Figure 4), which suggested the existence of stable cTnI-cTnC complex. However, the anti-cTnC mAb could not stain in two embryonic cell lines HEK 293 and L02.
Figure 4.
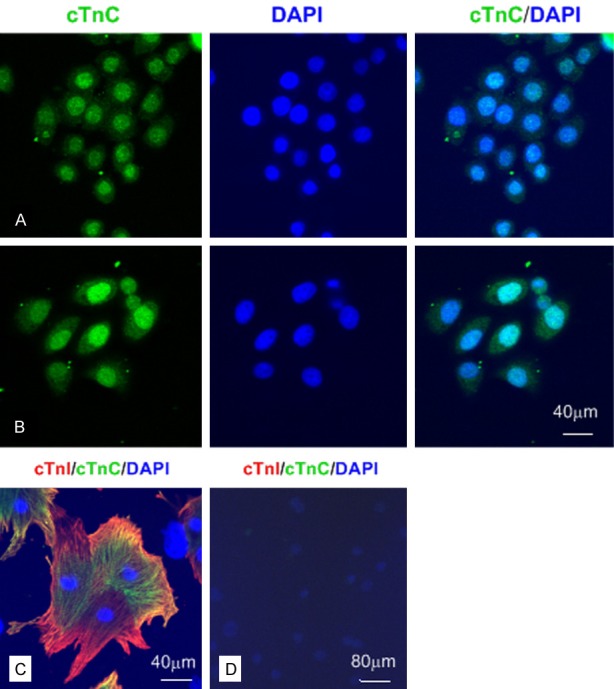
The anti-cTnC mAbs could stain BGC 823 and SPCA-1 cells by IF. Representative confocal images of different cells (A-C: bar = 40 μm; D: bar = 80 μm). A: The positive nucleus staining for cTnC in BGC 823 cells. B: The positive nucleus staining for cTnC in SPCA-1 cells. C: cTnI and cTnC were co-expressed in cytoplasm of cultured neonatal rat cardiomyocytes. D: cTnI and cTnC were negative staining in cultured rat endothelial cells.
Western blot
As can be seen in Figure 5, a ~25 kD band was detected in SPCA-1, BGC 823 and cardiomyocyte extracts using mouse anti-cTnI mAb XY15. Compared with that in cardiomyocyte group, the signals in SPCA-1 and BGC 823 groups were developed with longer time and weaker intensity.
Figure 5.
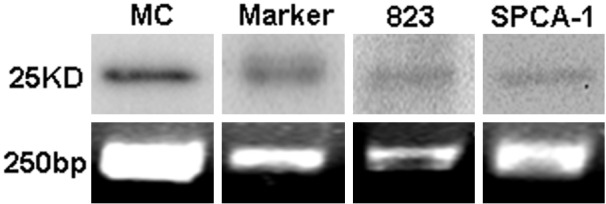
Western blot (upper) and DNA gel electrophoresis of real-time PCR products (lower). MC (cardiomyocyte extract) was regarded as positive control for cTnI in this experiment.
Real-time PCR
The average threshold cycles (Ct) value of negative control (without template) was 35.06 ± 1.44 (n = 4). The average Ct values for TNNI3 of cardiomyocytes, BGC 823 and SPCA-1 were 16.79 ± 3.44, 27.90 ± 0.52 and 27.67 ± 0.25 (n = 3), respectively. Meanwhile, the average Ct values for GAPDH of cardiomyocytes, BGC 823 and SPCA-1 were 17.90 ± 2.30, 15.71 ± 2.24 and 16.31 ± 1.99 (n = 3), respectively. By using the 2-ΔΔCt method, the relative gene expressions of TNNI3 in BGC 823 and SPCA-1 cells to cardiomyocytes were 1.0 × 10-4 and 1.8 × 10-4, respectively (the TNNI3 gene expression in cardiomyocytes was regarded as 1). We collected the PCR products and performed 2% agarose gel electrophoresis. The result showed that there were single bands about 250 bp in groups BGC 823 and SPCA-1, similar to those in the cardiomyocytes (Figure 5).
Discussion
Cardiac troponin I, a subunit of troponin complex expressed in cardiac muscle, is not expressed in skeletal muscle during development or during regenerative muscle diseases [7]. To date, cTnI has been identified as one of the most specific and sensitive biomarkers for myocardial injury [8,9]. Subsequently, cTnI measurement can be used as a valid diagnostic tool for early identification, assessment and monitoring of cardiotoxicity induced by radio-, chemo- and immuno-therapy for patients with cancer [10-12]. Until now, this is the only reported relationship between cancer and cTnI.
The expression of cTnI in cancer tissues has not been reported. In this study, we tested, for the first time, that immunopositive for cTnI was in 69.4% (34/49) NSCLC tissues evaluated, and was strong intensity in 35.3% (6/17) lung squamous cell carcinoma cases. The non-cancer-bearing lung tissues except tuberculosis showed negative staining for cTnI. Then, we tested the cTnI expression in human cancer cell lines SPCA-1 and BGC 823 by using seven different mAbs against human cTnI. The staining pattern was dependent on the epitope of the cTnI antibodies. The mAb 2F6.6 against N-terminal peptides of cTnI (13-36) stained cell membrane and nucleolus. Then, 19C7 (41-49) stained nucleus except nucleolus, and the other mAbs XY15, 16A11, XY13, 84 and XY10 (56-80, 86-90, 76-110, 117-126 and 181-210, respectively) stained cytoplasm with different intensity. The relative gene expressions of cTnI in SPCA-1 and BGC 823 were about ten thousand times less than that in cardiomyocytes. A ~25 kD band was identified by anti-cTnI mAb in SPCA-1 and BGC 823 extracts by western blot, as well as in cardiomyocyte extracts. We also noted that the cTnC staining was positive in nucleus of SPCA-1 and BGC 823 cells, which suggested the existence of stable cTnI-cTnC binding complex. In cardiac sarcomere, cTnI is a rod-like protein with a cardiac specific N-terminal extension region, an IT-arm region, the inhibitory region, the switch region, and the C-terminal mobile domain. The IT-arm region interacts with cTnC and cTnT [13]. We postulated that the whole length cTnI was expressed within cancer cells, and different regions were covered in nuclear and cytoplasm, respectively.
The function of cTnI in sarcomere inhibits the acto-myosin ATPase activity in the absence of activating levels of calcium, blocks actin-myosin interactions, and thereby mediates striated muscle relaxation. The new roles in the new-found location of nuclear cTnI might be related to maintain stable chromosomal integrity or development [14,15]. Moreover, cTnI can be covalently modified—by phosphorylation, for example—and these modifications typically modulate its function. The role and modifications of cTnI expressed in the membrane, nucleus and cytoplasm of cancer cells are unclear.
Here we found that the tumor tissues from some patients with NSCLC presented positive cTnI staining. On the one hand, the result suggested the potential use of cTnI as a marker or targeted therapy for cancer. On the other hand, careful attention must be paid to properly diagnose cardiotoxicity in the patients with anti-cancer therapy with cTnI elevation. In addition, it has been reported that a humoral and cellular autoimmune response against cTnI induced severe inflammation and fibrosis in the myocardium, thus was responsible for the pathogenesis of heart failure [16-19]. The relationship between cTnI-expressed tumor and heart failure in clinic needs more attention.
In conclusion, we report, for the first time, the expression of cTnI within some NSCLC tissues, SPCA-1, and BGC 823 using immunohistochemistry, western blot analysis, and real-time PCR. The function and location of cTnI in tumor tissues and cells need further investigation. These findings challenge the conventional view of cTnI as a cardiac-specific protein, enabling the potential use of cTnI as a diagnostic marker or targeted therapy for cancer.
Acknowledgements
This work has been funded by the National Natural Science Foundation of China (Nos. 81170220 and 81100156), Jiangsu Province Health Foundation (RC2011075) and Priority Academic Program Development of Jiangsu Higher Education Institutions. The authors would like to express their gratitude to Dr. Jack H. Ladenson (Division of Laboratory Medicine, Department of Pathology, Washington University School of Medicine, St. Louis, USA) for his generously providing the anti-cTnI mAbs 2F6.6 and 2B1.9. The authors thank Dr. Yi Zhu (Department of General Surgery, The First Affiliated Hospital of Nanjing Medical University), and Dr. Ping Liu and Dr. Wei Zhu (Department of Oncology, The First Affiliated Hospital of Nanjing Medical University), for their generously providing the cancer cell lines. The authors also thank Dr. Ling-Zhi Hong (Department of Oncology, Nanjing First Hospital, Nanjing Medical University) and Dr. Hong-Xia Li, Dr. Wei-Ming Zhang (Department of Pathology, The First Affiliated Hospital of Nanjing Medical University) for their assistance in obtaining NSCLC tissues and pathological analysis. The authors would like to thank Dr. Bin Zhou (Department of Genetics, Albert Einstein College of Medicine of Yeshiva University, New York, USA) for critical reading, advice and comments of the manuscript.
Disclosure of conflict of interest
None.
References
- 1.Bhavsar PK, Brand NJ, Yacoub MH, Barton PJ. Isolation and characterization of the human cardiac troponin I gene (TNNI3) Genomics. 1996;35:11–23. doi: 10.1006/geno.1996.0317. [DOI] [PubMed] [Google Scholar]
- 2.Solaro RJ. Multiplex kinase signaling modifies cardiac function at the level of sarcomeric proteins. J Biol Chem. 2008;283:26829–26833. doi: 10.1074/jbc.R800037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Layland J, Solaro RJ, Shah AM. Regulation of cardiac contractile function by troponin I phosphorylation. Cardiovasc Res. 2005;66:12–21. doi: 10.1016/j.cardiores.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann O, Zdunek S, Alkass K, Druid H, Bernard S, Frisen J. Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp Cell Res. 2011;317:188–194. doi: 10.1016/j.yexcr.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Chase PB, Szczypinski MP, Soto EP. Nuclear tropomyosin and troponin in striated muscle: new roles in a new locale? J Muscle Res Cell Motil. 2013;34:275–284. doi: 10.1007/s10974-013-9356-7. [DOI] [PubMed] [Google Scholar]
- 7.Bodor GS, Porterfield D, Voss EM, Smith S, Apple FS. Cardiac troponin-I is not expressed in fetal and healthy or diseased adult human skeletal muscle tissue. Clin Chem. 1995;41:1710–1715. [PubMed] [Google Scholar]
- 8.Bodor GS, Porter S, Landt Y, Ladenson JH. Development of monoclonal antibodies for an assay of cardiac troponin-I and preliminary results in suspected cases of myocardial infarction. Clin Chem. 1992;38:2203–2214. [PubMed] [Google Scholar]
- 9.Apple FS. Cardiac troponin: redefining the detection of myocardial infarction. Am Clin Lab. 2002;21:32–34. [PubMed] [Google Scholar]
- 10.Sawaya H, Sebag IA, Plana JC, Januzzi JL, Ky B, Tan TC, Cohen V, Banchs J, Carver JR, Wiegers SE, Martin RP, Picard MH, Gerszten RE, Halpern EF, Passeri J, Kuter I, Scherrer-Crosbie M. Assessment of echocardiography and biomarkers for the extended prediction of cardiotoxicity in patients treated with anthracyclines, taxanes, and trastuzumab. Circ Cardiovasc Imaging. 2012;5:596–603. doi: 10.1161/CIRCIMAGING.112.973321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colombo A, Cipolla C, Beggiato M, Cardinale D. Cardiac toxicity of anticancer agents. Curr Cardiol Rep. 2013;15:362. doi: 10.1007/s11886-013-0362-6. [DOI] [PubMed] [Google Scholar]
- 12.Colombo A, Cardinale D. Using cardiac biomarkers and treating cardiotoxicity in cancer. Future Cardiol. 2013;9:105–118. doi: 10.2217/fca.12.73. [DOI] [PubMed] [Google Scholar]
- 13.Solaro RJ, Rosevear P, Kobayashi T. The unique functions of cardiac troponin I in the control of cardiac muscle contraction and relaxation. Biochem Biophys Res Commun. 2008;369:82–87. doi: 10.1016/j.bbrc.2007.12.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asumda FZ, Chase PB. Nuclear cardiac troponin and tropomyosin are expressed early in cardiac differentiation of rat mesenchymal stem cells. Differentiation. 2012;83:106–115. doi: 10.1016/j.diff.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Sahota VK, Grau BF, Mansilla A, Ferrus A. Troponin I and Tropomyosin regulate chromosomal stability and cell polarity. J Cell Sci. 2009;122:2623–2631. doi: 10.1242/jcs.050880. [DOI] [PubMed] [Google Scholar]
- 16.Okazaki T, Tanaka Y, Nishio R, Mitsuiye T, Mizoguchi A, Wang J, Ishida M, Hiai H, Matsumori A, Minato N, Honjo T. Autoantibodies against cardiac troponin I are responsible for dilated cardiomyopathy in PD-1-deficient mice. Nat Med. 2003;9:1477–1483. doi: 10.1038/nm955. [DOI] [PubMed] [Google Scholar]
- 17.Goser S, Andrassy M, Buss SJ, Leuschner F, Volz CH, Ottl R, Zittrich S, Blaudeck N, Hardt SE, Pfitzer G, Rose NR, Katus HA, Kaya Z. Cardiac troponin I but not cardiac troponin T induces severe autoimmune inflammation in the myocardium. Circulation. 2006;114:1693–1702. doi: 10.1161/CIRCULATIONAHA.106.635664. [DOI] [PubMed] [Google Scholar]
- 18.Kaya Z, Goser S, Buss SJ, Leuschner F, Ottl R, Li J, Volkers M, Zittrich S, Pfitzer G, Rose NR, Katus HA. Identification of cardiac troponin I sequence motifs leading to heart failure by induction of myocardial inflammation and fibrosis. Circulation. 2008;118:2063–2072. doi: 10.1161/CIRCULATIONAHA.108.788711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jahns R. Autoantibodies against cardiac troponin I: friend or foe? Eur J Heart Fail. 2010;12:645–648. doi: 10.1093/eurjhf/hfq098. [DOI] [PubMed] [Google Scholar]