Abstract
Objective: The present study is to evaluate the effect of methylated p16 on the progression in patients with gastric cancer (GC), and develop a useful biomarker for predicting patient’s prognosis. Design and methods: Methylation status of p16 in GC, their corresponding para-cancerous histological normal tissues (PCHNTs), preoperative peritoneal washes (PPWs) and serum were assessed using real-time methylation specific-PCR (MSP). Results: The frequency of p16 methylation was significantly higher in GC tissues (85.9%; 79/92) than that in paired PCHNTs (12.0%; 11/92) (P<0.0001). p16 methylation correlated closely with lymph node metastasis, peritoneal metastasis, TNM stage, et al (all P<0.05). Both frequency of p16 methylation in PPWs and serum were 79.7% (63/92). The Aζ value of the receiver-operator characteristic curve for methylated p16 was 0.899 for serum and PPWs, compared to that in GC tissues. The patients with elevated methylated p16 levels in tumor tissues had poorer disease-free survival (DFS) rates than those without (P=0.042). There is a narrow significant difference in median survival time of more than 30 months between patients with and without preoperatively detectable methylated p16 in serum (P=0.057). Methylated p16 in PPWs revealed no significant association with survival (P=0.129). Cox regression analysis showed that serum methylated p16 DNAs was an independent risk factor for GC patients, with a remarkable decrease in DFS 30 months after surgical resection of the gastric tumor. Conclusions: Serum methylated p16 DNAs might serve as a potential biomarker for the progression and a prognostic factor in gastric cancer patients.
Keywords: Gastric carcinoma, p16 gene, methylation, peritoneal micrometastasis, prognosis
Introduction
Gastric cancer (GC) is one of the most widespread cancers and the second leading cause of cancer-related death worldwide [1]. The annual diagnosis of GC amounts over 400,000 in China, accounting for 42% of new cases worldwide in 2010 [1,2]. Occurrence of GC is a multistep carcinogenesis processes involving multiple factors which include H. pylori infection, carcinogen, oncogene activation, suppressor gene inactivation, gastric epithelium cell apoptosis, and failure of proliferation regulation [3,4]. Most cases with GC are diagnosed at an advanced stage due to lack of specific symptoms or signs, and there is no standard method for early detection, which results in poor prognosis [5]. Peritoneal dissemination is the most common cause of metastasis from malignancies in the abdominal cavity [6]. The prognosis of patients with peritoneal dissemination from GC is very poor, with a median survival time of approximately 3 months [7-9]. Peritoneal dissemination is established through a multistep process. Many metastasis-related factors such as adhesion molecules, matrix proteases, motility factors and angiogenic factors are involved in the formation of peritoneal dissemination [10]. The first step is the detachment of cancer cells from the serosal surface of the primary tumor and these detached cancer cells are called peritoneal free cancer cells. The second metastatic process is trans-lymphatic metastasis. Peritoneal free cancer cells migrate to the subperitoneal lymphatic sinus through the lymphatic orifices (stomata), and then progress to the peritoneal surface [6-9]. At present, there are no ideal diagnostic methods for peritoneal dissemination and the conventional cytological results are poor [11,12].
Accumulating data strongly suggests that several genetic and epigenetic alterations play an important role in the carcinogenesis and progression of GC [13]. DNA promoter methylation is one of the most important epigenetic alteration which influences gene repression without affecting genetic coding [14]. Our recent studies have been conducted on assessing the clinical implications of epigenetics in GC [15-17]. Free circulating, methylated tumor-derived DNAs are a good target for early detection as a serum marker [15-19]. p16, also known as MTS1 (multiple tumor suppressor 1), acts as a cell-cycle regulator, which codes as inhibitor of cyclin D-dependent protein kinase 4 (CDK4) and 6 (CDK6) and regulates the process from G1-phase to S-phase in cell cycle [20,21]. As an important tumor suppressor, the inactivation of p16 participates in tumorigenesis and development in human cancers [22-24], including GC [25]. The inactivation patterns of p16 gene is mainly through the deletion [23,24] and the promoter hypermethylation [26]. Several studies showed that dysfunction of p16 are frequent and is regulated through p16 promoter hypermethylation [23-26]. Although p16 promoter hypermethylation in tumor tissues is very specific for early detection of gastric cancer [25,27], no report is available about p16 promoter methylation in fluids and its potentiality as a biomarker. Further assessment of p16 promoter methylation will be useful to raise the sensitivity and provide a panel of serum markers for the purpose. In the present study, we examined p16 promoter methylation in paired tissues, preoperative peritoneal washes (PPWs) and serum samples from 92 patients with gastric cancer, and evaluated circulating methylated p16 as a potential biomarker for early diagnosis and peritoneal micrometastasis of gastric cancer patients.
Materials and methods
Clinical specimens
Tumor tissues, paired para-cancerous histological normal tissues (PCHNTs), PPWs and serum samples from 92 patients who had been first diagnosed as gastric adenocarcinoma were obtained from Zhejiang Province Cancer Hospital from January 2008 to December 2009. All cases had been histologically diagnosed by the preoperative endoscopy biopsy and the postoperative biopsy diagnosis. All of patients did not receive preoperative radiotherapy and chemotherapy. The study comprised 53 male cases and 39 female cases, ranging in age from 36-73 years with an average age of 53.9 years. Demographic, clinical and histopathological parameters of the cases were shown in Table 1. The growth pattern of tumor cells was marked according to Ming’s classification [28]. As a measure of prognosis, we analyzed the clinical data concerning disease-free survival (DFS), defined as the time from surgery date to first recurrence or death by GC or last contact. All recruited patients had been followed-up periodically until the due date. PPWs from 92 GC patients were collected, according to previously reported method [17]. The cells from PPWs were isolated, and immediately stored at -80°C until DNAs was extracted. Meanwhile, some sediment was smeared onto one or more glass slides and stained using the Papanicolau’s method. All cytological examinations were performed by three independent cytopathologists. Cytological findings were classified as positive or negative, according to the cell characteristics as previously reported [17]. Antral mucosa biopsy specimens from 88 non-tumor volunteers by gastroscopy were randomly collected as controls within the same period, including 54 men and 34 women, with an average age of 52.9 years old. Among these volunteers, 48 patients were diagnosed with chronic non-atrophic gastritis. Meanwhile, paired serum samples in 88 non-tumor volunteers were collected before endoscopy.
Table 1.
Clinicopathological correlations of p16 methylation in gastric cancer tissues, serums and preoperative peritoneal washes
| Clinicopathological parameters | Gastric cancer tissues | Serums | PPWs | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| U | M | χ2 | U | M | χ2 | U | M | χ2 | |
|
|
|
|
|||||||
| (p-value) | (p-value) | (p-value) | |||||||
| Gender | |||||||||
| Male | 7 | 46 | 0.088 | 13 | 40 | 2.833 | 14 | 39 | 1.510 |
| Female | 6 | 33 | (0.767) | 16 | 23 | (0.092) | 15 | 24 | (0.219) |
| Age (year) | |||||||||
| <60 | 10 | 58 | 0 | 22 | 46 | 0.083 | 22 | 46 | 0.083 |
| ≥60 | 3 | 21 | 1.000 | 7 | 17 | (0.773) | 7 | 17 | (0.773) |
| HP infection | |||||||||
| (-) | 8 | 42 | 0.315 | 18 | 32 | 1.018 | 17 | 33 | 0.312 |
| (+) | 5 | 37 | (0.574) | 11 | 31 | (0.313) | 12 | 30 | (0.577) |
| Lesion site | |||||||||
| Cardia | 4 | 26 | 0 | 8 | 22 | 0.486 | 8 | 22 | 0.486 |
| Body/Antrum | 9 | 53 | 1.000 | 21 | 41 | (0.486) | 21 | 41 | (0.486) |
| Tumor size | |||||||||
| <5 cm | 11 | 49 | 1.614 | 24 | 36 | 5.744 | 24 | 36 | 5.744 |
| ≥5 cm | 2 | 30 | (0.204) | 5 | 27 | (0.017) | 5 | 27 | (0.017) |
| Growth type | |||||||||
| Swell | 4 | 16 | 0.239 | 5 | 15 | 0.504 | 5 | 15 | 0.504 |
| Infiltration | 9 | 63 | (0.625) | 24 | 48 | (0.478) | 24 | 28 | (0.478) |
| Differentiation | |||||||||
| Well/Moderate | 11 | 51 | 1.233 | 22 | 40 | 1.383 | 21 | 41 | 0.486 |
| Poor | 2 | 28 | (0.267) | 7 | 23 | (0.240) | 8 | 22 | (0.486) |
| PLC | |||||||||
| (-) | 13 | 40 | 11.14 | 21 | 32 | 3.801 | 20 | 33 | 2.237 |
| (+) | 0 | 39 | (0.001) | 8 | 31 | (0.051) | 9 | 30 | (0.135) |
| Lymphatic invasion | |||||||||
| (-) | 13 | 54 | 4.163 | 24 | 43 | 2.111 | 23 | 44 | 0.900 |
| (+) | 0 | 25 | (0.041) | 5 | 20 | (0.146) | 6 | 19 | (0.343) |
| Vein invasion | |||||||||
| (-) | 11 | 58 | 0.269 | 24 | 45 | 1.360 | 24 | 45 | 1.360 |
| (+) | 2 | 21 | (0.604) | 5 | 18 | (0.244) | 5 | 18 | (0.244) |
| T stage | |||||||||
| T1/T2 | 9 | 29 | 4.870 | 18 | 20 | 7.532 | 17 | 21 | 5.238 |
| T3/T4 | 4 | 50 | (0.027) | 11 | 43 | (0.006) | 12 | 42 | (0.022) |
| Lymph node metastasis | |||||||||
| N0 | 13 | 33 | 15.139 | 19 | 27 | 4.079 | 18 | 28 | 2.467 |
| N1-3 | 0 | 46 | (<0.0001) | 10 | 36 | (0.043) | 11 | 35 | (0.116) |
| Distant metastasis | |||||||||
| M0 | 13 | 72 | 0.305 | 29 | 56 | 3.488 | 29 | 56 | 2.086 |
| M1 | 0 | 7 | (0.581) | 0 | 7 | (0.062) | 0 | 7 | (0.149) |
| Clinical stage | |||||||||
| I/II | 13 | 33 | 15.139 | 20 | 26 | 6.093 | 19 | 27 | 4.079 |
| III/IV | 0 | 46 | (<0.0001) | 9 | 37 | (0.014) | 10 | 36 | (0.043) |
PLC: peritoneal lavage cytology; PPWs: preoperative peritoneal washes; M: methylation; U: Unmethylation.
Analysis of H. pylori infection
Biopsies were obtained from all patients who had endoscopic examination. H. pylori status was determined by rapid Urease test and Giemsa staining methods. It was considered as H. pylori infection when both tests were positive, and the samples with single positive were excluded for statistical analysis [29].
DNA extraction, sodium bisulfite modification and real-time MSP
The collected target cells from tissues were treated with 40 μl of 200 μg/ml proteinase K (Sigma–Aldrich) at 42°C, for 72 hours. DNAs in serums were extracted using QIAamp DNAs Blood Mini Kit (Qiagen Co., Germany). DNAs were modified by sodium bisulfite using the EpiTect Bisulfite kit (Qiagen Inc.) following kit’s instructions. Modified DNAs were analyzed by real-time MSP on the ABI7500 PCR (ABI Co.) using the SYBR Premix Taq ExTaq Kit (TaKaRa Co. Ltd). p16 promoter methylation and unmethylation specific primers were designed using previous reference [24]. The percentage of methylated DNAs in the samples was calculated, and methylated DNAs were scored according to previous reports [15-17]. The cut off threshold for DNAs hypermethylation was set as 20% based on control normal samples and internal quality controls provided in the real-time MSP analysis.
Immunohistochemical analysis
The expression of p16 protein was determined by immunohistochemical analysis with p16 monoclonal antibody (Santa Cruz Biotechnology). The score of immunohistochemical staining is determined by three independent pathologists based on combining staining frequency and intensity as previously described [30].
Statistical analysis
SPSS 17.0 (SPSS, Chicago, IL) statistical software was adopted for data analysis. Counting data comparisons between groups were subjected to the χ2 test and Fisher’s exact test. Survival analysis was performed by means of the Kaplan-Meier method and significant levels were assessed by means of the log-rank test. A univariate analysis with the Cox regression model was used to determine identified prognostic factors, and multivariate analysis with the Cox regression model was used to estimate the prognostic effect of methylated genes and significant levels were assessed by means of Wald test. For all statistical analyses, P values <0.05 were considered to be statistical significance.
Results
Frequency of p16 promoter methylation in primary gastric cancer tissues, paired serum and PPW specimens
We first examined the frequency of p16 promoter methylation in GC tumors and corresponding normal tissues using the real-time MSP technique. Out of 92 primary tumor samples, 79 samples (85.9%) exhibited aberrant p16 promoter methylation, while only 11 of 92 (12.0%) paired para-cancerous histological normal tissues (PCHNTs) specimens exhibited it. There was a significant difference (P<0.0001) between them for p16 promoter methylation. Meanwhile, we examined p16 promoter methylation in non-cancer controls. Out of 48 (20.8%) patients with chronic non-atrophic gastritis, only 10 patients exhibited p16 promoter methylation. The frequency of p16 promoter methylation was found in one of the 40 (2.5%) healthy individuals. The frequency of p16 promoter methylation in GC tissues was significant higher than that in chronic non-atrophic gastritis or in healthy controls, respectively (both P<0.0001). No significant difference in p16 promoter methylation between the PCHNTs and non-cancer controls (P=0.435) (Figure 1) was observed. The results indicated that p16 promoter methylation may be a good marker to detect GC DNAs in paired body fluids because of its high methylation frequency in tumors.
Figure 1.
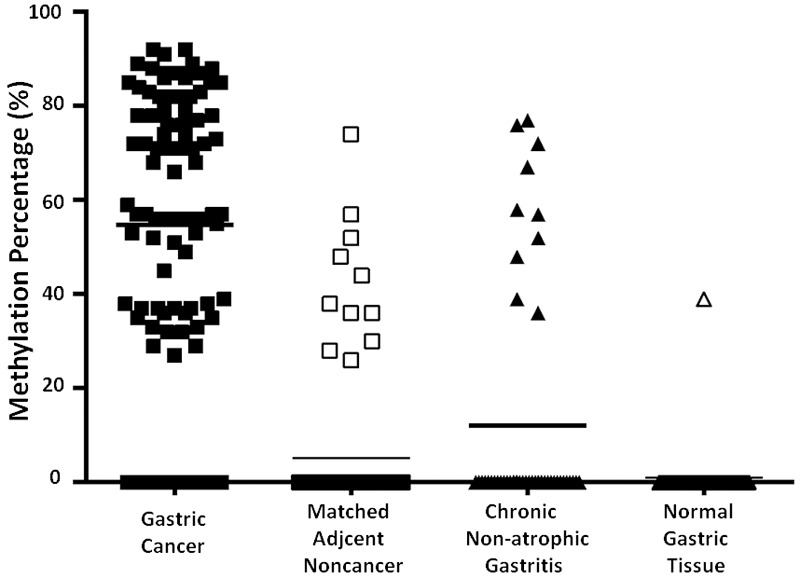
Summary of p16 hypermethylation in 272 samples including 92 gastric cancers (GC) and 92 para-cancerous histological normal tissue (PCHNT) from the same patients, 88 non-cancer volunteers. Data shows the frequency of p16 hypermethylation (DNAs methylation level ≥20%) in each group. The frequency of p16 promoter methylation in GC tumors is higher than that in corresponding normal tissues (P<0.0001). There is significant difference between GC tissues and patients with chronic non-atrophic gastritis or healthy individuals for p16 promoter methylation, respectively (both P<0.0001). There was not significant difference in p16 promoter methylation between in PCHNTs and non-cancer controls (P=0.435).
Subsequently, we examined p16 promoter methylation in the paired PPW and serum DNAs of patients with a p16 alteration in their primary tumors. Sixty-three of 79 (79.7%) patients exhibited the same alteration in their PPW DNAs, and sixty-three of 79 (79.7%) patients also demonstrated the similar alteration in paired serum DNAs. Of the 63 patients with a positive p16 promoter methylation in PPWs, 30 (47.6%) patients showed a positive cytology. And 30 of 39 (76.9%) patients with positive in peritoneal lavage cytology (PLC) demonstrated a p16 promoter methylation in PPWs. Results showed that the p16 promoter methylation in serums (γ=0.203, P=0.052) was almost correlated with the positive PLC, however, methylated p16 DNAs in PPWs (γ=0.156, P=0.138) was not correlated with the positive PLC.
Using the results of methylated p16 in GC tissues as the golden standard, the diagnostic value of PPWs or serums were determined by means of receiver-operating characteristic (ROC) curves, of which the Aζ value of ROC curve was 0.899 for both PPWs and serums, when compared to that in GC tissues (Figure 2).
Figure 2.
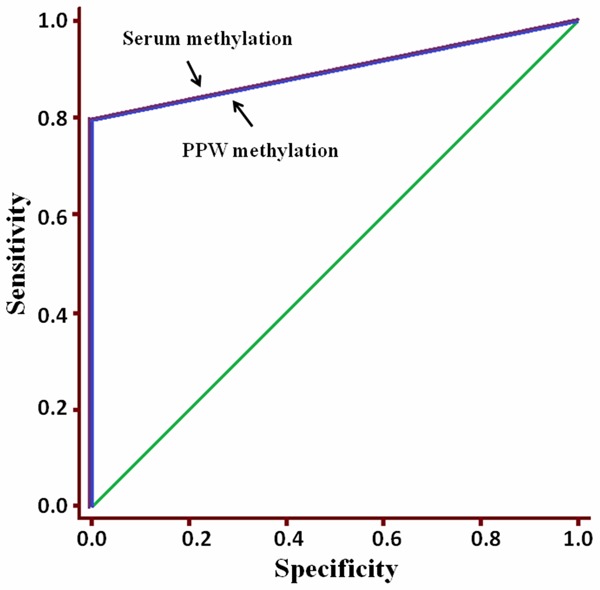
The diagnostic value of p16 hypermethylation in PPWs or serums was determined by means of receiver-operating characteristic (ROC) curves, of which the Aζ value of ROC curve was 0.899 for both (PPWs and serums) when compared to that in GC tissues. Both the value of p16 hypermethylation in PPWs/serums to diagnosis of GC progression were high.
p16 promoter methylation associated with malignant progression of gastric cancer
The frequency of p16 promoter methylation in GC tissues or PPWs or serums significantly correlated with tumor size, growth manner, histological differentiation, lymphatic invasion, venous invasion, invasive depth, lymph node metastasis, distant metastasis and clinical stage, respectively (all P<0.05) (Table 1). Especially, aberrant p16 methylation in body fluids associated with the positive results in PLC, which predicts peritoneal micrometastasis. 42.4% (39/ 92) GC patients showed a positive PLC was significantly related to the pathological findings. Overall, 94.9% of patients with a positive PLC had a T3/T4 tumor and 100% of the patients with a positive PLC had an N positive tumor (P<0.0001); in 76.9% of patients with a positive PLC, the tumor grade was low (P<0.0001). It indicated that the rate of positive PPW samples increases proportionally when the tumor invades into the deeper layers of the gastric wall or the lymph nodes, and when the tumor has lost differentiation. However, aberrant p16 promoter methylation in tumor tissues or body fluids have no relevance to gender, age at diagnosis, H. pylori infection, and tumor site (all P>0.05) (Table 1).
Down-regulation of p16 protein in primary gastric tumors
To check p16 protein level in GC tissues, we performed immunohistochemical analysis in the 92 GC tissues and their corresponding PCHNTs. The p16 protein expression was detected in high level in 98.9% (91/92) of PCHNTs (Figure 3A). However, high expression of p16 protein was only detected in 38.0% (35/92) of GC tissues and in majority of GC tissues the p16 protein level was low or absent (Figure 3B). The immunohistochemical findings are summarized in Table 2. The immunohistochemical staining score in GC tissues was significantly lower than that in PCHNTs (P<0.0001, Figure 3C). The loss of p16 protein significantly correlated with histological differentiation, lymphatic invasion, venous invasion, invasive depth, lymph node metastasis, distant metastasis and clinical stage (Table 2) (all P<0.05). However, it had no relationship to gender, age at diagnosis, growth manner, H. pylori infection, tumor size, and tumor site (all P>0.05) (Table 2). It was found that p16 protein level was significantly associated with p16 methylation level (γ=-0.926, P<0.0001).
Figure 3.
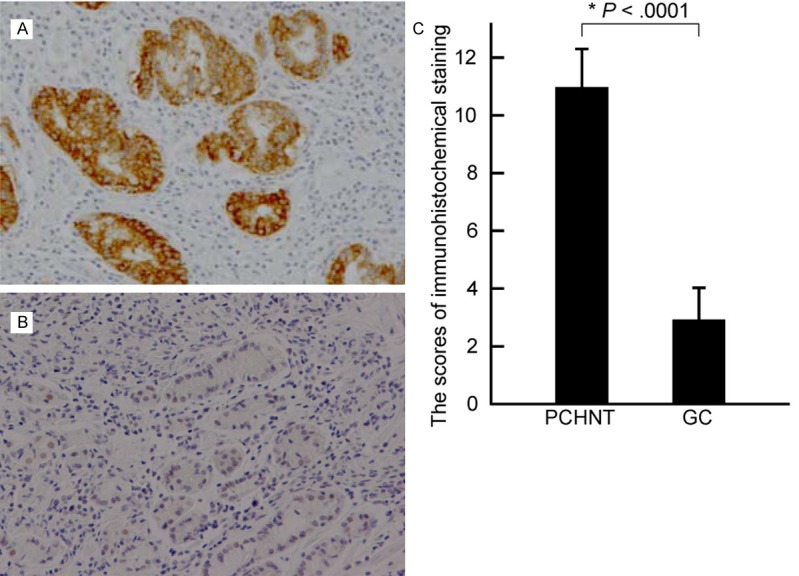
p16 protein expression was down-regulated in gastric cancer (GC) tissues. A. A representative positive, high expression of p16 protein in a PCHNT tissues; B. Absent of p16 expression in a poorly differentiated GC; original magnification ×200. C. Summary of the immunohistochemical staining results from 92 GCs and corresponding PCHNTs presented as ISS (immunohistochemical staining score) of the same samples.
Table 2.
Clinicopathological correlations of p16 protein expression in gastric cancer tissues
| Clinicopathological parameters | n | p16 expression | χ2 | P-value | |
|---|---|---|---|---|---|
|
| |||||
| Normal | Down | ||||
| Gender | |||||
| Male | 54 | 22 | 32 | 0.404 | 0.525 |
| Female | 38 | 13 | 25 | ||
| Age | |||||
| <60 | 68 | 23 | 45 | 1.969 | 0.161 |
| ≥60 | 24 | 12 | 12 | ||
| H. pylori | |||||
| (-) | 50 | 23 | 27 | 2.942 | 0.086 |
| (+) | 42 | 12 | 30 | ||
| Localization | |||||
| Cardia | 30 | 10 | 20 | 0.419 | 0.517 |
| Body/Antrum | 62 | 25 | 37 | ||
| Tumor size | |||||
| <5 cm | 60 | 26 | 34 | 2.048 | 0.152 |
| ≥5 cm | 32 | 9 | 23 | ||
| Growth manner | |||||
| Swell | 20 | 7 | 13 | 0.1 | 0.751 |
| Infiltration | 72 | 28 | 44 | ||
| Histological grade | |||||
| High/Medium | 62 | 29 | 33 | 6.149 | 0.013 |
| Low | 30 | 6 | 24 | ||
| Lymphatic invasion | |||||
| (-) | 67 | 31 | 36 | 7.077 | 0.008 |
| (+) | 25 | 4 | 21 | ||
| Venous invasion | |||||
| (-) | 69 | 31 | 38 | 5.549 | 0.018 |
| (+) | 23 | 4 | 19 | ||
| Invasive depth | |||||
| T1/T2 | 38 | 29 | 9 | 40.233 | 0.0001 |
| T3/T4 | 54 | 6 | 48 | ||
| Lymph node metastasis | |||||
| N0 | 46 | 25 | 21 | 10.376 | 0.001 |
| N1-3 | 46 | 10 | 36 | ||
| Distant metastasis | |||||
| M0 | 85 | 34 | 51 | 0.887 | 0.346 |
| M1 | 7 | 1 | 6 | ||
| TNM stage | |||||
| I/II | 46 | 30 | 16 | 28.822 | 0.0001 |
| III/IV | 46 | 5 | 41 | ||
Effect of methylated p16 on prognosis of GC patients
We performed survival analysis using the Kaplan-Meier method. At first, we examined the effect of methylated p16 in tumor tissues on the prognosis of GC patients. There was significantly difference between patients with preoperatively detectable and those patients with non-detectable (P=0.042, hazard ratio, 28.289; CI, 1.126-710.856), when detectable p16 promoter methylation as an unfavorable prognostic factor (Figure 4A). Subsequently, we analyzed the effect of methylated p16 in preoperative serums on the prognosis of GC patients. Results revealed a narrowly significant difference in median survival time of more than 30 months between patients with and without preoperatively detectable methylated p16 (P= 0.057; hazard ratio, 2.034; CI, 0.979-4.227) (Figure 4B). Finally, we found that in PPW specimens, methylated p16 showed no significant association with survival rate (P=0.129; hazard ratio, 1.724; CI, 0.853-3.485) (Figure 4C).
Figure 4.
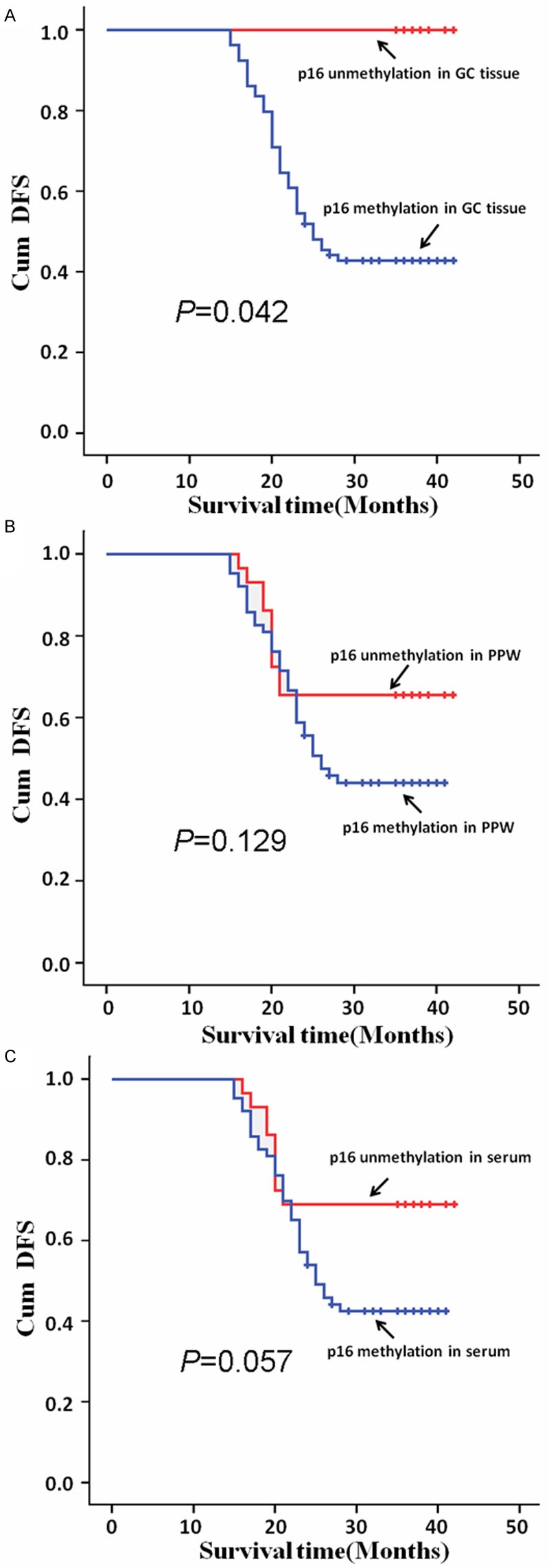
p16 hypermethylation in GC tissues and PPWs or serums correlated with patients’ prognosis. Cumulative disease-free survival (Cum DFS) curves are plotted against p16 DNAs hypermethylation level in GC tissues (A), in the PPWs (B) and in serums (C). In A-C, Kaplan-Meier analysis were used and P was 0.042, 0.129 and 0.057, respectively.
Disease-free survival (DFS) analyzed by a multivariate Cox regression model revealed that patients with methylated p16 DNAs in serum had an independent survival disadvantage (P<0.05; RR: 1.354, 95% CI: 1.003~1.826), only when the effect of TNM stages was eliminated. In addition, TNM stages could be considered as an independent influencing factor of prognosis in gastric cancer (P<0.0001; RR: 307.058, 95% CI: 21.190~4449.397). Using preoperative serum methylated p16 detection as a marker, we were able to discriminate between short (<2.5 years) and long survivors with quite a sensitivity and specificity. Preoperative detection levels of methylated p16 DNAs in serum was significantly higher in patients with peritoneal metastasis than that in those patients without peritoneal metastasis (P<0.0001).
Discussion
Aberrant hypermethylation of p16 has been studied in various malignancies tissues such as esophageal cancer [24], colorectal cancer [26], gastric cancer [25,27], et al. To explore the molecular mechanisms responsible for the p16 silence, we examined p16 promoter methylation in gastric cancer tissues (n=92), paired PCHNTs (n=92) and normal controls (n=88) using a real-time MSP technology. Results showed that the p16 hypermethylation was detected in 12.0% of PCHNTs, in 20.8% of patients with chronic non-atrophic gastritis and in 2.5% of normal gastric epithelial tissues from healthy individuals, in contrast with that were found to be present a fair frequency (85.9%) of promoter hypermethylation in GC tissues, which is in accord with the earlier reports that p16 promoter hypermethylation occur frequently in GC but not in normal gastric epithelial tissues [25,27]. Then, we confirmed that p16 protein expression was significantly down-regulated in primary gastric cancer tissues, and the down-regulation of p16 protein was significantly associated with tumor stages and metastasis, implicating loss of p16 function in tumor progression. It was found that p16 protein level significantly associates with p16 methylation level (γ=-0.926, P<0.0001). The frequency of p16 promoter methylation in tumor tissues significantly correlates with the progression of gastric cancer. Especially, aberrant p16 methylation in body fluids associated with the positive results in PLC, which predicts peritoneal micrometastasis. It indicated that the rate of positive PPW samples increases proportionally when the tumor invades the deeper layers of the gastric wall or the lymph nodes, and when the tumor has lost differentiation [17]. However, aberrant p16 promoter methylation in tumor tissues or body fluids have no relevance to gender, age at diagnosis, H. pylori infection, and tumor site, which is in consistence with previous report [25,27].
Recently, several studies have reported aberrant hypermethylation of fluids DNAs of cancer patients in diagnosis [15-19]. The hypermethylated status of p16 in body fluid DNAs of patients with malignancies and its clinical significance, have been described [31,32]. Nakayama et al found that p16 methylation score could sensitively reflect the recurrence status and may be useful for identifying the presence of recurrence during the follow-up of colorectal cancer patients [31]. Wakabayashi et al found p16 methylation in 12 (60%) of the 20 tissues with astrocytoma, but in only 1 of the 20 tissues with oligodendroglioma. Similar methylations were detected in the serum of 9 (75%) of the 12 patients with aberrant methylation in the tumor tissues. No methylated p16 sequences were detected in the peripheral serum of the patients having tumors without these methylation changes or in the 10 healthy controls. Additionally, p16 promoter methylation in the serum was observed in all brainstem astrocytoma cases, but not in other cases. It was suggested that p16 methylation has potential for use as a serum-based molecular diagnosis technique for diffuse glioma [32]. Our results showed that, of the 79 patients with the hypermethylated p16 in the primary tumors, 79.7% (64/79) paired serums was found to have detectable hypermethylated p16 DNAs available for MSP evaluation, respectively. However, no abnormal methylation was found in circulating DNAs, if this alteration was not present in the primary gastric tumor. The presence of detectable hypermethylated promoter p16 in serums indicates the presence of circulating tumor DNAs, which is closely related to the peritoneal metastasis and TNM stage (all P=0.051).
Preoperative measurement of methylated p16 promoter DNAs in serums may contribute to better estimate postoperative survival chances of GC patients. Here, we tried to elucidate whether circulating methylated p16 DNAs may reflect the clinical outcome of GC patients. Multivariate Cox proportional hazards regression analysis revealed that preoperative methylated p16 detection in serum is a significantly independent factor associated with outcome of GC patients (P<0.05; RR: 1.354, 95% CI: 1.003~1.826). Using preoperative circulating methylated p16 detection in serum as a marker, we were able to discriminate between short (<2.5 years) and long survivors with quite a sensitivity and specificity. Preoperative detection levels of methylated p16 DNAs in serum was significantly different between patients with and without peritoneal metastasis (P<0.0001), which it can be taken as a marker for peritoneal metastasis of GC. We could show that the level of methylated p16 DNAs in serum was significantly associated with outcome and that significantly increases the sensitivity and the specificity for the diagnosis of short versus long overall survival in GC patients. All these evidences suggest enhanced serum p16 hypermethylation predicts a poor prognosis for GC patients.
In summary, free-circulating methylated p16 DNAs in serum shows fine biological properties which can be compared favorably with the commonly used in tumor tissues and it may be used as a promising potential clinical applications in monitoring peritoneal metastasis, TNM stage, residual tumor and prognosis for GC patients after gastric surgery.
Acknowledgements
This research was supported by a grant from the program for New Century Excellent Talents in University, Ministry of Education, China (NCET-11-0949), a grant from the Science and Technology General Project of Zhejiang Province (no. 2009C33143), Key Research Projects of Medicine, Ministry of Health, China (No. WKJ2010-2-004), partly by a grant from the Backbone Talent of Zhejiang Provincial Medicine and Hygiene Platform Programs (no. 2011RCA009), a grant from the Scientific and Technological Innovations Fund of Henan Province Higher Education (no. 2009HAST1T001) and a grant from the Science and Technology Key Project of the Ministry of Education, China (no. 210130).
Disclosure of conflict of interest
None.
References
- 1.González CA, Sala N, Rokkas T. Gastric cancer: epidemiologic aspects. Helicobacter. 2013;18(Suppl 1):34–8. doi: 10.1111/hel.12082. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, Sun LL, Meng YL, Song GY, Hu JJ, Lu P, Ji B. Survival trends in gastric cancer patients of Northeast China. World J Gastroenterol. 2011;17:3257–62. doi: 10.3748/wjg.v17.i27.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamilton JP, Meltzer SJ. A review of the genomics of gastric cancer. Clin Gastroenterol Hepatol. 2006;4:416–25. doi: 10.1016/j.cgh.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita K, Sakuramoto S, Watanabe M. Genomic and epigenetic profiles of gastric cancer: potential diagnostic and therapeutic applications. Surg Today. 2011;41:24–38. doi: 10.1007/s00595-010-4370-5. [DOI] [PubMed] [Google Scholar]
- 5.Seshadri RA, Jayanand SB, Ranganathan R. Prognostic factors in patients with node-negative gastric cancer: an Indian experience. World J Surg Oncol. 2011;9:48. doi: 10.1186/1477-7819-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang M, Zhang H, Ma Y, Zhu G, Xue Y. Prognosis and surgical treatment of gastric cancer invading adjacent organs. ANZ J Surg. 2010;80:510–4. doi: 10.1111/j.1445-2197.2010.05376.x. [DOI] [PubMed] [Google Scholar]
- 7.Ali Z, Deng Y, Ma C. Progress of research in gastric cancer. J Nanosci Nanotechnol. 2012;12:8241–8. doi: 10.1166/jnn.2012.6692. [DOI] [PubMed] [Google Scholar]
- 8.Yonemura Y, Endo Y, Obata T, Sasaki T. Recent advances in the treatment of peritoneal dissemination of gastrointestinal cancers by nucleoside antimetabolites. Cancer Sci. 2007;98:11–8. doi: 10.1111/j.1349-7006.2006.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadeghi B, Arvieux C, Glehen O, Beaujard AC, Rivoire M, Baulieux J, Fontaumard E, Brachet A, Caillot JL, Faure JL, Porcheron J, Peix JL, François Y, Vignal J, Gilly FN. Peritoneal carcinomatosis from non-gynecologic malignancies: results of the EVOCAPE 1 multicentric prospective study. Cancer. 2000;88:358–63. doi: 10.1002/(sici)1097-0142(20000115)88:2<358::aid-cncr16>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 10.Chu DZ, Lang NP, Thompson C, Osteen PK, Westbrook KC. Peritoneal carcinomatosis in nongynecologic malignancy. A prospective study of prognostic factors. Cancer. 1989;63:364–7. doi: 10.1002/1097-0142(19890115)63:2<364::aid-cncr2820630228>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto M, Matsuyama A, Kameyama T, Okamoto M, Okazaki J, Utsunomiya T, Tsutsui S, Fujiwara M, Ishida T. Prognostic re-evaluation of peritoneal lavage cytology in Japanese patients with gastric carcinoma. Hepatogastroenterology. 2009;56:261–5. [PubMed] [Google Scholar]
- 12.Kodera Y. Gastric cancer with minimal peritoneal metastasis: is this a sign to give up or to treat more aggressively? Nagoya J Med Sci. 2013;75:3–10. [PMC free article] [PubMed] [Google Scholar]
- 13.Figueiredo C, Garcia-Gonzalez MA, Machado JC. Molecular pathogenesis of gastric cancer. Helicobacter. 2013;18(Suppl 1):28–33. doi: 10.1111/hel.12083. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5:553–68. doi: 10.2217/epi.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling ZQ, Lv P, Lu XX, Yu JL, Han J, Ying LS, Zhu X, Zhu WY, Fang XH, Wang S, Wu YC. Circulating Methylated XAF1 DNA Indicates Poor Prognosis for Gastric Cancer. PLoS One. 2013;8:e67195. doi: 10.1371/journal.pone.0067195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu XX, Yu JL, Ying LS, Han J, Wang S, Yu QM, Wang XB, Fang XH, Ling ZQ. Stepwise cumulation of RUNX3 methylation mediated by Helicobacter pylori infection contributes to gastric carcinoma progression. Cancer. 2012;118:5507–17. doi: 10.1002/cncr.27604. [DOI] [PubMed] [Google Scholar]
- 17.Yu QM, Wang XB, Luo J, Wang S, Fang XH, Yu JL, Ling ZQ. CDH1 methylation in preoperative peritoneal washes is an independent prognostic factor for gastric cancer. J Surg Oncol. 2012;106:765–71. doi: 10.1002/jso.23116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Mattos-Arruda L, Olmos D, Tabernero J. Prognostic and predictive roles for circulating biomarkers in gastrointestinal cancer. Future Oncol. 2011;7:1385–97. doi: 10.2217/fon.11.122. [DOI] [PubMed] [Google Scholar]
- 19.Vlassov VV, Laktionov PP, Rykova EY. Circulating nucleic acids as a potential source for cancer biomarkers. Curr Mol Med. 2010;10:142–65. doi: 10.2174/156652410790963295. [DOI] [PubMed] [Google Scholar]
- 20.Sperka T, Wang J, Rudolph KL. DNA damage checkpoints in stem cells, ageing and cancer. Nat Rev Mol Cell Biol. 2012;13:579–90. doi: 10.1038/nrm3420. [DOI] [PubMed] [Google Scholar]
- 21.Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130:1715–25. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu W, Sahin F, Iacobuzio-Donahue CA, Garcia-Carracedo D, Wang WM, Kuo CY, Chen D, Arking DE, Lowy AM, Hruban RH, Remotti HE, Su GH. Disruption of p16 and activation of Kras in pancreas increase ductal adenocarcinoma formation and metastasis in vivo. Oncotarget. 2011;2:862–73. doi: 10.18632/oncotarget.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guney S, Jardin F, Bertrand P, Mareschal S, Parmentier F, Picquenot JM, Tilly H, Bastard C. Several mechanisms lead to the inactivation of the CDKN2A (P16), P14ARF, or CDKN2B (P15) genes in the GCB and ABC molecular DLBCL subtypes. Genes Chromosomes Cancer. 2012;51:858–67. doi: 10.1002/gcc.21970. [DOI] [PubMed] [Google Scholar]
- 24.Tokugawa T, Sugihara H, Tani T, Hattori T. Modes of silencing of p16 in development of esophageal squamous cell carcinoma. Cancer Res. 2002;62:4938–44. [PubMed] [Google Scholar]
- 25.Lu ZM, Zhou J, Wang X, Guan Z, Bai H, Liu ZJ, Su N, Pan K, Ji J, Deng D. Nucleosomes correlate with in vivo progression pattern of de novo methylation of p16 CpG islands in human gastric carcinogenesis. PLoS One. 2012;7:e35928. doi: 10.1371/journal.pone.0035928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bihl MP, Foerster A, Lugli A, Zlobec I. Characterization of CDKN2A(p16) methylation and impact in colorectal cancer: systematic analysis using pyrosequencing. J Transl Med. 2012;10:173. doi: 10.1186/1479-5876-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou XP, Zhang B, Zhang XQ, Chen M, Cao J, Liu WJ. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Hum Pathol. 2009;40:1534–42. doi: 10.1016/j.humpath.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 28.Luebke T, Baldus SE, Grass G, Bollschweiler E, Thiele J, Dienes HP, Hoelscher AH, Moenig SP. Histological grading in gastric cancer by Ming classification: correlation with histopathological subtypes, metastasis, and prognosis. World J Surg. 2005;29:1422–7. doi: 10.1007/s00268-005-7795-z. discussion 1428. [DOI] [PubMed] [Google Scholar]
- 29.Tian XY, Zhu H, Zhao J, She Q, Zhang GX. Diagnostic performance of urea breath test, rapid urea test, and histology for Helicobacter pylori infection in patients with partial gastrectomy: a meta-analysis. J Clin Gastroenterol. 2012;46:285–92. doi: 10.1097/MCG.0b013e318249c4cd. [DOI] [PubMed] [Google Scholar]
- 30.Soslow RA, Dannenberg AJ, Rush D, Woerner BM, Khan KN, Masferrer J, Koki AT. COX-2 is expressed in human pulmonary, colonic, and mammary tumors. Cancer. 2000;89:2637–45. doi: 10.1002/1097-0142(20001215)89:12<2637::aid-cncr17>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama G, Kodera Y, Ohashi N, Koike M, Fujiwara M, Nakao A. p16INK4a methylation in serum as a follow-up marker for recurrence of colorectal cancer. Anticancer Res. 2011;31:1643–6. [PubMed] [Google Scholar]
- 32.Wakabayashi T, Natsume A, Hatano H, Fujii M, Shimato S, Ito M, Ohno M, Ito S, Ogura M, Yoshida J. p16 promoter methylation in the serum as a basis for the molecular diagnosis of gliomas. Neurosurgery. 2009;64:455–61. doi: 10.1227/01.NEU.0000340683.19920.E3. discussion 461-2. [DOI] [PubMed] [Google Scholar]


