Abstract
Cisplatin resistance is a major problem affecting ovarian carcinoma treatment. NF-E2-related factor 2 (Nrf2), a nuclear transcription factor, plays an important role in chemotherapy resistance. However, the underlying mechanism by which Nrf2 mediates cisplatin chemoresistance is unclear. Methods: The human ovarian carcinoma cell line, A2780, and its cisplatin-resistant variant, A2780cp were cultivated. Cell viability was determined with WST-8 assay. Western blot was applied to detect the expression of Nrf2, Nrf2 target genes, and autophagy-related proteins. RNA interference was used to knock down target genes. Annexin V and propidium iodide (PI) staining was utilized to quantify apoptosis. The ultrastructural analysis of autophagosomes was performed by transmission electron microscopy (TEM). Results: Nrf2 and its targeting genes, NQO1 and HO-1, are overexpressed in A2780cp cells compared with A2780 cells. Knocking down Nrf2 sensitized A2780cp cells to cisplatin treatment and decreased autophagy-related genes, Atg3, Atg6, Atg12 and p62 in both mRNA and protein levels. Furthermore, we demonstrated that in both cell lines cisplatin could induce the formation of autophagosomes and upregulate the expression of autophagy-related genes Atg3, Atg6 and Atg12. Treatment with an autophagy inhibitor, 3-Methyladenine (3-MA), or beclin 1 siRNA enhanced cisplatin-induced cell death in A2780cp cells, suggesting that inhibition of autophagy renders resistant cells to be more sensitive to cisplatin. Taken together, Nrf2 signaling may regulate cisplatin resistance by activating autophagy. Conclusions: Nrf2-activated autophagy may function as a novel mechanism causing cisplatin-resistance.
Keywords: Ovarian carcinoma, Nrf2, autophagy, cisplatin, chemoresistance
Introduction
Ovarian cancer is one of the most common gynecological malignances. The 5 year overall survival is approximately 30%, owing to advanced stage diagnosis and development of chemoresistance [1]. Cisplatin-based resistance has been a major obstacle for successful treatment of ovarian cancer. Currently, the underlying mechanisms can be classified into two categories. The first is reduced uptake, enhanced efflux, and increased inactivation of cisplatin, which involve steps preceding the binding of cisplatin to DNA or damage to DNA-cisplatin adducts; the other is an increased DNA damage repair program and prevention of apoptosis by a variety of signaling pathways that are elicited by DNA-cisplatin adducts [2]. However, this type of clinical issue is still not fully understood and optimal strategies to overcome cisplatin resistance need to be further explored.
Erythroid transcription factor NF-E2 (Nrf2) plays a critical role against oxidative and electrophilic stress [3]. Under normal conditions, Nrf2 interacts with Kelch-like ECH-associated protein 1 (Keap1), and is degraded by the ubiquitin-proteasome pathway to maintain Nrf2 at a low level. In response to electrophiles and oxidative stress, Nrf2 dissociates from Keap1, provoking the activation of Nrf2 [4,5]. Nrf2 accumulates in the nucleus and activates the induction of phase II detoxifying enzymes, including NAD(P)H: quinone oxidoreductase (NQO1) and antioxidant proteins, such as hemeoxygenase-1 (HO-1) [6]. Many investigations have shown that the Nrf2-Keap1 pathway protects cells from many diseases including neurodegenerative disease and cancers [7,8]. Recently, however, the dark side of Nrf2 was also revealed. Constitutive activation of Nrf2 is associated with tumorigenesis and resistance of chemotherapeutics in lung cancer, colon cancer, prostate cancer and type II endometrial cancer [9-12]. Whether Nrf2 plays a role in cisplatin chemoresistance of ovarian cancer, however, is unclear.
In autophagy, or “Type II Programmed Cell Death”, the cell digests damaged organelles and molecules to maintain cellular homeostasis [13]. Autophagy is regulated by a group of autophagy-related genes (Atgs) with the core machinery involving two ubiquitin systems: the Atg12-Atg5-Atg16 system and the Atg8-LC3 system [14]. In most cases, autophagy acts as a cytoprotective mechanism and dysfunction of autophagy has been shown to lead to diverse pathologies, including myopathies, aging and neurodegeneration [15]. In cancer this protective action can be deleterious because it may allow cancer cells to become resistant to anti-cancer treatment. In fact, some drug-resistant cancer cells exhibit increased activation of autophagy [16,17]. However, in other conditions, drug exposure impaired autophagy in resistant cancer cells [18].
Mammalian sequestosome 1 (p62/SQSTM1) is a multifunctional scaffolding protein, which binds LC3 and traffics proteins for autophagy degradation [19]. A recent study revealed that p62 was capable of binding to Keap1, and competes with Nrf2 for the Keap1 binding site [20]. Therefore, the Nrf2-Keap1 system is associated with autophagy by p62. Here, we hypothesized that the Nrf2-Keap1 signaling pathway may play a role in cisplatin resistance through regulation of autophagy in ovarian cancer. The illustration of the interaction between Nrf2 and autophagy pathway may help with the development of new potential therapeutic targets in ovarian cancer.
Methods
Cell lines
The cisplatin-sensitive human ovarian cancer cell line A2780 and its cisplatin-resistant clone A2780cp were obtained from Shanghai Key Laboratory of Female Reproductive Endocrine Related Diseases. A2780 cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco), supplemented with 10% fetal bovine serum (FBS, Biowest, South America Origin), 100 U/mL penicillin and 100 μg/mL streptomycin (Sigma-Aldrich, St. Louis, MO) at 37°C and 5% CO2 with high humidity. The cisplatin-resistant A2780cp cells were maintained in DMEM supplemented with 10% fetal bovine serum medium containing 1 ug/mL cisplatin to maintain resistance.
Reagents
Cisplatin was purchased from Hansoh Pharmaceutical Co., Ltd. (Lianyungang, Jiangsu, China). 3-Methylademine (3-MA) was purchased from Sigma-Aldrich and dissolved in sterile double distilled water at 65°C.
Cell viability assay
Cells were seeded at 1×104 cells per well in 96-well plates. The following day, different concentrations of cisplatin were added to the wells and incubated for 24 h. Each treatment was repeated in four wells. The measurement of viable cell mass was assessed by a Cell Counting Kit (Dojin Laboratory, Kumamoto, Japan). In brief, an aliquot of 10 μl of CCK-8 plus 100 μl DMEM was added to each well and incubated for 2 h. Absorbance was measured with a Microplate Reader (Model680, BioRad, USA) at a wavelength of 450 nm. Each experiment was repeated three times.
Western blot
Cells were washed with cold PBS twice and incubated in ice-cold lysis buffer (50 nM Tris pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS and complete mini-cocktail protease inhibitors) for 30 min at 4°C. After centrifugation at 12,000 rpm for 15 min, supernatants were collected. Protein concentration was determined using BCA protein assay kit (Pierce, Rockford, USA). For western blot analysis, 20 μg lysate proteins were separated by 10-15% w/v SDS-polyacrylamide gel (SDS-PAGE) and transferred to PVDF membranes (Millipore, Bedford, MA). Membranes were blocked with 5% non-fat milk in PBS for 1 h at room temperature. The membranes were incubated with rabbit monoclonal antibodies for Atg3, Atg5, beclin 1, Atg12 and Keap1 (1:1000) from Cell Signaling Technology; rabbit monoclonal antibody Nrf2 (1:20000) from Abcam; mouse monoclonal antibody NQO1 (1:2000) from Santa Cruz Biotechnology; rabbit polyclonal antibody p62 (1:500) from Proteintech; rabbit polyclonal antibody HO-1 (1:1000) from Enzo Life Science. HRP-conjugated β-actin monoclonal antibody (1:20000), HRP-conjugated anti-rabbit IgG (1:6000) and HRP-conjugated anti-mouse IgG1 (1:6000) from Sigma-Aldrich were used as secondary antibodies for an incubation period of 1.5 h. Membranes were washed three times with PBS-T between each antibody incubation. Protein bands were visualized using an enhanced chemiluminescence Western Blot analysis system (Pierce, Rockford, USA).
Quantitative real-time PCR analysis (qRT-PCR)
Total RNA was extracted from cultured cells using Trizol reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s protocol and quantified with Nanodrop 2000 (Thermo, Japan). First-strand cDNA synthesis and amplification were performed using reverse transcription reagents (Takara, Dalian, China) following the manufacturer’s instructions. The quantitative PCR reactions included 7.6 μl cDNA and 12.4 μl of SYBR Green Master Mix (Takara, Dalian, China) with a pair of primers. The reactions were monitored on a 7500 Real-Time PCR System with 7500 software, version 2.0.5 (Applied Biosystems, Foster City, CA). The levels of mRNA were calculated using the equation 2-ΔΔCT and normalized to human β-actin mRNA levels. The primers were synthesized by Sengon Bio Co. (Shanghai, China) and listed in Table 1. The real-time PCR condition was as follows: 1 cycle of initial denaturation (95°C for 10 min), 40 cycles of amplification (95°C for 15 s and 60°C for 60 s) and a cooling program (50°C for 5 s). Two independent PCR assays were performed.
Table 1.
Real-time Primers
| Gene | Primers |
|---|---|
| hNrf2 | forward (ACACGGTCCACAGCTCATC) |
| reverse (TGTCAATCAAATCCATGTCCTG) | |
| hKeap1 | forward (ATTGGCTGTGTGGAGTTGC) |
| reverse (CAGGTTGAAGAACTCCTCTTGC) | |
| hNQO1 | forward (ATGTATGACAAAGGACCCTTCC) |
| reverse (TCCCTTGCAGAGAGTACATGG) | |
| hHO-1 | forward (AACTTTCAGAAGGGCCAGGT) |
| reverse (CTGGGCTCTCCTTGTTGC) | |
| hp62 | forward (GAACTCCAGTCCCTACAGAT) |
| reverse (CGATGTCATAGTTCTTGGTC) | |
| hAtg3 | forward (GATTATGGTTGTTTGGCTAT) |
| reverse (TGGTCACATCTATGGGTT) | |
| hAtg5 | forward (TGGGCCATCAATCGGAAACTC) |
| reverse (TGCAGCCACAGGACGAAACAG) | |
| hAtg6 | forward (AGGAACTCACAGCTCCATTAC) |
| reverse (AATGGCTCCTCTCCTGAGTT) | |
| hAtg12 | forward (CCTCGGAACAGTTGTTTAT) |
| reverse (CAGGACCAGTTTACCATCAC) |
Nrf2 pathway and autophagy related genes Real-time Primers are listed out.
siRNA transfection
Nrf2 specific siRNA (sense, CCCGUUUGUAGAUGACAAUTT, antisense AUUGUCAUCUACAAACGGGTT), negative control siRNA (sense, UUCUCCGAACGUGUCACGUTT, antisense, ACGUGACACGUUCGGAGAATT) and the beclin 1 siRNA (sense, CGGCUCCUAUUCCAUCAAATT, antisense, UUUGAUGGAUAGGAGGCCGTT) were constructed by Genepharma (Shanghai, China). The transfection of siRNA was performed using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Briefly, a total of 20×104 cells were seeded into 6 well plates, and transfected the next day with a 100 nM final concentration of siRNA, using 5 μl Lipofectamine 2000. Cells were harvested 48 h after transfection for western blot analysis. To measure the effect of siRNA and cisplatin treatment together, the cells were treated with cisplatin for another 24 h before determining cell viability and apoptosis.
Transmission electron microscopy (TEM)
Cells were fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer for 2 h at 4°C, and then postfixed in 1% osmium tetroxide for 3 h. Samples were scraped and pelleted, dehydrated in a graded series of ethanol baths, and infiltrated and embedded in Epon resin. Ultrathin sections of 70 nM were cut in a Leica microtome (Leica, Deerfield, Ill), stained with uranyl acetate for 3 min, and examined in a JEOL JEM-1400 transmission electron microscopy (JEOL Ltd, Tokyo, Japan) at an accelerating voltage of 80 kv.
TUNEL assay
Cells were seeded at 30×104 cells per well on 6-well plates and fixed with 4% paraformaldehyde at room temperature for 1 h after adherence. Between each step, cells were thrice rinsed with PBS for 5 min each. Then 50 μl of TUNEL (In situ cell death detection kit, TMR red, Roche) reaction mixture (5 μl TdT enzyme + 45 μl dUTP) was added to the cells, and incubated at 37°C for 1 h. Cell nuclei were stained with 1 ml 10 μg/ml DAPI (Roche, USA) for 5 min at room temperature in the dark. The cells were observed under the fluorescence microscope for TUNEL and DAPI staining (Olympus IX51, Tokyo, Japan).
Apoptosis analysis
For assessment of apoptosis, the Annexin VFITC staining kit (BD Pharmalgen, CA, USA) was used. After treating the cells for 24 h, both floating and adherent cells were collected and centrifuged at 5,000 rpm for 5 min. Cells were resuspended in 500 μl binding buffer and stained with 5 μl Annexin V and 5 μl PI for 15 min at room temperature in the dark. The samples were analyzed by flow cytometry (FC500 MPL, Beckman coulter, USA). Twenty thousand events were measured and results were expressed as the percentage (Annexin V + with PI +/-) of apoptotic cells.
Statistical analysis
Data was presented as mean ± SD. Student’s t test was used to compare the difference between two groups (two-tailed; P<0.05 was considered significant). The analysis was performed using SPSS 16.0 software.
Results
Nrf2 pathway is activated in resistant ovarian cancer cells
To determine whether A2780cp cells are truly resistant to cisplatin, A2780 and A2780cp cells were treated with different concentrations of cisplatin for 24 h. The cell viability assay revealed that the percentage of surviving cells decreased significantly in a dose-dependent manner in both cell lines. However, the 50 percent inhibition concentration (IC50) for cisplatin in the A2780 and A2780cp cells was 6.5±0.81 (μg/ml) VS 39.4±8.9 (μg/ml), respectively (P<0.05) (Figure 1A). As shown in Figure 1B, the mRNA level of Nrf2 in A2780cp cells was 3.17 times fold, Keap1 2.51 times fold and HO-1 7.66 times fold higher than those in A2780 cells respectively (P<0.05). NQO1 mRNA expression showed no significant difference in both cell lines (P>0.05). Western blot analysis further confirm that all of Nrf2, Keap1, NQO1 and HO-1 in A2780cp cells were overexpressed compared with those in A2780 cells (Figure 1C).
Figure 1.
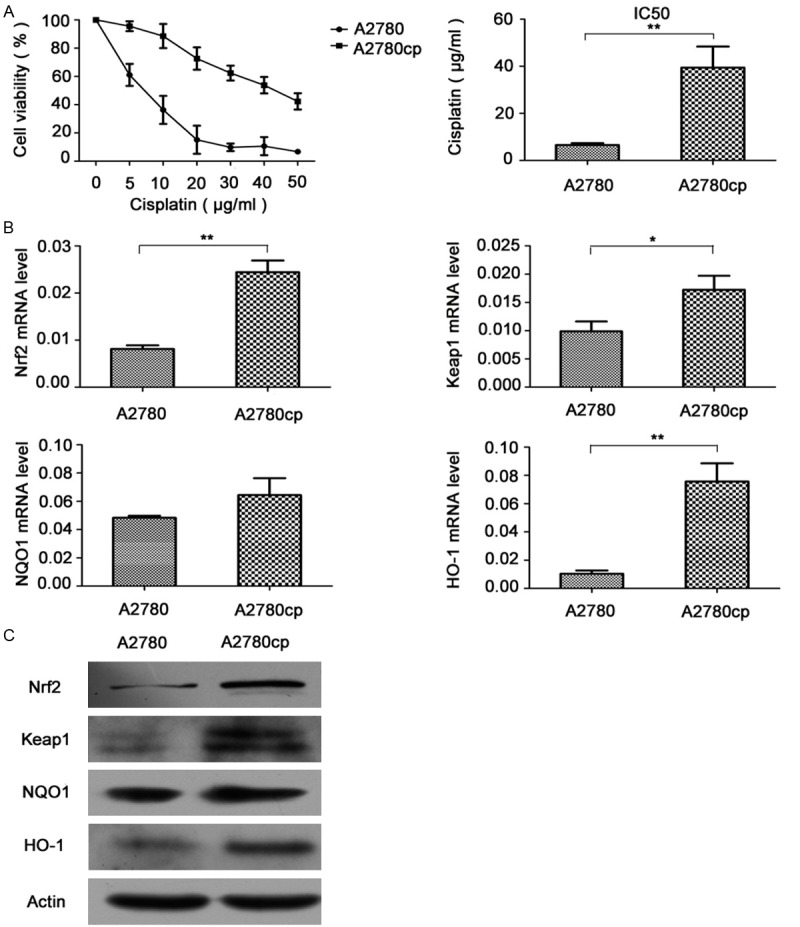
Expression of Nrf2 pathway genes in A2780 and A2780cp cell lines. A. Cells were treated with different concentrations (5 μg/ml-50 μg/ml) of cisplatin for 24 h. Cell viability was determined using a WST-8 assay. Data (mean ± SD) represent the mean value of three independent experiments. *P<0.05. B. RNA extracted from cells was subjected to mRNA level examination using a real-time PCR kit. Data (mean ± SD) represent the mean value of three independent experiments. *P<0.05, **P<0.01. C. Cell lysates were collected for western blot analyses for Nrf2, Keap1, NQO1 and HO-1 protein detection.
Knockdown of Nrf2 enhanced cisplatin-induced apoptosis in ovarian cancer cells
After small interfering RNA transfection (siRNA), Nrf2 protein level was knocked down by 57.45%, in accompany with the downregulation of NQO1 (44.60%) and HO-1 (62.71%) in contrast to control siRNA group. Keap1 level didn’t change significantly with Nrf2 decreasing (Figure 2A). As shown in Figure 2B, cell viability assay showed that the combined treatment (Nrf2 siRNA and cisplatin) enhanced cisplatin-induced cell death (41.48±3.42% VS 17.17± 0.39%, P<0.05). Much more apoptotic cells that got smaller, detached, and were stained red in TUNEL and DAPI staining were shown in combined treatment group. Furthermore, Annexin V/PI staining revealed that the apoptotic ratio of combined treatment group and cisplatin group were 49.2±8.52% VS 15.11±0.37% respectively (P<0.01) (Figure 2D).
Figure 2.
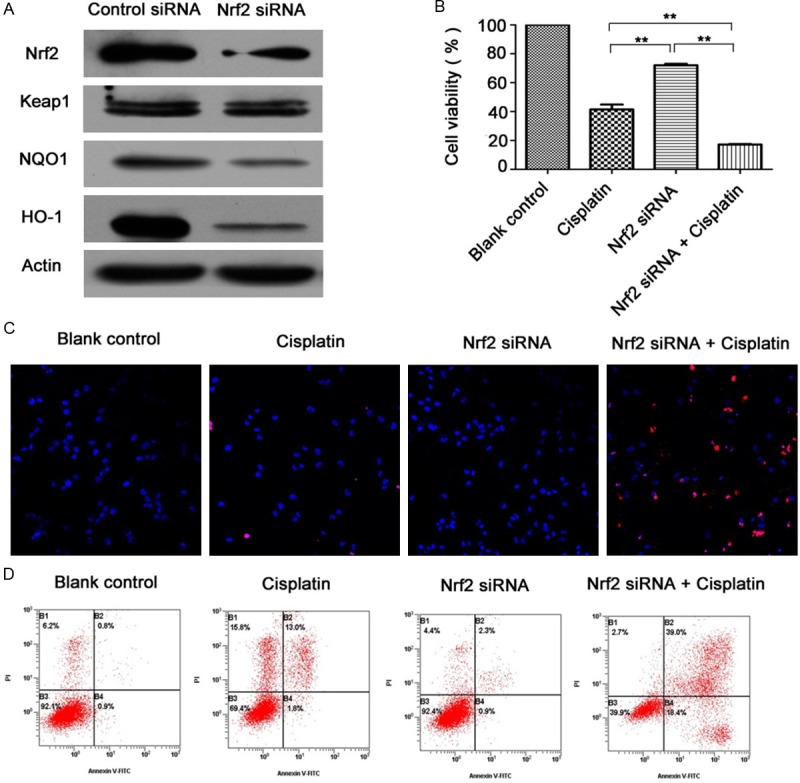
Effects of Nrf2 knockdown on cisplatin sensitivity in resistant A2780cp cells. A. After cells were transfected with Nrf2 siRNA (100 nM) for 48 h, cell lysates were collected for western blot analyses for Nrf2, Keap1, NQO1 and HO-1. B. Cells were transfected with Nrf2 siRNA (100 nM) for 48 h, and then treated with 40 μg/ml cisplatin for 24 h. Cell viability was determined using a WST-8 assay. Data (mean ± SD) represent the mean value of three independent experiments. **P<0.01. C. Cells were treated with TUNEL and DAPI staining and observed by fluoroscopy (magnification was 20×). D. Cells were subjected to Annexin V and PI staining. The apoptosis ratio is presented as a result of early-stage and advanced stage apoptosis.
Cisplatin induced autophagy in ovarian cancer cells
To illustrate the possible relationship between the Nrf2 pathway and autophagy in ovarian carcinoma cells, we treated A2780 and A2780cp cells with 6 μg/ml cisplatin for 6, 12 and 24 h. Interestingly, as shown in Figure 3A, at different time points, Atg3, Atg5 and Atg12 expressed at low level in A2780 cells relatively to those in A2780cp cells. The same came to p62, which acts as a connector between Nrf2 and autophagy. Atg6, also known as beclin 1, together with LC3 did not significantly change between both cells. Further, electron transmission microscopy analysis showed that more autophagosomes were observed in A2780cp cells treated with or without cisplatin compared with that in A2780 cells.
Figure 3.
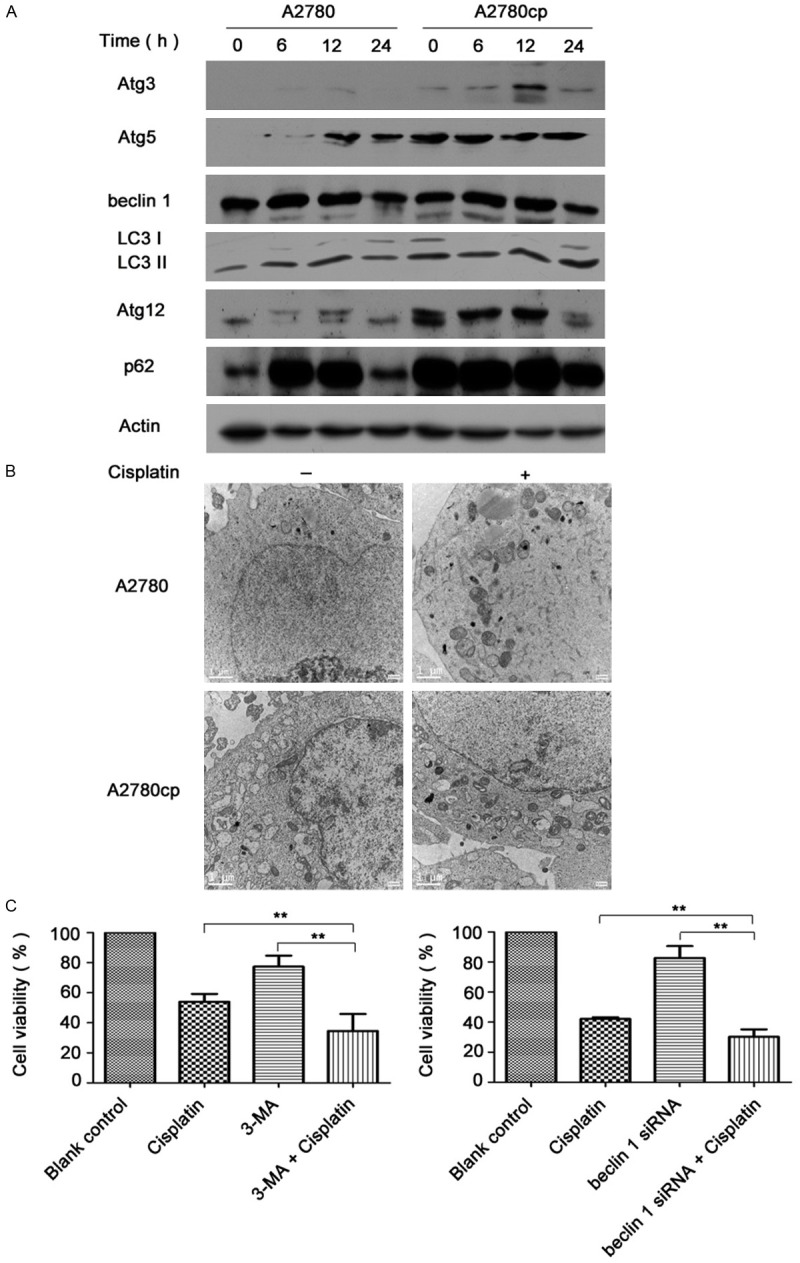
The role of autophagy in cisplatin resistance of ovarian cancer cells. A. After the cells were exposed to 6 μg/ml of cisplatin for different hours, cell lysates were collected to detect the protein levels of autophagy related genes. B. Cells were treated with 6 μg/ml cisplatin for 24 h and subjected to electron transmission microscopy for autophagosomes detection (Magnification was 10000×). Scale bars are 1 μm. C. A2780cp cells were pretreated with 3-MA for 1 h or transfected with beclin 1 siRNA (100 nM) for 48 h, and then treated with cisplatin (40 μg/ml) for 24 h. Cell viability was determined using a WST-8 assay (**P<0.01).
Next, A2780cp cells were pretreated with an autophagy inhibitor, 3-methyladenine (3-MA, 2.5 mmol) for 1 h or the cells were transfected with beclin 1 siRNA (100 nM), and then incubated with 40 μg/ml cisplatin for 24 h. The cell viability of 3-MA combined with cisplatin group and cisplatin group were 34.52±11.36% VS 53.94±5.25% respectively (P<0.01). Besides, beclin 1 siRNA combined with cisplatin group and cisplatin group showed cell viability as 30.18±5.01% VS 52.13±1.17%.
Nrf2 regulated autophagy in cisplatin-resistant ovarian cancer cells
After Nrf2 mRNA downregulation, we examined gene changes both at the mRNA level and protein level of the main autophagy related genes. A2780cp cells were pre-transfected with control siRNA or Nrf2 siRNA (100 nM) for 48 h. The mRNA results revealed that Atg3, Atg5, beclin 1, Atg12 and p62 decreased with Nrf2 knockdown (Figure 4A). Similar effects were detected at the protein level; Atg3, Atg5, beclin 1, Atg12 and p62 decreased significantly in contrast to control siRNA group (Figure 4B). Moreover, the number of autophagosomes in Nrf2 siRNA group also decreased in contrast to control siRNA group (Figure 4C).
Figure 4.
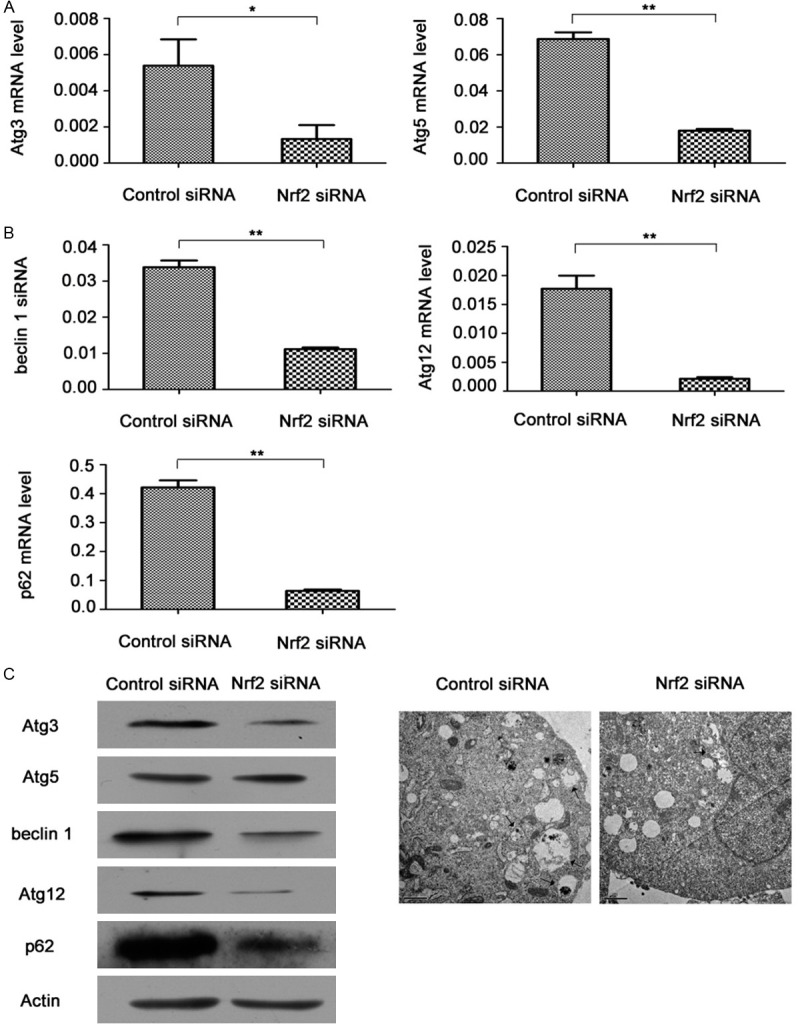
Effects of Nrf2 knockdown on autophagy level in A2780cp cells. A. mRNA was extracted from A2780cp cells after transiently transfecting them with control siRNA or Nrf2 siRNA (100 nM) for 48 h. After the mRNA was converted into cDNA, the cDNAs were subjected to real-time quantitative PCR for analysis of relative mRNA levels of Atg3, Atg5, beclin 1, Atg12, p62 and actin. Data was normalized to actin level. B. Cell lysates from A2780cp cells transfected with control siRNA or Nrf2 siRNA (100 nM) for 48 h were collected for western blot analysis for Atg3, Atg5, beclin 1, Atg12 and p62. C. Autophagosomes were detected by transmission electron microscopy after the cells were transfected with control siRNA or Nrf2 siRNA (100 nM) for 48 h (Magnification was 10000×). Scale bars are 1 μm. Arrows point to the autophagosomes.
Discussion
Exploring the mechanism of cisplatin resistance is an essential step to more successful treatment of ovarian cancer. Since its discovery, Nrf2 has been regarded as a cytoprotective transcription factor that plays a crucial role in antioxidant and detoxification processes. However, a number of studies have shown that persistent activation of Nrf2 is also involved in chemoresistance in a wide range of solid tumors. Stable overexpression of Nrf2 leads to enhanced resistance of cancer cells to chemotherapeutic agents including cisplatin, doxorubicin and etoposide [21]. A recent study found that in ovarian carcinoma cells, acquisition of doxorubicin resistance accompanied activation of the Nrf2 pathway [22]. Another paper demonstrated that transfection with Nrf2 siRNA into cisplatin resistant SK-OV ovarian cancer cells, enhanced cisplatin cytotoxicity [23]. These studies, however, failed to explore the underlying mechanisms about how the Nrf2 pathway participates in cisplatin resistance in ovarian cancer cells.
Here, we used the cisplatin sensitive A2780 and resistant A2780cp cells which provide an ideal pair of cell lines to study ovarian cancer resistance since the A2780cp cells are 6 times more resistant to cisplatin than the parental cell line (Figure 1A). To illustrate the role of the Nrf2 pathway in cisplatin resistance in ovarian cancer, we compared the level of Nrf2, Keap1, NQO1, and HO-1 in both ovarian cancer cells. The basal mRNA and protein expression of Nrf2, NQO1 and HO-1 were high in the resistant A2780cp cells, but low in the cisplatin-sensitive A2780 cells (Figure 1B and 1C). These data suggest that hyperactivation of the Nrf2 pathway may be involved in cisplatin resistance in ovarian cancer cells.
Next, we transfected A2780cp cells with Nrf2 siRNA and the expression of Nrf2, NQO1 and HO-1 decreased, indicating the dysfunction of Nrf2 pathway was in accompany with Nrf2 RNA down regulation. This confirmed that Keap1, as an upstream element, was not mediated by Nrf2 level directly, but NQO1, HO-1, locating at the downstream, decreased as Nrf2 down regulation. Furthermore, targeting Nrf2 rendered resistant A2780cp cells more susceptible to cisplatin induced cell death (Figure 2B) and apoptosis (Figure 2C and 2D). Moreover, it was reported that HO-1 was highly expressed in a variety of tumor tissue, and overexpression of HO-1 attenuated cisplatin induced apoptosis, while decreased expression by HO-1 siRNA enhanced the chemosensitivity of cisplatin [24,25]. Here, HO-1 was highly expressed in resistant ovarian cancer cells, and inhibition of Nrf2 resulted in the suppression of HO-1 at the same time, indicating that targeting both Nrf2 and HO-1 may be more efficient.
However, the underlying mechanism that involved in the ability of the Nrf2 pathway to enhance cisplatin cytotoxicity is not clear. As it is known, p62 is a multidomain adapter protein, which functions as a molecular hub to mediate various signaling pathways. The Nrf2 pathway, apoptosis and autophagy interact with p62, which binds LC3 in transitioning proteins for autophagosome degradation, and meanwhile competes for Keap1 binding with Nrf2 [26,27]. Previous studies revealed the tumorigenic suppressive role of p62 degradation by autophagy under normal conditions [28]. Interestingly, a recent paper found that p62 acted as a receptor for autophagic degradation, and knockdown of p62 could sensitize SKOV3/DDP ovarian cancer cells to cisplatin [29]. While in our study, p62 was highly expressed in cisplatin resistant cells, but low in sensitive cells at the time point of 0 h and 24 h with cisplatin treatment. While p62 expression at the time point of 6 h and 12 h in both cell lines was close to each other. Further, with Nrf2 siRNA transfection in A2780cp cells, we found that p62 decreased greatly in contrast to control siRNA transfection (Figure 4A and 4B). This indicated that p62 may be involved in cisplatin resistance, and Nrf2 knockdown resulted in p62 decrease in ovarian cancer cells.
What’s more, autophagy related genes were also downregulated as Nrf2 decreased. To determine whether the Nrf2 pathway has a connection with autophagy with cisplatin resistance, we examined the level of autophagy in both cell lines. The expression of autophagy related protein Atg3, Atg5 and Atg12 was high in A2780cp cells, but relatively low in A2780 cells. Beclin 1 and LC3 I/II protein level did not differ greatly in both cell lines. In transmission electron microscopy analysis, much more autophagosomes were detected in A2780cp cells than that in A2780 cells with or without cisplatin treatment. These data suggest that the level of autophagy in resistant cells were higher than that in sensitive cells. However, beclin 1 and LC3 I/II did not greatly differ in both cell lines, indicating autophagy may not act as a key role in cisplatin resistance here. We also inhibited autophagy in A2780cp cells with a pharmacological inhibitor 3-MA or beclin 1 siRNA transfection, which increased cisplatin-induced cell death significantly. Taken together, inhibition of autophagy contributed to reverse cisplatin resistance in ovarian cancer cells.
There is an increasing amount of evidence to confirm that autophagy activation participates in chemoresistance in cancer cells, and downregulation of autophagy sensitizes cancer cells to therapeutics [30-32]. However, few reports have studied the crosstalk between the Nrf2 signaling pathway and autophagy in cisplatin resistance of ovarian cancer. What we knew was that deficiency in autophagy resulted in the formation of cytoplasmic protein inclusions and overexpression of p62, which was mediated by Nrf2, but also contributed to the activation of Nrf2 [33]. Such hyperactivation of the Nrf2 signaling pathway seemed to be responsible for liver injury or the development of hepatic tumors [34,35]. In airway epithelial cells treated with cigarette smoke, overexpression of Nrf2 suppressed autophagy levels [36]. On the contrary, in renal cell carcinoma cell lines, it was reported that Nrf2 activation correlated with high basal autophagy [37]. These data demonstrate that the mechanism between Nrf2 signaling pathway and autophagy may differ depending on the type of tissue or cell line. Here, with the knock down of Nrf2 RNA level in resistant cells, and all of autophagy genes, Atg3, Atg5, beclin 1 and Atg12 decreased significantly in contrast to control siRNA group at mRNA level as well as at protein level (Figure 4A and 4B). Furthermore, autophagosomes in Nrf2 siRNA transfection group decreased compared to that in control siRNA group. These data suggest that downregulation of Nrf2 suppresses autophagy, and such regulation may serve as a mechanism to increase cisplatin-induced apoptosis.
Taken together, Nrf2 activates p62, which in turn upregulates autophagy in resistant ovarian cancer cells. Inhibition of Nrf2, suppresses p62, which then decreases the levels of autophagy. Therefore, inhibition of the Nrf2-p62-autophagy pathway in which Nrf2 plays a central role might sensitize cisplatin-resistant ovarian cancers to cisplatin treatment.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81302261) and Science and Technology Commission of Shanghai Municipality (Grant No. 09ZR1405000).
Disclosure of conflict of interest
None.
References
- 1.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–79. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 2.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–83. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 3.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–91. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26:221–9. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 7.Lee JM, Johnson JA. An important role of Nrf2-ARE pathway in the cellular defense mechanism. J Biochem Mol Biol. 2004;37:139–43. doi: 10.5483/bmbrep.2004.37.2.139. [DOI] [PubMed] [Google Scholar]
- 8.Baird L, Dinkova-Kostova AT. The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol. 2011;85:241–72. doi: 10.1007/s00204-011-0674-5. [DOI] [PubMed] [Google Scholar]
- 9.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, Gabrielson E, Feinstein E, Biswal S. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–84. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–46. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang T, Chen N, Zhao F, Wang XJ, Kong B, Zheng W, Zhang DD. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res. 2010;70:5486–96. doi: 10.1158/0008-5472.CAN-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akhdar H, Loyer P, Rauch C, Corlu A, Guillouzo A, Morel F. Involvement of Nrf2 activation in resistance to 5-fluorouracil in human colon cancer HT-29 cells. Eur J Cancer. 2009;45:2219–27. doi: 10.1016/j.ejca.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Cui J, Gong Z, Shen HM. The role of autophagy in liver cancer: molecular mechanisms and potential therapeutic targets. Biochim Biophys Acta. 2013;1836:15–26. doi: 10.1016/j.bbcan.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–6. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 15.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Donovan TR, O’Sullivan GC, McKenna SL. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy. 2011;7:509–24. doi: 10.4161/auto.7.6.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi CH, Jung YK, Oh SH. Autophagy induction by capsaicin in malignant human breast cells is modulated by p38 and extracellular signal-regulated mitogen-activated protein kinases and retards cell death by suppressing endoplasmic reticulum stress-mediated apoptosis. Mol Pharmacol. 2010;78:114–25. doi: 10.1124/mol.110.063495. [DOI] [PubMed] [Google Scholar]
- 18.Sirichanchuen B, Pengsuparp T, Chanvorachote P. Long-term cisplatin exposure impairs autophagy and causes cisplatin resistance in human lung cancer cells. Mol Cell Biochem. 2012;364:11–8. doi: 10.1007/s11010-011-1199-1. [DOI] [PubMed] [Google Scholar]
- 19.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 20.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–85. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, Wong PK, Zhang DD. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–43. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shim GS, Manandhar S, Shin DH, Kim TH, Kwak MK. Acquisition of doxorubicin resistance in ovarian carcinoma cells accompanies activation of the NRF2 pathway. Free Radic Biol Med. 2009;47:1619–31. doi: 10.1016/j.freeradbiomed.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Cho JM, Manandhar S, Lee HR, Park HM, Kwak MK. Role of the Nrf2-antioxidant system in cytotoxicity mediated by anticancer cisplatin: implication to cancer cell resistance. Cancer Lett. 2008;260:96–108. doi: 10.1016/j.canlet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Kim HJ, So HS, Lee JH, Lee JH, Park C, Park SY, Kim YH, Youn MJ, Kim SJ, Chung SY, Lee KM, Park R. Heme oxygenase-1 attenuates the cisplatin-induced apoptosis of auditory cells via down-regulation of reactive oxygen species generation. Free Radic Biol Med. 2006;40:1810–9. doi: 10.1016/j.freeradbiomed.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 25.Kim HR, Kim S, Kim EJ, Park JH, Yang SH, Jeong ET, Park C, Youn MJ, So HS, Park R. Suppression of Nrf2-driven heme oxygenase-1 enhances the chemosensitivity of lung cancer A549 cells toward cisplatin. Lung Cancer. 2008;60:47–56. doi: 10.1016/j.lungcan.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 26.Stepkowski TM, Kruszewski MK. Molecular cross-talk between the NRF2/KEAP1 signaling pathway, autophagy, and apoptosis. Free Radic Biol Med. 2011;50:1186–95. doi: 10.1016/j.freeradbiomed.2011.01.033. [DOI] [PubMed] [Google Scholar]
- 27.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–23. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 28.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu H, Su J, Xu Y, Kang J, Li H, Zhang L, Yi H, Xiang X, Liu F, Sun L. p62/SQSTM1 involved in cisplatin resistance in human ovarian cancer cells by clearing ubiquitinated proteins. Eur J Cancer. 2011;47:1585–94. doi: 10.1016/j.ejca.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 30.Qiao S, Tao S, Rojo DLVM, Park SL, Vonderfecht AA, Jacobs SL, Zhang DD, Wondrak GT. The antimalarial amodiaquine causes autophagic-lysosomal and proliferative blockade sensitizing human melanoma cells to starvation and chemotherapyinduced cell death. Autophagy. 2013;9:2087–102. doi: 10.4161/auto.26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ajabnoor GM, Crook T, Coley HM. Paclitaxel resistance is associated with switch from apoptotic to autophagic cell death in MCF-7 breast cancer cells. Cell Death Dis. 2012;3:e260. doi: 10.1038/cddis.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin X, Zhang N, Di W. Regulation of LC3-Dependent Protective Autophagy in Ovarian Cancer Cells by Protein Phosphatase 2A. Int J Gynecol Cancer. 2013;23:630–641. doi: 10.1097/IGC.0b013e3182892cee. [DOI] [PubMed] [Google Scholar]
- 33.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–91. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee MS, Tanaka K, Komatsu M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–84. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komatsu M. Potential role of p62 in tumor development. Autophagy. 2011;7:1088–90. doi: 10.4161/auto.7.9.16474. [DOI] [PubMed] [Google Scholar]
- 36.Zhu L, Barret EC, Xu Y, Liu Z, Manoharan A, Chen Y. Regulation of Cigarette Smoke (CS)-Induced Autophagy by Nrf2. PLoS One. 2013;8:e55695. doi: 10.1371/journal.pone.0055695. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Bray K, Mathew R, Lau A, Kamphorst JJ, Fan J, Chen J, Chen HY, Ghavami A, Stein M, DiPaola RS, Zhang D, Rabinowitz JD, White E. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS One. 2012;7:e41831. doi: 10.1371/journal.pone.0041831. [DOI] [PMC free article] [PubMed] [Google Scholar]


