Abstract
The Ski-interacting protein (SKIP) is a transcriptional cofactor distinct from other cofactors and is involved in regulation of many cancer-related proteins. However, its distribution and clinical significances in bladder cancer remains poorly understood. In this study, Quantitative real-time PCR and immunohistochemistry were performed to detect the expression of SKIP in clinical bladder cancer samples. In addition, the correlation of SKIP expression and clinicopathological features and clinical outcomes were analyzed. The expression levels of SKIP in clinical bladder cancer were much higher than that in paired adjacent noncancerous tissues. High expression of SKIP was closely related with histological grades and the poor prognosis of bladder cancer. Based on our data, we speculated that SKIP may be a potential prognostic marker in bladder cancer.
Keywords: SKIP expression, prognosis, bladder cancer
Introduction
Bladder cancer is the first and second most common malignancy of the urinary system in China and the USA, respectively [1]. Approximately 75% of patients with bladder cancer present as non-muscle invasive cancer (NMIBC) and the remaining 25% of BCs are muscle-invasive bladder cancer (MIBC) [2]. Although great progress has been made in the diagnosis and treatment of bladder cancer, tumor recurrence is still an annoying clinical problem. The reported recurrence rates of NMIBC after transurethral resection of bladder tumor (TUR-BT) are 10%~67% in 12 months [3,4] and the cumulative 5-yr mortality rate is up to 30.2% [5]. Currently, there are no reliable biomarkers to predict the risk of recurrence and the prognosis of bladder cancer [6].
SKIP was originally found as a binding partner of the viral oncogene v-Ski in a yeast two-hybrid system. It was demonstrated to interact with a highly conserved region of Ski, which is thought to be important for Ski’s transforming activity [7]. Previous studies have shown that SKIP can combine with pRb to form a highly stable complex, which effectively inhibit pRb-induced transcriptional repression and induce cell cycle activation [8]. SKIP also binds to Smad2,3 to enhance TGF-β-dependent transcription and may regulate cell growth and differentiation through the TGF-β pathway [9]. In addition, SKIP significantly overcomes both TGF-β1-induced uPA and MMP-9 promoter transactivation in the androgen-independent human prostate cancer cells [10]. Overall, these studies suggested that SKIP was a required transcriptional coactivator for many newly induced genes, which include interactions with VDR and RXR in a tripartite complex with SRCs [11], with androgen receptor in the nucleus and enhances AR-dependent transactivation, and so on [12]. Thus, SKIP appears to modulate a number of key signalling pathways involved in cell proliferation and differentiation, and as such may play a role in oncogenesis and tumor development. These properties of SKIP may become a novel therapeutic target in bladder cancer treatment.
In the present study, we detected the expression of SKIP protein in clinical low and high- grade urothelial carcinoma samples, and the mRNA expression of SKIP in both bladder cancer and adjacent noncancerous tissues. In addition, we investigated the relationship of the expression of SKIP and clinical prognosis and clinicopathological classification of bladder cancer. Thus we demonstrated that SKIP may be used as a potential marker for the prognosis of bladder cancer patients.
Materials and methods
Patients and tissue samples
A total of 70 formalin-fixed and paraffin-embedded specimens of bladder cancer were collected from the patients who suffered surgical resection without any radiotherapy and chemotherapy before the operations at the department of urology, the second hospital affiliated of Nanchang University from 2007 to 2012. 10 Paired fresh frozen samples include both bladder cancer and adjacent noncancerous tissues were collected for the mRNA analysis also and the adjacent noncancerous tissues were carefully checked without any evidences of Malignancy. The diagnosis of bladder cancer was confirmed by pathology. Of the 70 patients, 55 were men and 15 were women, aged (60.51±11.97) years (range 29-79) and 59 patients had primary NMIBC and 11 had MIBC. The tumors were graded according to the WHO 2004 criteria and staged by the 2009 TNM classification system. Pathological grades were classified to well (grade I; n=30), moderately (grade II; n=21), and poorly differentiated tumors (grade III; n=19). grade I and grade II were classified as low-grade urothelial carcinoma, grade III was classified as high-grade urothelial carcinoma. 70 cases of bladder cancer were divided into 19 cases of high-grade urothelial carcinoma and 51 cases of low-grade urothelial carcinoma according to the pathological results.
The mainly clinicopathological features of the patients are shown in Table 1. All bladder tissues samples were collected using protocols approved by the Ethics Committee of Cancer the Second Hospital Affiliated of Nanchang University, and written informed consent was obtained from every patient.
Table 1.
Expression of SKIP expression in bladder cancer patients according to clinicopathologic characteristics
| Expression of SKIP expression in bladder cancer patients according to clinicopathologic characteristics | ||||
|---|---|---|---|---|
|
| ||||
| NO | SKIP expression | p | ||
|
|
||||
| High N (%) | Low N (%) | |||
| Sex | 1.000 | |||
| Male | 55 | 17 | 38 | |
| Female | 15 | 5 | 10 | |
| Age (y) | 0.128 | |||
| ≤45 | 9 | 5 | 4 | |
| >45 | 61 | 17 | 44 | |
| Differentiation | 0.001* | |||
| Grade 1/2 | 51 | 10 | 41 | |
| Grade 3 | 19 | 12 | 7 | |
| Tumor types | 0.731 | |||
| NMIBC | 59 | 18 | 41 | |
| MIBC | 11 | 4 | 7 | |
| Distant metastasis | 0.448 | |||
| - | 61 | 18 | 43 | |
| + | 9 | 4 | 5 | |
| Recurrence | 0.004* | |||
| - | 38 | 6 | 32 | |
| + | 32 | 16 | 16 | |
Statistical analyses were performed by the Pearson χ2 test
P<0.05 was considered significant.
qRT-PCR analysis
Total RNA were extracted from 10 pairs of bladder tissues and adjacent noncancerous tissues using TRIZOL reagent (Invitrogen) according to the manufacturer’s Instructions. RNA was reverse-transcribed using SuperScript First Strand cDNA System (Invitrogen). The skip sense primer was Sense: 5’-GTGGAAATGCGTG-CCCAAGTAG-3’, and the antisense primer was 5’-CGAGGATTAGGAACACCGAGAG-3’. The GAPDH was used as an internal control, the sense primer was 5’-GTCACCAGGGCTGCTTTTAACTC-3’, and the antisense primer was 5’-CAGCATC-GCCCCACTTGATTTTG-3’. qRT-PCR was done using SYBR Green PCR master mix (Biotium) in a total volume of 20 μl on the CFX96 fast Real-time PCR system (BIO-RAD) as follows: 94°C for 3 min, 45 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s, then the relative levels of skip gene expression were detected according to the manufacturer’s instructions and the experiments were repeated in triplicatein the same reaction.
Immunohistochemistry (IHC)
Tissue sections (5-μm thick) of the paraffin-embedded samples from bladder cancer samples were cut and then put them on slides coated Poly-lysine. The expression of skip in bladder cancer samples were examined following standard immunohistochemistry protocol. Briefly, after deparaffinization and rehydration, the slides were sent into a boiling ethylenediamine tetraacetic acid (EDTA) buffer (pH=8.0) for Antigen retrieval. And then the sections were incubated in 3% hydrogen peroxide for 10 min at room temperature to quench endogenous peroxidase activity and 5% normal Bovine serum albumin for 20 min to block nonspecific conjugation. Subsequently, the slides were incubated with anti-SKIP rabbit polyclonal antibody (diluted 1:200; Santa Cruz) which was diluted with 1% FSG at 4°C overnight. All the sections were treated with biotinylated secondary antibody for 2 hours and then incubated with a streptavidin–horseradish peroxidase complex. After washed by intervening PBS, the sections were stained with DAB (3,3-diaminobenzidine). Finally, sections were rinsed with distilled water, counterstained with Mayer’s hematoxylin, dehydrated, and mounted. The expression of SKIP in clinical bladder cancer samples were evaluated by two independent, experienced clinical pathology experts.
Statistical analysis
Chi-square test was used to evaluate the association of the SKIP expression with clinical and pathological features. Survival curves (overall survival and recurrence-free survival) were evaluated by Kaplan-Meier curves and the differences between the groups were performed using the log ran test. The relationship between the expression of SKIP and pathological features for the prediction of recurrence was assessed by the Cox proportional hazards regression analysis. Statistical analysis was performed by using SPSS 13.0 for Windows XP (SPSS Inc., Chicago, IL, USA). Results were considered statistically significant if the P value was less than 0.05.
Results
The mRNA expression of SKIP in bladder cancer samples and adjacent noncancerous tissues
10 Paired fresh frozen samples include both bladder cancer and adjacent noncancerous tissues were collected for the SKIP mRNA analysis. SKIP mRNA expression in bladder cancer samples and adjacent noncancerous tissues were 0.332±0.179 and 0.059±0.060 respectively, the results of qRT-PCR analysis showed that SKIP mRNA expression in bladder cancer samples were significantly higher than that in adjacent noncancerous tissues. These results suggested that SKIP was correlated with the tumorigenesis of bladder cancer.
The protein expression of SKIP in low and high-grade urothelial carcinoma samples
In the present work, to investigate the potential roles of SKIP in bladder cancer, we examined the expression of SKIP in 70 paraffin embedded by immunohistochemistry. The representative immunostaining of SKIP in low and high-grade urothelial carcinoma samples were shown in Figure 2. We found that the majority high-grade urothelial carcinoma samples were high expression of SKIP in 12 cases (63.2%) and the low expression of SKIP in 7 cases (36.8%). However the minority low-grade urothelial carcinoma samples were high expression of SKIP only in 10 cases (19.6%), and the remaining were low expression of SKIP in 41 cases (80.4%) (Table 1). There were no significant differences in the expression of SKIP in the different pathological staging in bladder cancer. The overall survival (OS) and Recurrence-free survival (RFS) were closely related to the expression of SKIP in bladder cancer (Table 2). Taken together, these results suggested that SKIP was closely correlated with the prognosis of bladder cancer patients.
Figure 2.
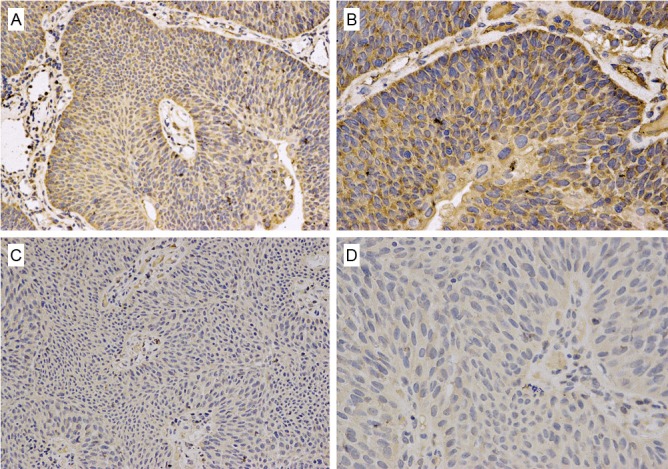
Representative immunostaining of SKIP in high-grade urothelial carcinoma samples (A, B), and low-grade urothelial carcinoma samples (C, D). High expression of SKIP (A, B) is showed in high-grade urothelial carcinoma samples and low expression of SKIP (C, D) is showed in low-grade urothelial carcinoma samples.
Table 2.
Multivariate Cox regression analysis of overall survival (OS) and Recurrence-free survival (RFS) in patients with bladder cancer
| Prognostic variables | OS | RFS | ||
|---|---|---|---|---|
|
|
||||
| Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P | |
| Age (>45 vs ≤45) | 1.117 (0.294-4.237) | 0.981 | 1.248 (0.430-3.619) | 0.683 |
| Differentiation (Grade 3 vs 1/2) | 1.019 (0.214-4.859) | 0.871 | 1.133 (0.487-2.634) | 0.772 |
| SKIP (+ vs -) | 6.338 (1.333-30.131) | 0.020* | 2.697 (1.161-6.264) | 0.021* |
Statistical analyses were performed by the Cox regression analysis
P<0.05 was considered significant;
Prognostic role of SKIP in bladder cancer.
Correlation of SKIP expression with the clinical features of bladder cancer
To investigate the clinicopathological significance of SKIP expression in bladder cancer, we evaluated the association of SKIP expression with clinicopathological characteristics. Patients with bladder cancer were divided into two groups: high SKIP expression and low SKIP expression according to the mean value of SKIP expression. As shown in Table 1, we found that the expression of SKIP was closely correlated with bladder cancer recurrence (p=0.004) and poor differentiation (p=0.001), but it was no obvious association with age (p=0.142), Tumor types (p=0.731), and Distant metastasis (p=0.448). Based on the results, we supposed that the SKIP expression was significantly correlated with clinical prognosis. Clinical follow-up was performed and the survival analysis suggested a closely correlation between SKIP expression and both the recurrence-free survival and overall survival of bladder cancer patients. The multivariate Cox regression analysis shown that SKIP expression was an independent prognostic factor in patients with bladder cancer (Table 2 and Figure 1).
Figure 1.
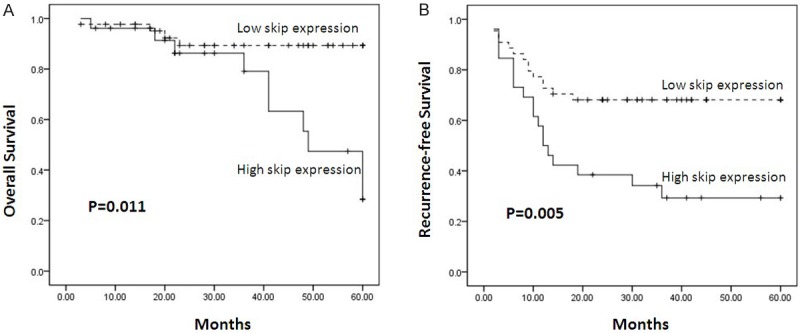
Patients were divided into two groups: high SKIP expressers (n=51) and low SKIP expressers (n=19). Kaplan-Meier analysis of overall survival (A) and recurrence-free survival (B) in relation to skip expression in 70 bladder cancer patients showed a highly significant separation between curves.
Discussion
Clinically, pTNM stage and tumor histopathological grade are used to evaluate the prognosis of patients with bladder cancer, including both bladder cancer recurrence and overall survival [13]. However, the exact molecular mechanisms of the progression and recurrence of bladder cancer are unclear, and there is lack of valid and reliable biomarkers to predict bladder cancer recurrence [14,15]. In this study, we observed that the mRNA expression of SKIP in bladder cancer samples and the paired normal bladder tissues. The SKIP mRNA expression in bladder cancer samples were significantly higher than that in adjacent noncancerous tissues, we speculated that the skip may play an important role in bladder cancer. 19 cases of high-grade urothelial carcinoma and 51 cases of low-grade urothelial carcinoma were collected in this clinical research. Postoperative follow-up survey and skip immunohistochemistry were performed to investigate the relationships between the expression of SKIP in bladder cancer samples and clinicopathological features of bladder cancer, as well as prognosis. Our statistical analysis showed that the expression of SKIP was closely correlated with overall survival and recurrence of bladder cancer. Patients with high expression of SKIP exhibited relatively short recurrence-free survival and poor overall survival time, and patients with low SKIP expression had relatively longer survival time compared with the patients who had high SKIP expression. the expression of SKIP and clinicalopathological parameters were examined by the multivariate Cox regression analysis. The results revealed that the expression of SKIP was an independent prognostic factor for a poor overall survival (OS) and Recurrence-free survival (RFS) of bladder cancer patients. The above results strongly revealed that SKIP may play an important role in bladder cancer progression, and it may serve as a valuable biomarker to predict the prognosis of patients with bladder cancer.
Previous studies showed that SKIP may play an essential role in the regulation of many cancer-related proteins [16,17] and a number of key signalling pathways [10,18,19]. Leong et al. reported that SKIP can bind to smad2 and smad3 proteins and augment transforming growth factor-β dependent transcription. It was speculated that SKIP may play an important role in regulating cell growth and differentiation through the TGF-β pathway [9]. Jonine et al. demonstrated that overexpression of SKIP has specific inhibitory effects on BMP-2-induced differentiation in C2C12 cells. It is implicated SKIP may serve as a novel regulator of the differentiation programming induced by TGF-β signals [20]. Interestingly, Victor et al reported that ectopic expression of SKIP in Human PC-3 prostate cancer cells inhibited both TGF-β1-induced uPA and MMP-9 promoter transactivation, which were closely correlated with the poor tumour differentiation, invasive stage of cancer, poor patient prognosis, metastasis to secondary organs, and shorter survival time in cancer. SKIP was suggested as a novel therapeutic target in prostate cancer treatment [10].Guoliang et al. revealed that High SKIP expression is correlated with poor prognosis and cell proliferation of hepatocellular carcinoma. The SKIP could serve as a potential prognostic marker and therapeutic target of HCC [21]. The above studies and our results strongly suggested that overexpression of SKIP was closely correlated with poor prognosis in patients with bladder cancer, and the detailed molecular mechanisms of SKIP on Biological behavior of bladder cancer were unclear and need to be further elucidated.
In conclusion, this present study showed a significantly higher mRNA expression of SKIP in bladder cancer samples than that in the paired normal bladder tissues, and the protein expression of SKIP in high-grade urothelial carcinoma samples were higher than that in low-grade urothelial carcinoma samples. We found a significantly correlation of SKIP expression and overall survival (OS) and Recurrence-free survival (RFS). Moreover, bladder cancer patients with high SKIP expression are more inclined to early recurrence than those with low SKIP expression. These results indicate that SKIP can be use as a potential biomarker for prediction of the progression and recurrence of patients with bladder cancer.
Disclosure of conflict of interest
None.
References
- 1.Zhu Z, Wang X, Shen Z, Lu Y, Zhong S, Xu C. Risk of bladder cancer in patients with diabetes mellitus: an updated meta-analysis of 36 observational studies. BMC Cancer. 2013;13:310. doi: 10.1186/1471-2407-13-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Sylvester RJ, van der Meijden AP, Oosterlinck W, Witjes JA, Bouffioux C, Denis L, Newling DW, Kurth K. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49:466–465. doi: 10.1016/j.eururo.2005.12.031. discussion 475-467. [DOI] [PubMed] [Google Scholar]
- 4.Tolley DA, Parmar MK, Grigor KM, Lallemand G, Benyon LL, Fellows J, Freedman LS, Hall RR, Hargreave TB, Munson K, Newling DW, Richards B, Robinson MR, Rose MB, Smith PH, Williams JL, Whelan P. The effect of intravesical mitomycin C on recurrence of newly diagnosed superficial bladder cancer: a further report with 7 years of follow up. J Urol. 1996;155:1233–1238. [PubMed] [Google Scholar]
- 5.Giannarini G, Kessler TM, Thoeny HC, Nguyen DP, Meissner C, Studer UE. Do patients benefit from routine follow-up to detect recurrences after radical cystectomy and ileal orthotopic bladder substitution? Eur Urol. 2010;58:486–494. doi: 10.1016/j.eururo.2010.05.041. [DOI] [PubMed] [Google Scholar]
- 6.Tilki D, Singer BB, Shariat SF, Behrend A, Fernando M, Irmak S, Buchner A, Hooper AT, Stief CG, Reich O, Ergun S. CEACAM1: a novel urinary marker for bladder cancer detection. Eur Urol. 2010;57:648–654. doi: 10.1016/j.eururo.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 7.Dahl R, Wani B, Hayman MJ. The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene. 1998;16:1579–1586. doi: 10.1038/sj.onc.1201687. [DOI] [PubMed] [Google Scholar]
- 8.Prathapam T, Kuhne C, Banks L. Skip interacts with the retinoblastoma tumor suppressor and inhibits its transcriptional repression activity. Nucleic Acids Res. 2002;30:5261–5268. doi: 10.1093/nar/gkf658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leong GM, Subramaniam N, Figueroa J, Flanagan JL, Hayman MJ, Eisman JA, Kouzmenko AP. Ski-interacting protein interacts with Smad proteins to augment transforming growth factor-beta-dependent transcription. J Biol Chem. 2001;276:18243–18248. doi: 10.1074/jbc.M010815200. [DOI] [PubMed] [Google Scholar]
- 10.Villar V, Kocic J, Santibanez JF. Skip Regulates TGF-beta1-Induced Extracellular Matrix Degrading Proteases Expression in Human PC-3 Prostate Cancer Cells. Prostate Cancer. 2013;2013:398253. doi: 10.1155/2013/398253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leong GM, Subramaniam N, Issa LL, Barry JB, Kino T, Driggers PH, Hayman MJ, Eisman JA, Gardiner EM. Ski-interacting protein, a bifunctional nuclear receptor coregulator that interacts with N-CoR/SMRT and p300. Biochem Biophys Res Commun. 2004;315:1070–1076. doi: 10.1016/j.bbrc.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Abankwa D, Millard SM, Martel N, Choong CS, Yang M, Butler LM, Buchanan G, Tilley WD, Ueki N, Hayman MJ, Leong GM. Ski-interacting protein (SKIP) interacts with androgen receptor in the nucleus and modulates androgen-dependent transcription. BMC Biochem. 2013;14:10. doi: 10.1186/1471-2091-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gospodarowicz MK. Staging of bladder cancer. Semin Surg Oncol. 1994;10:51–59. doi: 10.1002/ssu.2980100109. [DOI] [PubMed] [Google Scholar]
- 14.Shariat SF, Karakiewicz PI, Ashfaq R, Lerner SP, Palapattu GS, Cote RJ, Sagalowsky AI, Lotan Y. Multiple biomarkers improve prediction of bladder cancer recurrence and mortality in patients undergoing cystectomy. Cancer. 2008;112:315–325. doi: 10.1002/cncr.23162. [DOI] [PubMed] [Google Scholar]
- 15.Bolenz C, Lotan Y. Molecular biomarkers for urothelial carcinoma of the bladder: challenges in clinical use. Nat Clin Pract Urol. 2008;5:676–685. doi: 10.1038/ncpuro1259. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Zhang L, Jones KA. SKIP counteracts p53-mediated apoptosis via selective regulation of p21Cip1 mRNA splicing. Genes Dev. 2011;25:701–716. doi: 10.1101/gad.2002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W, Angelis K, Danielpour D, Haddad MM, Bischof O, Campisi J, Stavnezer E, Medrano EE. Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type beta transforming growth factor. Proc Natl Acad Sci U S A. 2000;97:5924–5929. doi: 10.1073/pnas.090097797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YF, Chiu HH, Wu CH, Wang JY, Chen FM, Tzou WH, Shin SJ, Lin SR. Retinoblastoma protein (pRB) was significantly phosphorylated through a Ras-to-MAPK pathway in mutant K-ras stably transfected human adrenocortical cells. DNA Cell Biol. 2003;22:657–664. doi: 10.1089/104454903770238139. [DOI] [PubMed] [Google Scholar]
- 19.Zhou S, Fujimuro M, Hsieh JJ, Chen L, Miyamoto A, Weinmaster G, Hayward SD. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC To facilitate NotchIC function. Mol Cell Biol. 2000;20:2400–2410. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueroa JD, Hayman MJ. Differential effects of the Ski-interacting protein (SKIP) on differentiation induced by transforming growth factor-beta1 and bone morphogenetic protein-2 in C2C12 cells. Exp Cell Res. 2004;296:163–172. doi: 10.1016/j.yexcr.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Liu G, Huang X, Cui X, Zhang J, Wei L, Ni R, Lu C. High SKIP expression is correlated with poor prognosis and cell proliferation of hepatocellular carcinoma. Med Oncol. 2013;30:537. doi: 10.1007/s12032-013-0537-4. [DOI] [PubMed] [Google Scholar]


