Abstract
Although increasing studies have indicated that Nucleotide-oligomerization domain-containing protein 1 (NOD1) signaling could play an important role in gastrointestinal tumorigenesis, the protein expression and function of NOD1 signaling have not been understood well in oral squamous cell carcinoma (OSCC) progression. The objective of this study is, thus, to examine protein expression of NOD1 signaling immunohistochemically in normal, premalignant and malignant specimens of oral cavity, and to take insights into the association between the protein expression of NOD1 signaling pathway and OSCC precession. In this study immunohistochemical expression of NOD1, Receptor-interacting protein 2 (RIP2), Caspase12, human β Defensin1, 2 and 3 (hBD1, 2, 3) was examined in 15 normal controls, 30 cases of oral leukoplakia (OLK) and 60 cases of OSCC. The immunostaining score was assessed by 2 pathologists, respectively. We found that the expression of NOD1, RIP2, Caspase12, hBD1, 2, and 3 decreased along with the progression of OSCC. NOD1 expression was correlated significantly to tumor differentiation, lymph node metastasis, and tumor size. Our results also showed the correlation of hBD2, 3 to lymph node metastasis of OSCC. These results suggest that the dysfunction of NOD1 signaling pathways could be associated with OSCC development and progression. NOD1, RIP2 and Caspase12 could be used as potentially novel biomarkers for oral carcinogenesis.
Keywords: NOD1, RIP2, Caspase12, human β Defensin, oral squamous cell carcinoma
Introduction
Innate immunity constitutes the first line of defense against infection by microbes. As two main classes of innate immune receptors, the Toll-like receptors (TLR) and NOD-like receptors (NLR) serve as pattern recognition receptors that recognize conserved structures of pathogens and endogenous compounds or cellular damage known as “danger signals”. Although the importance of innate immune signals in sensing microbes has been established, the molecular machineries whereby innate immunity communicates with oncogenic stresses and regulates tumorigenesis remain elusive [1].
TLRs have been implicated in the tumorigenesis, but the role of NLRs in the tumorigenesis remains unclear. As a fundamental member of NLR family, nucleotide-oligomerization domain-containing protein 1 (NOD1) plays an important role in the induction of innate immune and inflammatory responses. Several crucial proteins, NOD1, Receptor-interacting protein 2 (RI-P2), Caspase12 and human β Defensin1, 2, 3 (hBD1, 2, 3) constitute or regulate NOD1 signaling pathway. da Silva Correia and colleagues [2] were the first to report that NOD1 could play an important role in controlling tumorigenesis. The disruption of NOD1 functions rendered MCF-7 breast cancer cells to resist apoptotic cell death and promote in vivo tumor growth. Using an established mouse model system of colitis-associated colon tumorigenesis, study data of Chen et al. suggested that NOD1 deficiency results in the increased development of both colitis-associated and Apc tumor suppressor-related colon tumors [3]. Moreover, NOD1/CARD4 and NOD2/CARD15 gene polymorphisms may be associated with altered risk of various tumors [4]. Recently a study has shown that gene polymorphisms of NOD1 may be associated with Helicobacter pylori-induced premalignant gastric lesions and gastric cancer in the Chinese population [5].
RIP2 is not only a crucial constituent of NOD1 signaling but also an important regulator of cellular proliferation, apoptosis and differentiation. In a most recent study, Guirado et al. found the association of RIP2 polymorphisms and urothelial cancer risk [6].
Caspase12 is a crucial molecule associated with endoplasmic reticulum (ER) stress-induced apoptosis and inflammasome activation. Increasing reports indicate Caspase-12 is a pivotal regulator of tissue homeostasis and innate immunity, including NOD1 signaling [7-9]. Dupaul-Chicoine et al. showed that Caspase-12 deficiency resulted in exaggerated epithelial cell compensatory proliferation due to the resistance to acute and chronic colitis and could enhance azoxymethane and dextran sulfate sodium induced colon tumorigenesis [10].
hBD1, 2, 3 are small secreted peptides and expressed in mucosal tissues. Significantly altered expression of hBDs has been observed in some tumors, such as renal cancer, prostate cancer, lung cancer, cervical cancer, cutaneous squamous cell carcinoma (CSCC), oral squamous cell carcinoma (OSCC), and salivary gland tumor [11-22]. Therefore, hBDs have been considered as potential cancer biomarkers and antitumour molecules [23].
In the gastrointestinal tract, NOD1 signaling has been linked to gastric cancer and colon cancer, but the expression and role of NOD1 signaling pathway in oral cancer have not been clarified. Therefore, we hypothesized there were potential links between expressional changes of crucial proteins in NOD1 signaling pathway and the tumorigenesis of oral cavity. The aim of our study was to investigate the potentiality of crucial proteins in NOD1 signaling pathway as novel biomarkers for oral squamous cell carcinoma progression.
Materials and methods
Patients and tissue sampling
In total, 105 specimens from 105 individuals (60 OSCC, 30 OLK, 15 normal controls) were included in the study. Before the study, informed consent was obtained from each subject. All had been undergone histopathological examination and surgical resection at the Institute and Hospital of Stomatology, Nanjing University Medical School between 2011 and 2012. Medical records were reviewed to collect patient data including age, gender, tumor differentiation, lymph node metastasis, tumor size, and pathological grade of OLK. No patient underwent preoperative radiotherapy, chemotherapy, or any other treatment. Normal specimens of oral mucosa were from the normal controls who received orthognathic surgery or surgical removal of completely impacted third molar. This study was approved by the Ethics Committee of the Institute and Hospital of Stomatology, Nanjing University Medical School.
Immunohistochemistry
Serial tissue sections (4 μm) were sliced from paraffin-embedded formalin-fixed normal, preneoplastic or neoplastic tissues and immunohistochemical staining was performed. Briefly, tissue sections were deparaffinized, hydrated, and stained using specific primary antibodies, biotin-conjugated secondary antibodies, and horse radish peroxidase (HRP)-conjugated avidin. Specific antibody interactions were detected with the HRP substrate 3, 3’-diaminobenzidine (DAB). Subsequently, sections were washed and counterstained with hematoxylin. Appropriate positive controls were concurrently performed and negative controls were obtained by omission of the primary antibodies from the staining procedure. Primary antibodies used were rabbit polyclonal anti-NOD1 (1:200; Abcam), mouse monoclonal anti-RIP2 (1:50 dilution; Abcam), rabbit polyclonal anti-Caspase12 (1:800 dilution; Abcam), mouse monoclonal anti-hBD1 (1:300 dilution; Abcam), rabbit polyclonal anti-hBD2 (1:500 dilution; Abcam), and rabbit polyclonal anti-hBD3 (1:400 dilution; Novus).
Immunostaining scoring
The immunostaining results were assessed by 2 pathologists, respectively. The immunoreactivity measurement was represented by the intensity and percentage of positive staining. Final immunohistochemical score was determined from a combination of intensity and percentage and categorized as - (no staining or the percentage of positive staining ≤5% of epithelial cells), +1 (weak, the percentage of positive staining >5% and ≤25% of epithelial cells), +2 (moderate, the percentage of positive staining >25% and ≤50% of epithelial cells), and +3 (strong, the percentage of positive staining >50% of epithelial cells) at 10 randomly selected 400×magnified fields. In order to avoid artificial effects, significant folding, staining artifacts or blurriness was excluded.
Statistical analysis
Statistical analysis was performed using SPSS 13.0 (SPSS, Chicago, IL). The Kruskal-Wallis test was used to evaluate the differences of immunohistochemical score data between multiple groups. The Fisher’s exact test was used to evaluate the differences of immunohistochemical score data between two groups. P value less than 0.05 was considered to be statistically significant.
Results
Clinicopathological characteristics of patient cohort
Table 1 summarizes patient’s clinicopathological characteristics. The ages of the patients and normal controls ranged from 19 to 84 years (mean, 55.1 years). In 30 patients with OLK, 13 patients are male and 17 are female. The age range of OLK patients is from 28 to 75 years (mean, 56.3 years). In total, 60 patients with OSCC, including 32 males and 28 females, were analyzed in this retrospective study. The ages of the patients ranged from 41 to 84 years (mean, 59.9 years). The tumor sizes ranged from 0.5 to 4 cm (mean, 2.1 cm). The histological tumor type of all 60 patients was squamous cell carcinoma.
Table 1.
Characteristics of tissue samples used for immunohistochemical analysis
| Variables | Frequency | % |
|---|---|---|
| Age | 55.1* | |
| Gender | ||
| Male | 49 | 46.7 |
| Female | 56 | 53.3 |
| Diagnostic category | ||
| Healthy control | 15 | 14.3 |
| OLK | 30 | 28.6 |
| OSCC | 60 | 57.1 |
| OLK pathological grade | ||
| No dysplasia | 9 | 30.0 |
| Mild dysplasia | 13 | 43.3 |
| Moderate dysplasia | 7 | 23.3 |
| Severe dysplasia | 1 | 3.4 |
| OSCC differentiation | ||
| Well | 40 | 66.7 |
| Moderate | 12 | 20.0 |
| Poor | 8 | 13.3 |
| Tumor size | ||
| <2 cm | 24 | 40.0 |
| ≥2 cm | 36 | 60.0 |
| Lymph node metastasis | ||
| Negative | 44 | 73.3 |
| Positive | 16 | 26.7 |
| Neoplasm metastasis | ||
| Negative | 60 | 100 |
| Positive | 0 | 0 |
mean value.
The expression of NOD1, RIP2, Caspase12 and hBD1, 2, 3
We examined expression of NOD1, RIP2, Caspase12 and hBD1, 2, 3 in normal mucosa, OLK and OSCC specimens by IHC. Representative immunohistochemical expression of NOD1, RIP2, Caspase12 and hBD1, 2, 3 are presented in Figure 1. IHC analysis showed that the expression of NOD1, RIP2, Caspase12 and hBD1, 2, 3 decreased gradually with the progression of OSCC. In the normal squamous epithelium and OLK tissues, moderate to strong staining intensity was observed, while no or weak staining intensity was noted in malignant epithelium frequently. As Figure 1 shown, NOD1, Caspase12, hBD1 and hBD2 expression are observed in the cytoplasm and nucleus. RIP2 and hBD3 expression was mainly observed in the cytoplasm, with some cases also demonstrating nucleus staining. The staining intensity of NOD1, RIP2 and Caspase12 in cancer nest areas was weaker than that in surrounding dysplastic epithelium areas (Figure 2).
Figure 1.
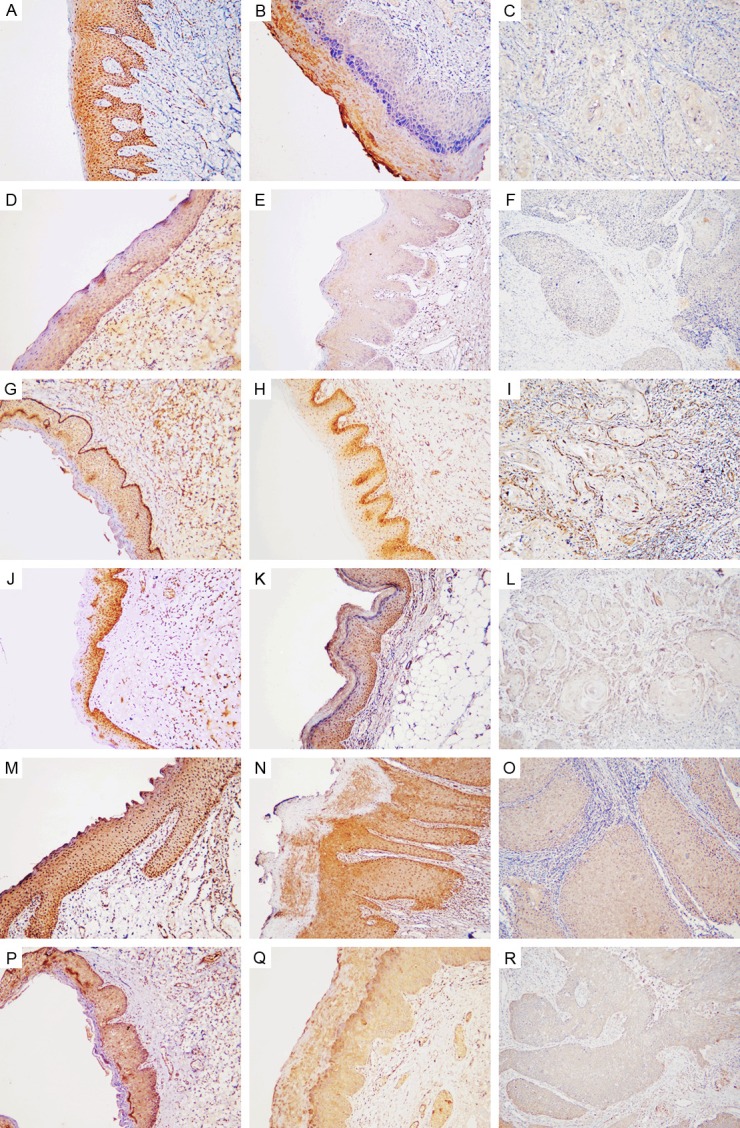
Representative immunohistochemical expression for NOD1, RIP2, Caspase12, hBD1, hBD2 and hBD3. NOD1 is stained in cytoplasm and nucleus shown with strong staining in the whole epithelium of normal oral specimens (A), moderate cytoplasm staining mainly in the granular layers and cuticular layer of OLK tissues (B), and weak staining in OSCC (C). RIP2 is stained in cytoplasm shown with moderate staining in normal oral epithelium (D), weak staining in OLK (E), and no staining in OSCC (F). Caspase12 (G-I), hBD1 (J-L) and hBD2 (M-O) are shown with cytoplasmic and nuclear staining, while hBD3 is mainly shown with cytoplasmic staining (P-R). Representative Caspase12 expression is shown with extremely strong staining intensity in the basal layer and strong staining intensity in the spinous layer of normal oral epithelium (G), strong staining intensity in OLK (H) and weak staining intensity in OSCC (I). hBD1 is detected in the squamous epithelium shown with strong staining in normal oral specimens (J), strong staining in OLK (K), and absent staining in OSCC (L). hBD2 is detected in the squamous epithelium shown with strong staining in normal oral specimens (M), strong staining in OLK (N), and weak staining in OSCC (O). hBD3 is detected in the squamous epithelium shown with strong staining in normal oral specimens (P), strong staining in OLK (Q), and absent staining in OSCC (R). Original magnification: 100×.
Figure 2.
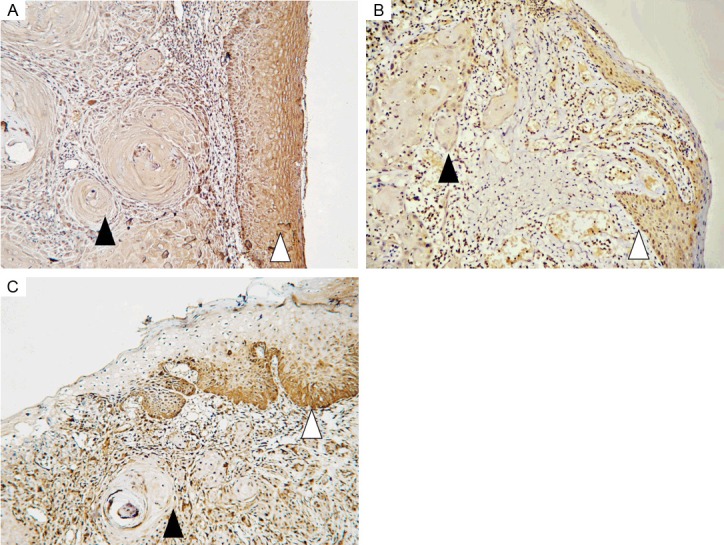
Representative immunohistochemical staining for NOD1 (A), RIP2 (B) and Caspase12 (C) at the border between cancer nest area of OSCC and surrounding dysplastic epithelium area. Closed arrowheads indicate cancer nest areas, while open arrowheads indicate dysplastic epithelium areas. Original magnification: 100×.
Relationship between immunostaining score results and clinicopathological parameters
Immunostaining score results of NOD1, RIP2, Caspase12 and hBD1, 2, 3 were described in Tables 2, 3, 4, 5 and 6. NOD1, RIP2, Caspase12 and hBD1, 2, 3 expression correlated significantly to diagnostic category (Table 2). NOD1 expression was also correlated significantly to tumor differentiation (P = 0.008), lymph node metastasis (P = 0.014), and tumor size (P = 0.027) (Tables 3, 4 and 5). However, there was no statistically significant difference in NOD1 expression with regard to dysplastic grade of OLK (Table 6). RIP2 and hBD1 expression was not correlated significantly to tumor differentiation, lymph node metastasis, tumor size, and dysplastic grade of OLK (Tables 3, 4, 5 and 6). Statistically significant correlation was found between Caspase12 expression and tumor differentiation (P = 0.049), while there was no association between Caspase12 expression and lymph node metastasis, tumor size and dysplastic grade of OLK (Tables 3, 4, 5 and 6). There was statistically significant difference between hBD2 expression and tumor differentiation (P = 0.031), lymph node metastasis (P = 0.004), dysplastic grade of OLK (P = 0.002) (Tables 3, 4 and 6). However, there was no association between hBD2 expression and tumor size (Table 5). hBD3 expression was correlated significantly to lymph node metastasis (P = 0.039) and tumor size (P = 0.011), while there was no statistically significant difference in hBD3 expression with regard to tumor differentiation and dysplastic grade of OLK (Tables 3, 4, 5 and 6).
Table 2.
Association of immunostaining scores of NOD1, RIP2, Caspase12 and hBD1, 2, 3 and OSCC progression
| Variables | Diagnostic category | N | n (%) | p value | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| - | +1 | +2 | +3 | ||||
| NOD1 | HC | 15 | 0 (0) | 2 (14) | 5 (33) | 8 (53) | 0.006 |
| OLK | 30 | 1 (3) | 17 (57) | 9 (30) | 3 (10) | ||
| OSCC | 60 | 20 (33) | 14 (23) | 8 (14) | 18 (30) | ||
| RIP2 | HC | 15 | 1 (7) | 5 (33) | 9 (60) | 0 (0) | <0.001 |
| OLK | 30 | 8 (26) | 18 (60) | 2 (7) | 2 (7) | ||
| OSCC | 60 | 45 (75) | 9 (15) | 6 (10) | 0 (0) | ||
| Caspase12 | HC | 15 | 0 (0) | 0 (0) | 2 (13) | 13 (87) | <0.001 |
| OLK | 30 | 1 (3) | 2 (7) | 17 (57) | 10 (33) | ||
| OSCC | 60 | 6 (10) | 23 (38) | 24 (40) | 7 (12) | ||
| hBD1 | HC | 15 | 0 (0) | 0 (0) | 3 (20) | 12 (80) | <0.001 |
| OLK | 30 | 0 (0) | 8 (27) | 12 (40) | 10 (33) | ||
| OSCC | 60 | 4 (7) | 36 (60) | 14 (23) | 6 (10) | ||
| hBD2 | HC | 15 | 0 (0) | 1 (7) | 3 (20) | 11 (73) | <0.001 |
| OLK | 30 | 0 (0) | 0 (0) | 2 (7) | 28 (93) | ||
| OSCC | 60 | 2 (3) | 8 (13) | 30 (50) | 20 (34) | ||
| hBD3 | HC | 15 | 0 (0) | 0 (0) | 4 (27) | 11 (73) | <0.001 |
| OLK | 30 | 1 (3) | 6 (20) | 12 (40) | 11 (37) | ||
| OSCC | 60 | 7 (12) | 27 (45) | 14 (23) | 12 (20) | ||
Table 3.
Association of immunostaining scores of NOD1, RIP2, Caspase12 and hBD1, 2, 3 and tumor differentiation of OSCC
| Variables | Tumor differentiation | N | n (%) | p value | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| - | +1 | +2 | +3 | ||||
| NOD1 | Well | 40 | 10 (25) | 8 (20) | 6 (15) | 16 (40) | 0.008 |
| Moderate | 12 | 4 (33) | 4 (33) | 2 (17) | 2 (17) | ||
| Poor | 8 | 6 (75) | 2 (25) | 0 (0) | 0 (0) | ||
| RIP2 | Well | 40 | 30 (75) | 4 (10) | 6 (15) | 0 (0) | 0.168 |
| Moderate | 12 | 7 (58) | 5 (42) | 0 (0) | 0 (0) | ||
| Poor | 8 | 8 (100) | 0 (0) | 0 (0) | 0 (0) | ||
| Caspase12 | Well | 40 | 4 (10) | 15 (38) | 17 (42) | 4 (10) | 0.049 |
| Moderate | 12 | 0 (0) | 4 (33) | 5 (42) | 3 (25) | ||
| Poor | 8 | 2 (25) | 4 (50) | 2 (25) | 0 (0) | ||
| hBD1 | Well | 40 | 0 (0) | 26 (65) | 12 (30) | 2 (5) | 0.053 |
| Moderate | 12 | 3 (25) | 3 (25) | 2 (17) | 4 (33) | ||
| Poor | 8 | 1 (13) | 7 (87) | 0 (0) | 0 (0) | ||
| hBD2 | Well | 40 | 2 (5) | 3 (8) | 17 (42) | 18 (45) | 0.031 |
| Moderate | 12 | 0 (0) | 3 (25) | 7 (58) | 2 (17) | ||
| Poor | 8 | 0 (0) | 2 (25) | 6 (75) | 0 (0) | ||
| hBD3 | Well | 40 | 3 (8) | 17 (42) | 8 (20) | 12 (30) | 0.080 |
| Moderate | 12 | 4 (33) | 3 (25) | 5 (42) | 0 (0) | ||
| Poor | 8 | 0 (0) | 7 (88) | 1 (12) | 0 (0) | ||
Table 4.
Association of immunostaining scores of NOD1, RIP2, Caspase12 and hBD1, 2, 3 and lymph node metastasis of OSCC
| Variables | Lymph node metastasis | N | n (%) | p value | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| - | +1 | +2 | +3 | ||||
| NOD1 | Negative | 44 | 10 (23) | 10 (23) | 8 (18) | 16 (36) | 0.014 |
| Positive | 16 | 10 (62) | 4 (25) | 0 (0) | 2 (13) | ||
| RIP2 | Negative | 44 | 33 (75) | 7 (16) | 4 (9) | 0 (0) | 0.895 |
| Positive | 16 | 12 (75) | 2 (13) | 2 (12) | 0 (0) | ||
| Caspase12 | Negative | 44 | 4 (9) | 18 (41) | 18 (41) | 4 (9) | 0.666 |
| Positive | 16 | 2 (13) | 5 (31) | 6 (37) | 3 (19) | ||
| hBD1 | Negative | 44 | 3 (7) | 27 (61) | 10 (23) | 4 (9) | 0.959 |
| Positive | 16 | 1 (6) | 9 (56) | 4 (25) | 2 (13) | ||
| hBD2 | Negative | 44 | 2 (5) | 3 (7) | 27 (61) | 12 (27) | 0.004 |
| Positive | 16 | 0 (0) | 5 (31) | 3 (19) | 8 (50) | ||
| hBD3 | Negative | 44 | 6 (14) | 16 (36) | 10 (23) | 12 (27) | 0.039 |
| Positive | 16 | 1 (6) | 11 (69) | 4 (25) | 0 (0) | ||
Table 5.
Association of immunostaining scores of NOD1, RIP2, Caspase12 and hBD1, 2, 3 and tumor size of OSCC
| Variables | Tumor size | N | n (%) | p value | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| - | +1 | +2 | +3 | ||||
| NOD1 | <2 cm | 24 | 10 (42) | 4 (16) | 0 (0) | 10 (42) | 0.027 |
| ≥2 cm | 36 | 10 (28) | 10 (28) | 8 (22) | 8 (22) | ||
| RIP2 | <2 cm | 24 | 20 (84) | 2 (8) | 2 (8) | 0 (0) | 0.510 |
| ≥2 cm | 36 | 25 (70) | 7 (19) | 4 (11) | 0 (0) | ||
| Caspase12 | <2 cm | 24 | 2 (8) | 6 (25) | 14 (59) | 2 (8) | 0.139 |
| ≥2 cm | 36 | 4 (11) | 17 (47) | 10 (28) | 5 (14) | ||
| hBD1 | <2 cm | 24 | 2 (8) | 14 (59) | 6 (25) | 2 (8) | 1.000 |
| ≥2 cm | 36 | 2 (6) | 22 (61) | 8 (22) | 4 (11) | ||
| hBD2 | <2 cm | 24 | 0 (0) | 1 (4) | 15 (63) | 8 (33) | 0.191 |
| ≥2 cm | 36 | 2 (6) | 7 (19) | 15 (42) | 12 (33) | ||
| hBD3 | <2 cm | 24 | 3 (13) | 5 (21) | 8 (33) | 8 (33) | 0.011 |
| ≥2 cm | 36 | 4 (11) | 22 (61) | 6 (17) | 4 (11) | ||
Table 6.
Association of immunostaining scores of NOD1, RIP2, Caspase12 and hBD1, 2, 3 and dysplasia grade of OLK
| Variables | OLK dysplasia | N | n (%) | p value | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| - | +1 | +2 | +3 | ||||
| NOD1 | No | 9 | 1 (11) | 5 (56) | 1 (11) | 2 (22) | 0.322 |
| Mild | 13 | 0 (0) | 7 (54) | 6 (46) | 0 (0) | ||
| Moderate | 7 | 0 (0) | 5 (71) | 2 (29) | 0 (0) | ||
| Severe | 1 | 0 (0) | 0 (0) | 0 (0) | 1 (100) | ||
| RIP2 | No | 9 | 3 (33) | 4 (45) | 2 (22) | 0 (0) | 0.204 |
| Mild | 13 | 4 (31) | 9 (69) | 0 (0) | 0 (0) | ||
| Moderate | 7 | 1 (14) | 5 (72) | 0 (0) | 1 (14) | ||
| Severe | 1 | 0 (0) | 0 (0) | 0 (0) | 1 (100) | ||
| Caspase12 | No | 9 | 0 (0) | 1 (11) | 6 (67) | 2 (22) | 0.180 |
| Mild | 13 | 1 (8) | 1 (8) | 8 (61) | 3 (23) | ||
| Moderate | 7 | 0 (0) | 0 (0) | 3 (43) | 4 (57) | ||
| Severe | 1 | 0 (0) | 0 (0) | 0 (0) | 1 (100) | ||
| hBD1 | No | 9 | 0 (0) | 1 (11) | 6 (67) | 2 (22) | 0.480 |
| Mild | 13 | 0 (0) | 5 (38) | 3 (24) | 5 (38) | ||
| Moderate | 7 | 0 (0) | 1 (14) | 3 (43) | 3 (43) | ||
| Severe | 1 | 0 (0) | 1 (100) | 0 (0) | 0 (0) | ||
| hBD2 | No | 9 | 0 (0) | 0 (0) | 1 (11) | 8 (89) | 0.002 |
| Mild | 13 | 0 (0) | 0 (0) | 0 (0) | 13 (100) | ||
| Moderate | 7 | 0 (0) | 0 (0) | 0 (0) | 7 (100) | ||
| Severe | 1 | 0 (0) | 0 (0) | 1 (100) | 0 (0) | ||
| hBD3 | No | 9 | 0 (0) | 1 (11) | 2 (22) | 6 (67) | 0.153 |
| Mild | 13 | 0 (0) | 3 (23) | 6 (46) | 4 (31) | ||
| Moderate | 7 | 1 (14) | 2 (29) | 3 (43) | 1 (14) | ||
| Severe | 1 | 0 (0) | 0 (0) | 1 (100) | 0 (0) | ||
Discussion
In the present study, we found a statistically significant association between the diminishing expression of NOD1 and OSCC progression. Moreover NOD1 expression was also correlated significantly to tumor differentiation, lymph node metastasis, and tumor size. To the best of our knowledge, the present study showed the involvement of NOD1 signaling pathway in oral carcinogenesis for the first time.
The etiology of cancer is highly complicated. The development of cancer has been associated with microbial infection, repeating injury, chronic inflammation, cigarette smoke and genetics factors [6]. NOD1 have an important role not only in host defense against pathogens but also in maintaining tissue homeostasis by regulating cell apoptosis, the inflammatory and tissue repair responses to injury. The dysregulation of NOD1 signaling could result in the persistent infection and chronic inflammation due to insufficient pathogen clearance, while chronic inflammation can initiate and promote cancer development [2]. In the gastrointestinal tract, NOD1 has been linked to gastric cancer and colon cancer [3,5]. Our results show that decreased expression of NOD1 could play a crucial role in oral carcinogenesis.
In this study, we found the expression of RIP2 diminished paralleled with oral carcinogenesis. The results suggest RIP2 appeared to be involved in the progression of OSCC. NOD1 is thought to recruit the downstream effector RIP2. Activation of the effector kinase RIP2 induces the NF-κB and the MAP kinase pathways, resulting in activation of inflammatory genes and production of antimicrobial polypeptide, such as hBD1, 2, 3. Therefore RIP2 plays an important role in mediating inflammation and innate immune. A recent study indicates that RIP2 polymorphism could promote a bladder chronic inflammation, leading to the development of bladder cancer [6]. Apart from the crucial role in inflammatory response to infection, RIP2 can mediate NOD1-dependent apoptosis [24]. A previous study indicates that differential expression of RIP2 can be associated with abnormal growth and differentiation behavior of skeletal myoblasts [25]. Therefore our results show that abnormal expression of RIP2 in OLK and OSCC could impair innate immune and disturb cell apoptosis, proliferation and differentiation which promote the carcinogenesis of oral mucosa.
In this study, we found that the expression of Caspase-12 is significantly associated with OSCC progression. Considerable data confirm that Caspase12 plays a crucial role in ER stress-mediated apoptosis and inflammasome activation [9]. Therefore Caspase12 downregulation could contribute to the apoptosis resistance and chronic inflammation, both of which could promote the OSCC progression. In addition, our results revealed significant correlation between Caspase12 expression and tumor differentiation. Some conditions, such as hypoxia and nutrient deprivation, are frequently encountered in tumor tissues and induce ER stress of cancer cells. Therefore, the role of ER stress in tumor progression is being actively researched. The expression of some other ER stress-related proteins, such as CHOP and BIP, had been correlated to histological grade or tumor size of lung cancer [26]. Our results and others indicate that the expression of ER stress markers could be related to clinicopathologic factors of human cancers.
In the present study, we found that the expression of hBD1, 2, 3 is correlated to OSCC progression. The results are consistent with the previous study [18,21]. hBDs play important roles in innate immune and adaptive immune, such as antimicrobial activity, antitumor effect, chemoattractive effect and immunomodulation. Decreased production of hBDs has negative influence on antimicrobial activity, wound healing, and immunoregulatory functions, all of which are associated with cancer progression.
Study data on hBDs expression in OSCC are in conflict. Loss of hBD1, hBD2 and hBD3 expression has been shown in OSCC [21]. Several studies have reported that hBD1, hBD2 and hBD3 gene expression were significantly downregulated in OSCC [18,21]. Decreased expression of hBD1 has also been suggested in OLK, a common premalignant lesion of oral mucosa [18]. Oppositely, elevated expression of hBD2, 3 has been reported in OSCC [17,19,20]. Overexpression hBD3 has also been observed in premalignant cells of oral mucosa [20]. According to accumulated evidences, hBDs could play a complex and poorly understood role in cancer pathogenesis either promoting or suppressing tumor cell growth [14].
A study showed that there was significantly lower hBD2 expression in poorly differentiated lung cancer than that in moderately differentiated lung cancer [14]. In accordance with the previous study, our results indicated there was the correlation of hBD2 to tumor differentiation of OSCC. The other finding in our study was the correlation of hBD2, 3 to lymph node metastasis of OSCC. Study data have shown that hBD2 may play a role in tumor angiogenesis and cancer metastasis [27]. Several studies suggested that hBD3 enhanced lymphatic invasion of squamous cell carcinoma of the head and neck [28,29]. However, a recent study showed that hBD3 suppressed head and neck cancer cell migration [30]. Except hBD2, we did not find the association of NOD1, RIP2, Caspase12, hBD1 and hBD3 expression with dysplastic degree of OLK. This is a limitation in our study. One explanation for the limitation could be small number of OLK specimens, especially for OLK specimen with severe dysplasia. The other explanation could be that differential protein expression in neoplastic cells is more notable than that in dysplastic cells.
Biological markers of cancer have become increasingly important for the clinician to diagnose, predict and monitor progression, and assess the therapeutic effect of interventions on underlying pathogenic mechanisms [31]. The present study and strong evidences from others suggest that hBDs may be useful markers of OSCC. The current data indicate that NOD1, RIP2, Caspase12 may serve as a novel and potential biomarker for development and progression of OSCC. Further studies are needed to investigate the molecular mechanisms of NOD1 signaling pathway in the development of OSCC.
Acknowledgements
This work was supported by the National Natural Scientific Foundation of China (No.81070839), Jiangsu Province’s Outstanding Medical Academic Leader program (No.LJ201110), and Key Project supported by Medical Science and technology development Foundation, Nanjing Department of Health (No.ZKX10030).
Disclosure of conflict of interest
The authors declare no conflicts of interest.
References
- 1.Jinushi M. The role of innate immune signals in antitumor immunity. Oncoimmunology. 2012;1:189–194. doi: 10.4161/onci.1.2.18495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Da SCJ, Miranda Y, Austin-Brown N, Hsu J, Mathison J, Xiang R, Zhou H, Li Q, Han J, Ulevitch RJ. Nod1-dependent control of tumor growth. Proc Natl Acad Sci U S A. 2006;103:1840–1845. doi: 10.1073/pnas.0509228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen GY, Shaw MH, Redondo G, Nunez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 2008;68:10060–10067. doi: 10.1158/0008-5472.CAN-08-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutikhin AG. Role of NOD1/CARD4 and NOD2/CARD15 gene polymorphisms in cancer etiology. Hum Immunol. 2011;72:955–968. doi: 10.1016/j.humimm.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Wang P, Zhang L, Jiang JM, Ma D, Tao HX, Yuan SL, Wang YC, Wang LC, Liang H, Zhang ZS, Liu CJ. Association of NOD1 and NOD2 genes polymorphisms with Helicobacter pylori related gastric cancer in a Chinese population. World J Gastroenterol. 2012;18:2112–2120. doi: 10.3748/wjg.v18.i17.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guirado M, Gil H, Saenz-Lopez P, Reinboth J, Garrido F, Cozar JM, Ruiz-Cabello F, Carretero R. Association between C13ORF31, NOD2, RIPK2 and TLR10 polymorphisms and urothelial bladder cancer. Hum Immunol. 2012;73:668–672. doi: 10.1016/j.humimm.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Bian ZM, Elner SG, Elner VM. Regulated expression of caspase-12 gene in human retinal pigment epithelial cells suggests its immunomodulating role. Invest Ophthalmol Vis Sci. 2008;49:5593–5601. doi: 10.1167/iovs.08-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeBlanc PM, Yeretssian G, Rutherford N, Doiron K, Nadiri A, Zhu L, Green DR, Gruenheid S, Saleh M. Caspase-12 modulates NOD signaling and regulates antimicrobial peptide production and mucosal immunity. Cell Host Microbe. 2008;3:146–157. doi: 10.1016/j.chom.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Scott AM, Saleh M. The inflammatory caspases: guardians against infections and sepsis. Cell Death Differ. 2007;14:23–31. doi: 10.1038/sj.cdd.4402026. [DOI] [PubMed] [Google Scholar]
- 10.Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, Meunier C, Turbide C, Gros P, Beauchemin N, Vallance BA, Saleh M. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Donald CD, Sun CQ, Lim SD, Macoska J, Cohen C, Amin MB, Young AN, Ganz TA, Marshall FF, Petros JA. Cancer-specific loss of beta-defensin 1 in renal and prostatic carcinomas. Lab Invest. 2003;83:501–505. doi: 10.1097/01.lab.0000063929.61760.f6. [DOI] [PubMed] [Google Scholar]
- 12.Sun CQ, Arnold R, Fernandez-Golarz C, Parrish AB, Almekinder T, He J, Ho SM, Svoboda P, Pohl J, Marshall FF, Petros JA. Human beta-defensin-1, a potential chromosome 8p tumor suppressor: control of transcription and induction of apoptosis in renal cell carcinoma. Cancer Res. 2006;66:8542–8549. doi: 10.1158/0008-5472.CAN-06-0294. [DOI] [PubMed] [Google Scholar]
- 13.Shestakova T, Zhuravel E, Bolgova L, Alekse-enko O, Soldatkina M, Pogrebnoy P. Expression of human beta-defensins-1, 2 and 4 mRNA in human lung tumor tissue: a pilot study. Exp Oncol. 2008;30:153–156. [PubMed] [Google Scholar]
- 14.Shestakova T, Zhuravel E, Bolgova L, Zaitsev S, Efanova O, Soldatkina M, Pogrebnoy P. Immunohistochemical analysis of beta-defensin-2 expression in human lung tumors. Exp Oncol. 2010;32:273–276. [PubMed] [Google Scholar]
- 15.Hubert P, Herman L, Maillard C, Caberg JH, Nikkels A, Pierard G, Foidart JM, Noel A, Boniver J, Delvenne P. Defensins induce the recruitment of dendritic cells in cervical human papillomavirus-associated (pre)neoplastic lesions formed in vitro and transplanted in vivo. Faseb J. 2007;21:2765–2775. doi: 10.1096/fj.06-7646com. [DOI] [PubMed] [Google Scholar]
- 16.Scola N, Gambichler T, Saklaoui H, Bechara FG, Georgas D, Stucker M, Glaser R, Kreuter A. The expression of antimicrobial peptides is significantly altered in cutaneous squamous cell carcinoma and precursor lesions. Br J Dermatol. 2012;167:591–597. doi: 10.1111/j.1365-2133.2012.11110.x. [DOI] [PubMed] [Google Scholar]
- 17.Sawaki K, Mizukawa N, Yamaai T, Yoshimoto T, Nakano M, Sugahara T. High concentration of beta-defensin-2 in oral squamous cell carcinoma. Anticancer Res. 2002;22:2103–2107. [PubMed] [Google Scholar]
- 18.Wenghoefer M, Pantelis A, Dommisch H, Reich R, Martini M, Allam JP, Novak N, Berge S, Jepsen S, Winter J. Decreased gene expression of human beta-defensin-1 in the development of squamous cell carcinoma of the oral cavity. Int J Oral Maxillofac Surg. 2008;37:660–663. doi: 10.1016/j.ijom.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Kesting MR, Loeffelbein DJ, Hasler RJ, Wolff KD, Rittig A, Schulte M, Hirsch T, Wagenpfeil S, Jacobsen F, Steinstraesser L. Expression profile of human beta-defensin 3 in oral squamous cell carcinoma. Cancer Invest. 2009;27:575–581. doi: 10.1080/07357900802620851. [DOI] [PubMed] [Google Scholar]
- 20.Kawsar HI, Weinberg A, Hirsch SA, Venizelos A, Howell S, Jiang B, Jin G. Overexpression of human beta-defensin-3 in oral dysplasia: potential role in macrophage trafficking. Oral Oncol. 2009;45:696–702. doi: 10.1016/j.oraloncology.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 21.Joly S, Compton LM, Pujol C, Kurago ZB, Guthmiller JM. Loss of human beta-defensin 1, 2, and 3 expression in oral squamous cell carcinoma. Oral Microbiol Immunol. 2009;24:353–360. doi: 10.1111/j.1399-302X.2009.00512.x. [DOI] [PubMed] [Google Scholar]
- 22.Kesting MR, Stoeckelhuber M, Kuppek A, Hasler R, Rohleder N, Wolff KD, Nieberler M. Human beta-defensins and psoriasin/S100A7 expression in salivary glands: anti-oncogenic molecules for potential therapeutic approaches. Biodrugs. 2012;26:33–42. doi: 10.2165/11597570-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 23.Droin N, Hendra JB, Ducoroy P, Solary E. Human defensins as cancer biomarkers and antitumour molecules. J Proteomics. 2009;72:918–927. doi: 10.1016/j.jprot.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Da SCJ, Miranda Y, Leonard N, Hsu J, Ulevitch RJ. Regulation of Nod1-mediated signaling pathways. Cell Death Differ. 2007;14:830–839. doi: 10.1038/sj.cdd.4402070. [DOI] [PubMed] [Google Scholar]
- 25.Ehlers S, Mueck T, Adams S, Landuzzi L, Lollini PL, Munz B. RIP2 regulates growth and differentiation of normal myoblasts and of rhabdomyosarcoma cells. Eur J Cell Biol. 2008;87:163–172. doi: 10.1016/j.ejcb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Kim KM, Yu TK, Chu HH, Park HS, Jang KY, Moon WS, Kang MJ, Lee DG, Kim MH, Lee JH, Chung MJ. Expression of ER stress and autophagy-related molecules in human non-small cell lung cancer and premalignant lesions. Int J Cancer. 2012;131:E362–E370. doi: 10.1002/ijc.26463. [DOI] [PubMed] [Google Scholar]
- 27.Kawsar HI, Ghosh SK, Hirsch SA, Koon HB, Weinberg A, Jin G. Expression of human beta-defensin-2 in intratumoral vascular endothelium and in endothelial cells induced by transforming growth factor beta. Peptides. 2010;31:195–201. doi: 10.1016/j.peptides.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Mburu YK, Abe K, Ferris LK, Sarkar SN, Ferris RL. Human beta-defensin 3 promotes NF-kappaB-mediated CCR7 expression and anti-apoptotic signals in squamous cell carcinoma of the head and neck. Carcinogenesis. 2011;32:168–174. doi: 10.1093/carcin/bgq236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shuyi Y, Feng W, Jing T, Hongzhang H, Haiyan W, Pingping M, Liwu Z, Zwahlen RA, Hongyu Y. Human beta-defensin-3 (hBD-3) upregulated by LPS via epidermal growth factor receptor (EGFR) signaling pathways to enhance lymphatic invasion of oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2011;112:616–625. doi: 10.1016/j.tripleo.2011.02.053. [DOI] [PubMed] [Google Scholar]
- 30.Wang K, Wang JH, Baskaran H, Wang R, Jurevic R. Effect of human beta-defensin-3 on head and neck cancer cell migration using micro-fabricated cell islands. Head Neck Oncol. 2012;4:41. doi: 10.1186/1758-3284-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCully ML, Fairhead T, Blake PG, Madrenas J. The future of RIP2/RICK/CARDIAK as a biomarker of the inflammatory response to infection. Expert Rev Mol Diagn. 2008;8:257–261. doi: 10.1586/14737159.8.3.257. [DOI] [PubMed] [Google Scholar]


