Abstract
WNT3A has been regarded as an activator of the canonical Wnt signaling pathway. It has been found Wnt signaling pathway is closely related with embrionic development and Hirschsprung disease (HSCR). A common haplotype consisting of minor SNPs alleles located in the WNT3A gene has been described as a risk factor for various genetic disorders. However, whether WNT3A contributes to the onset of HSCR has not been identified. The present study aims to detect the interactions of genetic variations in the WNT3A gene and examine the biological expression levels with Hirschsprung disease (HSCR) in the Chinese people. We analyzed WNT3A gene (rs61743220, rs192966556 and rs145882986) variants in the whole blood samples from HSCR patients and normal children (control groups). WNT3A expression was also examined by quantitative real-time PCR (qRT-PCR), western blotting and immunostaining. Consequently, when rs192966556 and rs145882986 alleles of the WNT3A gene lack the SNPs, they are especially associated with a greater risk of HSCR (OR [95% confidence interval] = 1.791, p = 0.001; OR [95% confidence interval] = 1.556, p = 0.003, respectively). The mRNA and protein expressions of WNT3A were higher in the aganglionic colon segment tissues than in the normal ganglionic segments tissues. Immunostaining indicates that the staining of WNT3A was much stronger (brown) in the aganglionic colon segment tissues than that in the normal ganglionic colon segment tissues (colorless or light yellow) in the mucous layer and muscular layer. Although preliminary, these results suggest that WNT3A may play an important role in the pathogenesis of HSCR.
Keywords: Hirschsprung disease, WNT3A, polymorphism, gene and protein, expression
Introduction
Hirschsprung disease (HSCR) is a congenital disorder of the enteric nervous system (ENS) and is characterized by the absence of intestinal ganglion cells in myenteric and submucosal plexuses. Its incidence is approximately 1/ 5,000 human live births, and has a male preponderance of 4:1 [1]. At present, the cause of HSCR still remains unclear, but there is a common understanding that HSCR is a complex disease influenced by multiple genetic and environmental factors. Recent studies have shown that the genetic etiology of the neurocristopathy is complex and many genes may be involved in the development of HSCR [2-4]. It is well known that RET and EDNRB are primary genes in the etiology of HSCR. The RET mutations are associated with the development of HSCR in 50% of cases in familial series, but only 3% of sporadic HSCR cases carry RET mutations. Besides RET and EDNRB, other genes have been identified in sporadic affected individuals, such as endothelin 3 (EDN3) [5], glial derived neurotrophic factor (GDNF) [6], sex determining region Y-box 10 (SOX10) [7], paired-like homeobox 2B (PHOX2B) [8] and ZFHX1B (SIP1) [9]. Thus, HSCR has become a model of a complex polygenic disorder in which the interaction of different genes need to be elucidated.
Wnt signaling is one of the most critical signaling pathways in many aspects of development [10,11]. Wnt signaling is subdivided into the canonical β-catenin dependent and the non-canonical β-catenin independent branch. Canonical Wnt signaling is characterized by the stabilization and subsequent nuclear transport of β-catenin resulting in the activation of transcriptional responses. Non-canonical Wnt signaling is more diverse and includes several different signaling modes that regulate cell behavior. Genes of the Wnt family encode structurally related, cystein-rich glycoproteins involved in cell-cell signalling for a wide variety of developmental processes [12-15]. Wnt can act either on the cell that secrets them (autocrine feedback) or on neighboring cells by paracrine signaling. Wnt signaling pathway plays rather important roles in the process of the embryonic development. It has been shown that the Wnt signaling pathway plays important role in intestinal cell proliferation [16].
WNT3A, a canonical Wnt ligand, has been regarded as an activator of the canonical Wnt signaling pathway. WNT3A has been reported to induce the accumulation of β-catenin and activation of the canonical Wnt signaling pathway [17]. Mice that are homozygous for a hypomorphic wnt3a allele display vertebral defects, a short tail due to loss of caudal vertebrae, deficient cloacal development and incomplete uro-rectal septation [18]. Moreover, studies involving human pluripotent stem cells have shown that WNT3A is required for hindgut specification [19]. Thus, WNT3A is a pivotal component of the mesoderm gene that regulates network with ramifications for embryonic development. The objective of this study was to investigate whether genetic variations in the WNT3A gene are associated with HSCR and examine the biological expression levels of WNT3A in the Chinese people. The single nucleotide polymorphisms (SNPs) in WNT3A gene (rs61743220, rs 192966556 and rs145882986) were selected and analyzed in a group of patients with HSCR and matched control samples. In this study, we further detected differential expressions of WNT3A by qRT-PCR, western blot and immunostaining.
Materials and methods
Patients and specimens
This study was approved by Ethics Committee of China Medical University (Ethical Number: 2013PS07K). Blood samples of 200 HSCR patients were collected from the Department of Pediatric Surgery, Shengjing Hospital of China Medical University. Patients with familial constipation and a history of other congenital gastrointestinal tract malformations were excluded from the study. The patients ranged from 0.5 to 3.5 years old including 156 males and 44 females (average age, 1.5 ± 0.3 years), and were recruited as the HSCR groups. An additional 200 healthy children that matched with the HSCR group in age and gender were used as control groups (average age, 2 ± 0.5 years). The control groups had no history of constipation. Tissue samples (aganglionic and normal ganglionic colon segment tissues) were obtained from 50 HSCR patients (41 males and 9 females) with pathologically confirmed HSCR pre- or post-operatively at Shengjing Hospital of China Medical University. Age ranged from 0.5 to 4.5 years old with an average of 1.5 years. None of these cases received any preoperative treatment or had a history of HSCR’s enterocolitis or active enterocolitis at the time of surgery. Tissue samples were collected immediately after operation and were stored at -80°C until use.
Genomic DNA extraction
Venous blood (200 μl) was obtained from the study participants using EDTA as an anticoagulant. Genomic DNA of peripheral blood white blood cells (WBCs) was extracted according to the QIAamp® DNA Blood Mini Kit Handbook. For the present study, the absorbance value at 260/280 nm (A 260/A 280) ranged from 1.6 to 2.0, which met the requirements for further experiments.
Polymerase chain reaction (PCR), DNA sequence analysis and statistical analysis
We have chosen rs61743220, rs192966556 and rs145882986 for WNT3A gene as our polymorphism analysis loci based on the values of MAF. Specific primers to WNT3A gene were designed using DNASTAR primer design program and synthesized by Invitrogen Co. (Shanghai) (Table 1). The primers have no homology with other genes as determined by BLAST analysis on homology.
Table 1.
Primer sequences and conditions for polymorphism analysis
| SNP locus | Sequence (5’-3’) | Location & Amplicon | Annealing temperature |
|---|---|---|---|
| rs61743220 | F: ATA CAC CAC CCA ACC TCA CG | 306 bp | 57°C |
| R: AGA CAC CAT CCC ACC AAA CT | ‘A/G’ | ||
| rs192966556 | F: AGA GCA AAG GGT CTG TAG C | 237 bp | 55°C |
| R: CAC TCC TGG ATG CCA AT | ‘C/T’ | ||
| rs145882986 | F: CAG GCA AGG GCT GGA AGT | 316 bp | 59°C |
| R: GCT GTG GTG TAG CTG GAT GG | ‘C/T’ |
Genomic DNA from peripheral blood was obtained with QIAamp Blood kits (Takara, Dalian, China) using standard methods [20]. Basically, 150 ng of genomic DNA was amplified in a 50 μl reaction that contained 20 pmol each primer and 2U Taq DNA polymerase (TaKaRa Biotechnology Co.). Conditions for PCR amplification in rs61743220, rs192966556 and rs145882986 of the WNT3A gene fragments are shown in Table 1. A 50 μl reaction system included 10 × PCR buffer solutions, 1 mmol/L of MgCl2, 0.1 mmol/L of dNTP, 10 pmol/L of each of the upstream and downstream primers, 100 ng of DNA template, and 1U of Taq DNA polymerase. PCR condition was: denaturing at 95°C for 3 min, then 35 cycles of denaturing at 95°C for 30 sec, annealing at 55~59°C for 30 sec and elongation at 72°C for 1 min, and finally incubation at 72°C for 7 min. PCR products were electrophoresed on 1.5% agarose gel, stained with ethidium bromide (EB), and visualized with an automatic gel documentation system (TaKaRa Biotechnology Co., Ltd. Japan). The PCR products were sent to Genomics Institute Co. (Beijing, China) for sequence analysis.
qRT-PCR
Total RNA was extracted from the aganglionic and normal ganglionic colon segment tissues with HSCR by using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA syntheses involved 3 μg RNA with the TaKaRa RNA PCR kit (Takara, Dalian, China). The primers of WNT3A used in PCR were as follows: Human WNT3A, sense: GTT CGG GAG GTT TGG G, and Human WNT3A, antisense: CCA GGA AAG CGG ACC AT. The qRT-PCR was performed with a 12.5 μl reaction system in triplicate for each specimen in the presence of SYBR green PCR Master mix (Takara Biotechnology Co.) in a Lightcycler (Roche Molecular Biochemicals, Co.). The housekeeping gene β-actin (Takara, DR3783) was used as an endogenous control. The reaction program was: 5 min pre-denaturation at 95°C and 40 cycles of 5 s of denaturation at 95°C, 30 s of annealing at 56.4°C for WNT3A. After the termination of PCR, the production was analyzed by the Lightcycler system automatically. The amplification process was followed by a melting curve analysis and CT value was recorded. The average CT value was the extreme CT value of the sample. The expression difference of the gene was calculated by the 2-ΔΔct method [21].
Western blot analysis
Approximately 50 mg specimen was minced to small pieces using surgical blades and sonicated in protein lysis buffer. Protein concentrations were measured by the Bradford method, and specimens were adjusted to the same protein concentration, aliquoted and stored at -80°C. Equal amounts of total proteins from tissues were separated on SDS-polyacrylamide gels and then electro-transfer to PVDF membranes (Millipore, USA). The blots were incubated with rabbit polyclonal WNT3A antibody (1:1000, Polyclonal rabbit anti-WNT3A, Millipore Co. Catalog Number 09-162) overnight at 4°C; washed and then incubated with horseradish peroxidase-linked secondary antibodies (1:2000) for 2 h at room temperature and detected using an enhanced chemiluminescence (ECL, Pierce Biotechnology, Rockford, IL, USA) kit. The grayscale values of the WNT3A band were normalized to the values of the corresponding β-actin band to determine the expression level of the protein. The experiments were repeated 3 times independently.
Hematoxylin and eosin staining and immunostaining staining
Diagnosis of HSCR was based on hematoxylin and eosin (H&E) staining of ganglion cells. It is confirmed by the review of surgical pathological reports from resections following each biopsy diagnosis of HSCR whether the diagnosis was correct or not. Aganglionic segment and ganglionic segment were obtained according to H&E staining (Figure 1). And the segments were fixed in 10% neutral-formalin and embedded in paraffin.
Figure 1.
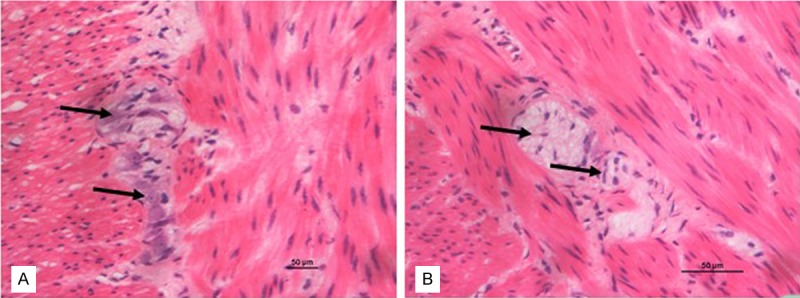
Photomicrographs of aganglionic and ganglionic intestine by H&E. A: Ganglionic colon segment tissue. B: Aganglionic colon segment tissue. A and B: × 400 (the bar = 50 μm).
Immunostaining was performed on 5 μm sections obtained from formalin-fixed, paraffin embedded blocks using Biotin streptavidin complex method. For antigen retrieval, slides were incubated in a microwave oven for 10 minutes in 0.01 mol/L citrate buffer (pH = 6), followed by cooling at room temperature. After incubated in 10% normal goat serum in PBS for 30 min to block nonspecific binding sites, samples were incubated at 4°C overnight with antibody against WNT3A (1:500, Millipore Co.). Samples washed and then incubated with anti-rabbit IgG-peroxidase antibody for 20 minutes at 37°C. Diaminobenzidine (DAB) color developed under the light microscope, and the sections were counterstained. The specimens were photographed using a digitized microscope camera (Nikon E800, Japan). Negative controls were performed by equivalent PBS instead of rabbit WNT3A. Two pathologists independently reviewed the immunostaining-stained slides and agreed on diagnoses by consensus.
Statistical analysis
The Statistical Program for Social Sciences, version 13.0 (SPSS, Chicago, IL), was used for statistical analysis. Chi-square tests were performed to determine whether each polymorphism was in Hardy-Weinberg equilibrium within control group and patients group. A t test was used to compare the WNT3A expression level between aganglionic and normal ganglionic colon segments. Statistical significance was determined by using Student’s t test, and P < 0.05 was considered statistically significant.
Results
PCR amplification in rs61743220, rs192966556 and rs145882986 of WNT3A gene
PCR amplification was successfully performed. The amplified segments of the WNT3A gene in rs61743220, rs192966556 and rs145882986 were consistent in size with their predicted lengths of 306 bp, 237 bp and 316 bp, respectively. The amount of amplified products was Figure 1. Photomicrographs of aganglionic and ganglionic intestine by H&E. A: Ganglionic colon segment tissue. B: Aganglionic colon segment tissue. A and B: × 400 (the bar = 50 μm). Figure 2. Agarose gel electrophoresis of PCR products of the WNT3A gene. M, Marker DL 2000; lanes 1-6, PCR amplification products of rs61743220; lanes 7-12, PCR amplification products of rs192966556; lanes 13-18, PCR amplification products of rs145882986. large and no non-specific bands appeared (Figure 2).
Figure 2.
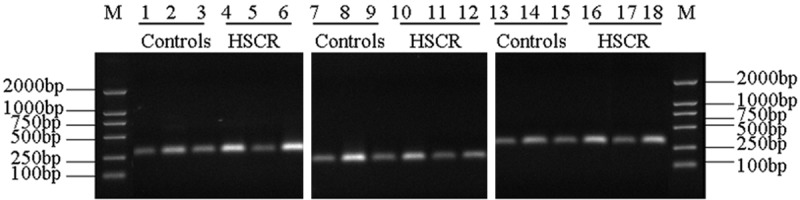
Agarose gel electrophoresis of PCR products of the WNT3A gene. M, Marker DL 2000; lanes 1-6, PCR amplification products of rs61743220; lanes 7-12, PCR amplification products of rs192966556; lanes 13-18, PCR amplification products of rs145882986.
Distribution of WNT3A allele and genotype frequencies in patients with HSCR and controls
Genotype distributions in the 3 SNPs were in accordance with the Hardy-Weinberg equilibrium (Figure 3). As show in the Tables 2 and 3, allele frequencies revealed a significant negative association of the rs192966556 SNP with HSCR (P = 0.001). CC homozygosity was positively associated with HSCR (P = 0.005). The allele frequencies revealed a significant negative association of the rs145882986 SNP with HSCR (P = 0.003). CC homozygosity was positively associated with HSCR (P = 0.012). However, comparison of rs61743220 A and G allelic frequencies showed no significant difference. (P > 0.05).
Figure 3.
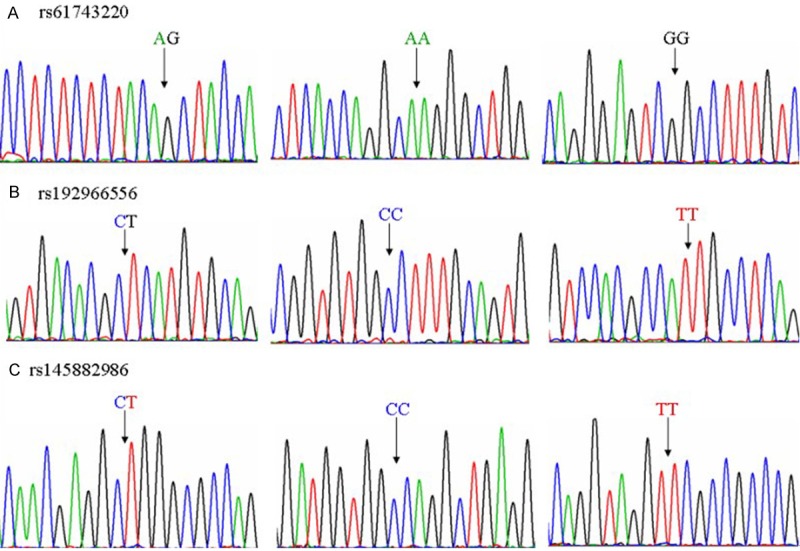
The sequencing results of different genotypes of rs61743220, rs192966556 and rs145882986. A: AG, AA and GG sequencing results of rs61743220. B: CT, CC and TT sequencing results of rs145882986. C: CT, CC and TT sequencing results of rs rs145882986. The arrow denotes the SNP location.
Table 2.
Allele and genotype frequency distribution in patients with HSCR and controls
| type | HSCR | Controls | X2 | P | OR (95% CI) | |
|---|---|---|---|---|---|---|
| rs61743220 | AG | 97 | 101 | - | - | - |
| AA | 77 | 71 | 0.313 | 0.576 | 0.886 (0.578-1.356) | |
| GG | 26 | 28 | 0.012 | 0.913 | 1.034 (0.566-1.889) | |
| A | 251 | 243 | - | - | - | |
| G | 149 | 157 | 0.339 | 0.561 | 1.088 (0.818-1.448) | |
| rs192966556 | CT | 73 | 91 | - | - | - |
| CC | 108 | 73 | 7.925 | 0.005 | 0.542 (0.353-0.832) | |
| TT | 19 | 36 | 1.679 | 0.195 | 1.520 (0.805-2.869) | |
| C | 289 | 237 | - | - | - | |
| T | 111 | 163 | 15.009 | 0.001 | 1.791 (1.332-2.408) | |
| rs145882986 | CT | 78 | 96 | - | - | - |
| CC | 101 | 72 | 6.381 | 0.012 | 0.579 (0.379-0.886) | |
| TT | 21 | 32 | 0.448 | 0.504 | 1.238 (0.662-2.316) | |
| C | 280 | 240 | - | - | - | |
| T | 120 | 160 | 8.791 | 0.003 | 1.556 (1.161-2.085) |
Table 3.
Allele and genotype frequency distribution in HSCR
| Group | Case (n) | Genotype frequency (%) | Allele frequency | ||||
|---|---|---|---|---|---|---|---|
| AG | AA | GG | A | G | |||
| rs61743220 | HSCR | 200 | 97 (48.50) | 77 (38.50) | 26 (13.00) | 251 (62.75) | 149 (37.25) |
| Controls | 200 | 101 (50.50) | 71 (35.50) | 28 (14.00) | 243 (60.75) | 157 (39.25) | |
| X2 = 0.398 P = 0.819 | X2 = 0.339 P = 0.561 | ||||||
| CT | CC | TT | C | T | |||
| rs192966556 | HSCR | 200 | 73 (36.50) | 108 (54.00) | 19 (9.50) | 289 (72.25) | 111 (27.75) |
| Controls | 200 | 91 (45.50) | 73 (36.50) | 36 (18.00) | 237 (59.25) | 163 (40.75) | |
| X2 = 13.998 P = 0.001 | X2 = 15.009 P = 0.001 | ||||||
| CT | CC | TT | C | T | |||
| rs145882986 | HSCR | 200 | 78 (39.00) | 101 (50.50) | 21 (10.50) | 280 (70.00) | 120 (30.00) |
| Controls | 200 | 101 (50.50) | 72 (36.00) | 32 (16.00) | 240 (60.00) | 160 (40.00) | |
| X2 = 9.006 P = 0.011 | X2 = 8.791 P = 0.003 | ||||||
Sequence variants of rs192966556 and rs145882986
By sequencing the genotype of rs192966556 and rs145882986 in patients with HSCR, we found, CT genotype in the rs192966556 polymorphism lacked one ‘T’ at codon 341 and the TT genotype of the rs192966556 polymorphism had an extra ‘T’ at codon 343. CT genotype in the rs145882986 polymorphism lacked one ‘T’ at codon 187 and the CC genotype of the rs145882986 polymorphism also lacked one ‘C’ at codon 434 (Figure 4).
Figure 4.
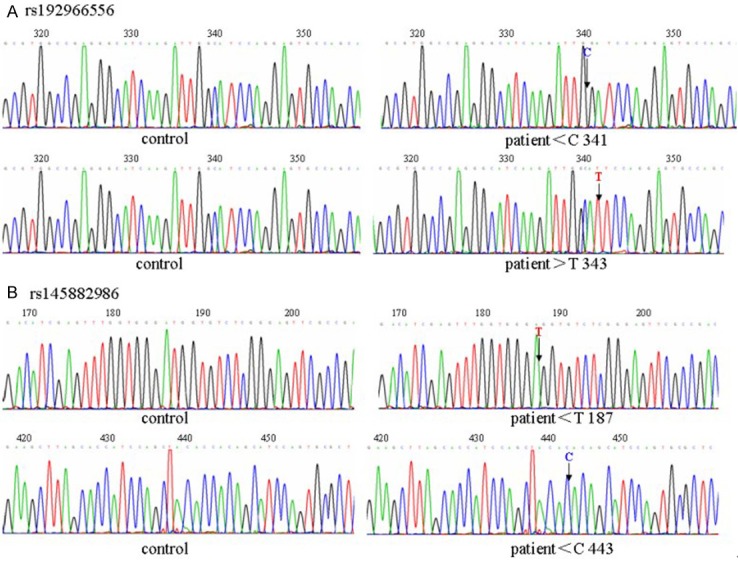
A novel mutation in the WNT3A gene. A: Sequencing analysis of rs192966556 in the patient and the control group. Arrows denote the codon point. B: Sequencing analysis of rs145882986 in the patient and the control group. Arrows denote the codon point.
qRT-PCR analysis
The OD value of RNA calculated by A260/A280 was from 1.8 to 2.0. In the course of the qRT-PCR, the amplification curve was received by fluorescent threshold and cycle, a fair reproducibility of each sample and basically coincident efficacy amplification were demonstrable. It was showed that the mRNA levels of WNT3A was 2.6 fold higher in aganglionic colon segment tissues than that in normal ganglionic segments tissues by the qRT-PCR (n = 50, P < 0.03) (Table 4).
Table 4.
The relative quantity of Wnt3a mRNA in two segments
| segment | Wnt3a average Ct value | β-actin average Ct value | ∆Ct | ∆∆Ct | times of gene (compared to normal segment) |
|---|---|---|---|---|---|
| normal | 31.53 ± 2.41 | 23.94 ± 1.95 | 7.59 | 0 | 1 |
| aganglionic | 30.27 ± 1.92 | 24.06 ± 1.80 | 6.21 | -1.38 | 2.6 |
Expression of WNT3A protein
The expression of WNT3A protein was evaluated by western blotting with specific antibodies in the same group of 50 HSCR patients. Consistent with the results of qRT-PCR, significant increases of WNT3A was detected in aganglionic colon segments compared to the matched normal ganglionic colon segments (Figure 5). The protein levels of WNT3A was 34.56 ± 3.21 in the aganglionic colon segment, whose value was much higher than that measured in the normal ganglionic colon segment (16.37 ± 2.46, P < 0.05).
Figure 5.
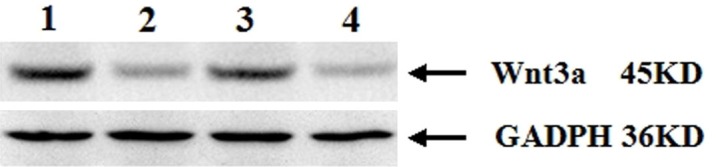
The expression of WNT3A proteins in Western blot. WNT3A is detected as an approximately 45-kDa band on Western blots of protein extracted from both the ganglionic and aganglionic colon segment tissue. Immunoblot shows a remarkable WNT3A signal protein in the aganglionic tissues but weak in the ganglionic tissue. Lines 1 and 3: the WNT3A proteins of the aganglionic segment; lines 2 and 4: the WNT3A proteins of the ganglionic segment.
Immunostaining results of WNT3A
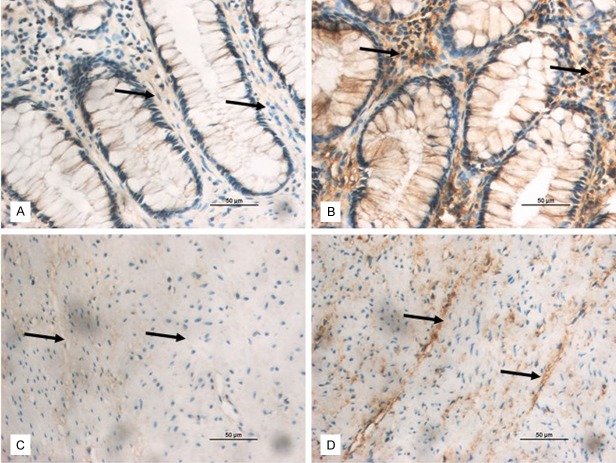
The aganglionic and ganglionic colon segments were first defined by the absence of the focal colon ganglion cells through H&E staining (Figure 1). Immunostaining indicates that the positive reaction mainly located in the mucous layer and muscular layer of colon segment. The brown yellow depositions in the aganglionic colon segment tissues were far more rich and widespread in submucosa, and were reticulodromous in the circular muscular layer; while those in the ganglionic colon segment tissues were punctiform. WNT3A immunostaining showed the tendency of regional increase from ganglionic colon segment to aganglionic colon segment (Figure 6).
Figure 6.
Expression of WNT3A detected by immunostaining in aganglionic and ganglionic colon segment tissue. The brown yellow depositions in aganglionic segment were far more rich and widespread in mucous layer and muscular layer, while those in the ganglionic segment were punctiform. A: Mucous layer of ganglionic colon segment tissue. B: Mucous layer of aganglionic colon segment tissue. C: Muscular layer of ganglionic colon segment tissue. D: Muscular layer of aganglionic colon segment tissue. A-D: × 400 (the bar = 50 μm).
Discussion
HSCR disease is the most common congenital gut motility disorder and is characterized by an absence of enteric neurons in terminal regions of the gut, leading to tonic contraction of the affected segment, intestinal obstruction and massive distension of the proximal bowel (megacolon). The mechanism of motility dysfunction in HSCR is still unclear although colonic motility dysfunction is a main manifestation. Despite certain achievements have been made in identifying some of the genetic basis of HSCR disease, the cause of HSCR remains unclear. Wnt signaling is one of the most critical signaling pathways in many aspects of development [14,22]. It is reported that the WNT8A gene is involved in the susceptibility to HSCR and plays an important role in the occurrence of HSCR [23]. Besides, dishevelled gene, a critical mediating site in the Wnt signaling pathway, also plays an important role in the pathogenesis of HSCR [24]. WNT3A, a canonical Wnt ligand, plays an important role in the embryonic development of the gut. It has been reported that WNT3A is initially required at late egg cylinder stages for somite development, but by late primitive streak/tailbud stages, formation of all mesodermal precursors is dependent, directly or indirectly, on WNT3A signaling. The abnormal expression of WNT3A affects the development of somites and tailbud formation [25]. Mice that are homozygous for a hypomorphic WNT3A allele display vertebral defects, a short tail due to loss of caudal vertebrae, deficient cloacal development and incomplete uro-rectal septation [18]. Besides, studies involving human pluripotent stem cells have shown that WNT3A is required for hindgut specification [19].
Although the main susceptibility genes stated above and candidate SNPs of WNT3A have been extensively investigated in HSCR patients from different ethnic groups, but for the impact and role, the disease still remains obscure. In this study, we performed a comprehensive genetic study for WNT3A gene in sporadic HSCR patients in the northeast area of China. We examined SNPs of WNT3A (rs61743220, rs192966556 and rs145882986) in 200 patients with HSCR and 200 controls. Associations between specific genotypes and the development of HSCR were examined by the logistic regression analysis to calculate OR and 95% confidence intervals (CI) (Tables 2 and 3). In conclusion, the results clearly suggest that those susceptible factors related to the WNT3A polymorphic were predisposed for HSCR.
As HSCR is a multifactorial congenital disorder, the cumulative genetic effects that result in an individual phenotypic variation play a crucial role in its development. Therefore, it is important to assess whether WNT3A polymorphisms are associated with HSCR susceptibility. The aim of the present study was to examine polymorphic markers of the WNT3A gene to determine their association with the risk and development of HSCR in Chinese individuals. DNA was extracted from whole blood samples, and WNT3A polymorphisms were analyzed by PCR. Associations between specific genotypes and the development of HSCR were examined by logistic regression analysis to calculate the odds ratio (OR) and 95% confidence intervals (CI). The risk of HSCR increased as the number of putative high-risk genotypes increased for the combined genotypes of WNT3A heterozygosity. In conclusion, the results obtained in this study clearly suggest that the susceptible factor related to different WNT3A polymorphisms is predisposing risk factor for HSCR. We observed that the WNT3A gene polymorphisms (rs192966556 and rs145882986) are associated with an increased risk of HSCR in our study sample. The differences in genotypes and allele distributions of rs192966556 and rs145882986 between various clinical classifications were statistically significant. Moreover, sequence analysis revealed that the WNT3A gene may influence the risk of this common developmental anomaly.
In the study, we also investigate the differential expressions in mRNA and protein levels between the aganglionic and the normal ganglionic of colon tissues from HSCR patients in order to obtain more information about bowel motility disturbance. We analyzed the aganglionic and the normal ganglionic colon segment tissues derived from 50 patients with sporadic HSCR and found that the expression of WNT3A in aganglionic colon segments was higher than that in the normal ganglionic colon segments (Table 4), and the differences were statistically significant (P < 0.05). The same protein expression result was further confirmed that significant increase of WNT3A was detected in aganglionic colon segments compared to the normal ganglionic colon segments. Immunohistochemical staining of WNT3A was brown in aganglionic colon segment tissues, while was pale yellow or colorless in normal ganglionic colon segment tissues.
For the possible reasons about the higher expression levels of WNT3A in the aganglionic tissues compared with the ganglionic tissues, we postulate that aberrant Wnt signalling may contribute to neurological disorders resulting from the higher expressions of WNT3A. It has been identified that the WNT8A gene is involved in the susceptibility to HSCR and plays an important role in the occurrence of HSCR [21]. As a Wnt ligand, WNT3A binds to seven-pass transmembrane receptors of the Frizzled (Fzd) family and co-receptors, low density lipoprotein-related protein (LRP) 5 and 6, leading to the inhibition of the APC/Axin/CK1/GSK3b destruction complex and stabilization and translocation of β-catenin to the nucleus where it interacts with TCF/Lef family transcription factors to regulate the transcription of target genes [14,26]. These target genes may eventually stimulate more synapse formation by increasing synaptic assembly to promote the normal development of the ganglionosis. Alternatively, WNT3A might also promote HSCR via non-canonical Wnt signaling mechanisms [17].
In summary, our study demonstrates that polymorphic variants of WNT3A might be involved in HSCR etiology. Through the differential expression, we detect and characterize WNT3A as a differentially expressed gene in aganglionic and normal ganglionic colon segments with HSCR. However, the precise role the WNT3A plays in HSCR awaits further investigation. Our study may provide more insights into HSCR.
Acknowledgements
This work was supported by grants from Shenyang Science and Technology Plan Project (grant#: F13-318-1-01).
Disclosure of conflict of interest
There is no interest of conflicts about this paper.
References
- 1.Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED, Chakravarti A. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 2.Lantieri F, Griseri P, Ceccherini I. Molecular mechanisms of RET-induced Hirschsprung pathogenesis. Ann Med. 2006;38:11–19. doi: 10.1080/07853890500442758. [DOI] [PubMed] [Google Scholar]
- 3.Arighi E, Borrello MG, Sariola H. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev. 2005;16:441–467. doi: 10.1016/j.cytogfr.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Tam PKH, Garcia-Barcelo M. Molecular genetics of Hirschsprung’s disease. Semin Pediatr Surg. 2004;13:236–248. doi: 10.1053/j.sempedsurg.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Bidaud C, Salomon R, Van CG, Pelet A, Attié T, Eng C, Bonduelle M, Amiel J, Nihoul-Fékété C, Willems PJ, Munnich A, Lyonnet S. Endothelin-3 gene mutations in isolated and syndromic Hirschsprung disease. Eur J Hum Genet. 1997;5:247–251. [PubMed] [Google Scholar]
- 6.Angrist M, Bolk S, Halushka M, Lapchak PA, Chakravarti A. Germline mutations in glial cell line-derived neurotrophic factor (GDNF) and RET in a Hirschsprung disease patient. Nat Genet. 1996;14:341–344. doi: 10.1038/ng1196-341. [DOI] [PubMed] [Google Scholar]
- 7.Pan ZW, Lou J, Luo C, Yu L, Li JC. Association analysis of the SOX10 polymorphism with Hirschsprung disease in the Han Chinese population. J Pediatr Surg. 2011;46:1930–1934. doi: 10.1016/j.jpedsurg.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 8.Kwon MJ, Lee GH, Lee MK, Kim JY, Yoo HS, Ki CS, Chang YS, Kim JW, Park WS. PHOX2B mutations in patients with Ondine-Hirschsprung disease and a review of the literature. Eur J Pediatr. 2011;170:1267–1271. doi: 10.1007/s00431-011-1434-5. [DOI] [PubMed] [Google Scholar]
- 9.Gregory-Evans CY, Vieira H, Dalton R, Adams GGW, Salt A, Gregory-Evans K. Ocular coloboma and high myopia with Hirschsprung disease associated with a novel ZFHX1B missense mutation and trisomy 21. Am J Med Genet A. 2004;131:86–90. doi: 10.1002/ajmg.a.30312. [DOI] [PubMed] [Google Scholar]
- 10.Goodrich LV, Strutt D. Principles of planar polarity in animal development. Development. 2011;138:1877–1892. doi: 10.1242/dev.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien AJ, Conrad WH, Moon RT. A Wnt survival guide: from flies to human disease. J Invest Dermatol. 2009;129:1614–1627. doi: 10.1038/jid.2008.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nusse R, Varmus HE. Wnt genes. Cell. 1992;69:1073–1087. doi: 10.1016/0092-8674(92)90630-u. [DOI] [PubMed] [Google Scholar]
- 13.Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 14.Dierick H, Bejsovec A. Cellular mechanisms of wingless/Wnt signal transduction. Curr Top Dev Biol. 1999;43:153–190. doi: 10.1016/s0070-2153(08)60381-6. [DOI] [PubMed] [Google Scholar]
- 15.Sokol SY. Wnt signaling and dorso-ventral axis specification in vertebrates. Curr Opin Genet Dev. 1999;9:405–410. doi: 10.1016/S0959-437X(99)80061-6. [DOI] [PubMed] [Google Scholar]
- 16.Kolligs FT, Bommer G, Goke B. Wnt/beta-catenin/tcf signaling: a critical pathway in gastrointestinal tumorigenesis. Digestion. 2002;66:131–144. doi: 10.1159/000066755. [DOI] [PubMed] [Google Scholar]
- 17.Nalesso G, Sherwood J, Bertrand J, Pap T, Ramachandran M, De BC, Pitzalis C, Dell’accio F. WNT-3A modulates articular chondrocyte phenotype by activating both canonical and noncanonical pathways. J Cell Biol. 2011;193:551–64. doi: 10.1083/jcb.201011051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van DVC, Bialecka M, Neijts R, Young T, Rowland EJ, Stringer EJ, van RC, Meijlink F, Nóvoa A, Freund JN, Mallo M, Beck F, Deschamps J. Concerted involvement of Cdx/Hoxgenes and Wnt signaling in morphogenesis of the caudal neural tube and cloacal derivatives from the posterior growth zone. Development. 2011;138:3451–3462. doi: 10.1242/dev.066118. [DOI] [PubMed] [Google Scholar]
- 19.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai Y, Wang Z, Dai W, Li QZ, Chen GL, Cong N, Guan MX, Li H. A six-generation Chinese family in haplogroup B4C1C exhibits high penetrance of 1555A > G-induced hearing loss. BMC Med Genet. 2010;11:129. doi: 10.1186/1471-2350-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Merkel CE, Karner CM, Carroll TJ. Molecular regulation of kidney development: is the answer blowing in the Wnt? Pediatr Nephrol. 2007;22:1825–1838. doi: 10.1007/s00467-007-0504-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao H, Chen D, Liu XM, Wu M, Mi J, Wang WL. Polymorphisms and expression of the WNT8A gene in Hirschsprung’s disease. Int J Mol Med. 2013;32:647–652. doi: 10.3892/ijmm.2013.1433. [DOI] [PubMed] [Google Scholar]
- 24.Chen D, Mi J, Wu M, Wang WL, Gao H. The expression of dishevelled gene in Hirschs-prung‘s disease. Int J Clin Exp Pathol. 2013;6:1791–1798. [PMC free article] [PubMed] [Google Scholar]
- 25.Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 26.Sethi JK, Vidal-Puig A. Wnt signalling and the control of cellular metabolism. Biochem J. 2010;427:1–17. doi: 10.1042/BJ20091866. [DOI] [PMC free article] [PubMed] [Google Scholar]