Abstract
The FGF/FGFR-system plays an important role in embryogenesis, tissue homeostasis and carcinogenesis. Mutational activation of FGFR2 resulting in aberrant FGFR2 signaling activation is known from both hereditary germ line alterations and somatic mutations in various malignancies (e.g. breast, gastric or ovarian cancer). FGFR2 mutations are mainly located within the hinge between Ig-like domains (exon 7), around the 3rd Ig-like domains and within the kinase domain. For bladder cancer only sparse data on FGFR2 mutations are available. Most interestingly a case of early-onset papillary carcinoma of the bladder showing a FGFR2 p.Pro253Arg mutation in exon 7 in a patient with Apert Syndrome was reported recently. To further evaluate the importance of FGFR2 exon 7 alterations in bladder cancer a cohort of 254 bladder tumors (cohort 1: unselected cases: n=139; cohort 2: early-onset bladder cancer cases (age at time of diagnosis ≤45 years): n=115) was analyzed. Sections from formalin-fixed, paraffin-embedded bladder tumors were used for DNA isolation. After precise microdissection exon 7 of the FGFR2 gene was analyzed by direct Sanger sequencing. All cases could be analyzed successfully. Mutations in exon 7 of FGFR2 could not be detected in any of the cases. All tumors showed wild type sequence. Our data demonstrate that the recently reported association between early-onset papillary carcinoma of the bladder with germ line FGFR2 p.Pro253Arg mutation could not be found in our cohorts of sporadic bladder tumors. These results indicate that FGFR2 gene mutations might only play a minor role in bladder carcinogenesis.
Keywords: FGFR2, mutation, bladder cancer, early-onset, Apert Syndrome, sequencing
Introduction
Aberrant expression or uncontrolled mutational activation of multiple FGF family members and their four corresponding receptors (FGFR) are found in multiple cancers. FGFRs are transmembrane glycoproteins with a conserved structure comprising an extracellular part with two to three immunoglobulin-like domains (IgI-III), a transmembrane domain and an intracellular split tyrosine-kinase domain. Signal transduction of the FGF/FGFR system is quite complex. Complexity levels are built up by different affinity of FGFRs for specific FGFs and the fact that FGFRs are subjected to alternative splicing [1,2].
Deregulated FGF/FGFR signaling is also known in tumors of the genito-urinary system, especially in bladder, prostate (PCa) and renal cell carcinoma (RCC). For PCa and RCC mainly overexpression of FGFRs was reported, activating mutations seem to play no important roles [1-5].
In bladder cancer very high frequencies of alterations in FGFR3 were reported. Activating mutations have first been reported more than twelve years ago and could be shown commonly in most benign urothelial lesions and low-grade and low-stage tumors [1,6]. In addition, overexpression of both mutated and wild type FGFR3 was reported in bladder tumors while the underlying molecular mechanisms still remain unclear. Recently, constitutively activated fusion genes generated via chromosomal re-arrangements were identified as a new mechanism of uncontrolled FGFR3 activation [1,7].
Interestingly, a case of early-onset low-grade papillary bladder cancer in a patient with Apert Syndrome and a germ line FGFR2 p.Pro253Arg mutation was described for the first time recently [8]. The specific mutation was also detectable in the bladder tumor with concurrent FGFR3 wild type sequence. In general, only sparse data on FGFR2 mutation analysis in bladder cancer are available. Ricol and colleagues investigated the entire coding region of FGFR2 in 33 bladder tumors. Two tumors showed deletions (exon 13 or exon 17), but no single point mutations could be detected [9]. This lack of comprehensive data prompted us to perform a mutation analysis of FGFR2 exon 7 containing codon 253 in a large series of bladder tumors from patients with both early-onset and regular-onset disease.
Materials and methods
Bladder cancer tissue samples
Overall, 254 archival formalin-fixed, paraffin-embedded bladder tumors (cohort 1: unselected cases: n=139; cohort 2: early-onset bladder cancer cases (age at time of diagnosis ≤45 years): n=115) achieved by transurethral resections of the bladder were analyzed. The tumors were diagnosed according to the WHO classification of bladder tumors and staged according the TNM system [10,11]. Clinical and histopathological characteristics of patients and tumors are shown in Table 1. Prior IRB approval was obtained for the study.
Table 1.
Characteristics of the study cohort
| Unselected Tumor Group | Early Onset Tumor Group | |||
|---|---|---|---|---|
| Number | n=139 | n=115 | ||
| Age (years) | Median: 70 | Range: 48-95 | Median: 40 | Range: 4-45 |
| Mean: 70.3 | Mean: 36.4 | |||
| Stage | Papilloma | n=1 | n=14 | |
| Ta | n=73 | n=58 | ||
| T1 | n=37 | n=18 | ||
| ≥T2 | n=27 | n=17 | ||
| unknown | n=1 | n=8 | ||
| Grading | G1 | n=34 | n=32 | |
| G2 | n=54 | n=43 | ||
| G3 | n=36 | n=15 | ||
| unknown | n=15 | n=25 | ||
| Gender | Male | n=108 | n=81 | |
| female | n=30 | n=29 | ||
| n=1 | n=5 |
Microdissection and DNA isolation
DNA was extracted from paraffin-embedded tumor tissues as described previously [12]. In brief, 5 μm histologic tissue sections were deparaffinized and stained with 0.1% methylene blue for 15 seconds. Manual microdissection was used to obtain tumor cells with a purity of at least 85%. Prior to microdissection the areas with highest tumor cell density were marked on a representative hematoxylin and eosin stained section by an experienced surgical resecpathologist (AH, JG). Non-malignant tissue was removed from the section by scraping off with a sterile needle. Tumor tissue was then collected with a separate sterile needle and transferred into a sterile tube. DNA isolation was performed using the High Pure PCR Template Preparation Kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer’s instructions.
FGFR2 exon 7 mutation analysis
Exon 7 of the FGFR2 gene was amplified by PCR using primers (sense: 5’- GGT CTC TCA TTC TCC CAT CCC -3’; antisense: 5’- CCC AGT TGT GGG TAC CTT TAG -3’) obtained from Metabion (Martinsried, Germany) in a total volume of 25 μl containing approx. 100 ng DNA, 0.2 mM dNTP (Promega), 0.18 μM primers and 0.0025 U/μl GoTaq (Promega, Mannheim, Germany). The thermal cycling conditions were as follows: initial denaturation for 3 min at 94°C, 45 cycles of denaturation at 94°C for 1 min, annealing at 61°C for 1 min, elongation at 72°C for 1 min and final primer extension at 72°C for 10 min. Gradient PCR was used for optimization of cycling conditions. After amplification, PCR-products (size: 280 bp) were purified using the Qiagen Dye Ex 2.0 TM Spin Kit according to the manufacturer’s conditions. Sequence analysis was performed (primers: sense: 5’- TCA TTC TCC CAT CCC CAC -3’; antisense: 5’- CCC AGT TGT GGG TAC CTT TAG -3’) using Applied Biosystems Big Dye Terminator v1.1 Cycle Sequencing Kit and an Applied Biosystems ABI 3500 Genetic Analyzer.
Results
Specificity and sensitivity of the sequencing analysis
As no positive control containing the FGFR2 p.Pro253Arg mutation was available we aimed the testing for the specificity and sensitivity of our methodical approach before screening the tumor samples. For this reason we used DNA from the endometrium adenocarcinoma cell line MFE-280 with a known FGFR2 p.Ser252Trp mutation [13]. This mutation is adjacent to the FGFR2 p.Pro253Arg mutation reported in the bladder tumor from the patient with Apert Syndrome. The expected FGFR2 mutation could clearly be detected in the MFE-280 cells (Figure 1B) using our sequencing method. In order to test the sensitivity of our method we spiked DNA from MFE-280 cells with DNA from the cell line UROtsa representing normal urothelium without any alterations [14]. We were able to detect the FGFR2 p.Ser252Trp mutation in DNA from MFE-280 cells containing more than 85% wild type DNA from UROtsa cells (Figure 2).
Figure 1.
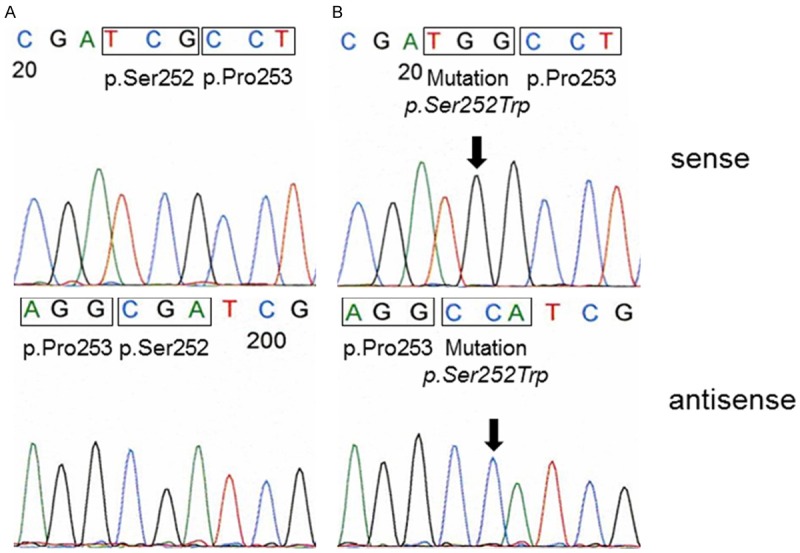
A. Representative example of FGFR2 codon p.Ser252 and p.Pro253 sequence analysis of a DNA from a bladder tumor. Both codons show wild type sequence. B. FGFR2 sequence analysis of DNA from cell line MFE-280. The p.Ser252Trp mutation is clearly detectable (arrow). Codon p.Pro253 shows a wild type sequence.
Figure 2.
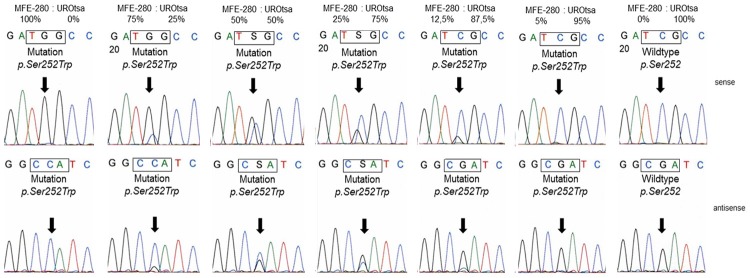
Sequencing analysis of FGFR2 codon p.Ser252 and p.Pro253 in MFE-280 DNA spiked with DNA from UROtsa cells. Arrows indicate the FGFR2 p.Ser252Trp mutation in DNA from MFE-280 cells. The p.Ser252Trp mutation is still detectable in a background of >85% wild type DNA.
FGFR2 exon 7 sequence analysis in bladder tumors
All 254 bladder tumors could be analyzed successfully. Mutations in exon 7 of the FGFR2 gene were found in 0/254 tumors. All cases showed a wild type sequence (Figure 1A).
Discussion
In the presented study we analyzed exon 7 of the FGFR2 gene to evaluate the frequency of the recently published p.Pro253Arg mutation in bladder cancer [8]. In addition, as the reported FGFR2 mutated tumor occurred in a patient with early disease onset, we included a cohort of tumors from early-onset bladder cancer patients in our study. In our two cohorts no mutations could be detected at all. These data indicate that exon 7 mutations play no role in bladder tumorigenesis in general.
One explanation for missing the mutation could be working with material highly contaminated with wild-type stromal cells. Great care was taken during microdissection for enrichment of tumor cells. In addition, testing the sensitivity of our method showed that a mutation could clearly be detected in a background of >80% wild type DNA. Therefore we do not think that methodical problems hampered our analysis.
FGFR2 mutations were frequently found in other cancers (e.g. breast cancer, gastric cancer, endometrial cancer) [15]. Most of these mutations clustered around the hinge region and the third Ig-like domain of FGFR2. Only in endometrial cancer FGFR2 mutations clustered within the kinase domain [16]. Therefore we did not expand our analyses to further exons as the probability for discovering mutations outside the known hot spots was low. Moreover, four studies presenting whole exome or whole genome sequencing data from bladder tumors were published most recently [17-20]. Overall 204 tumors were investigated in these studies. None of the tumors showed a FGFR2 mutation which strengthens our findings.
The general role of FGFR2 in bladder cancer is still not clear. The splice variant FGFR2-IIIb is expressed in normal urothelium. About 30% of analyzed tumors showed a decreased FGFR2 expression, and this decreased expression on both mRNA and protein level was associated with poor prognosis (tumor-specific death) [21]. The chromosomal region of the FGFR2 gene (chromosome 10q25.3-26) was described as affected by loss of heterozygosity in approx. 27% of analyzed cases without any correlation to decreased FGFR2 expression. Re-expression of FGFR2 resulted in lower proliferation rates and a decreased tumorigenic potential in nude mice [9]. Based on these data FGFR2 appears to act as a tumor-suppressor with protective properties in bladder cancer but conclusive data are still missing.
In conclusion, the results of our study on FGFR2 exon 7 sequencing analysis in combination with previously data suggest no role of mutational activation of FGFR2 in bladder carcinogenesis. Expression levels of FGFR2 might influence both tumor aggressiveness and patient’s outcome but the underlying mechanisms need to be clarified in more detail.
Acknowledgements
The present work was performed in fulfillment of the requirements for obtaining the degree “Dr. med.” (M.D.). We acknowledge support by Deutsche Forschungsgemeinschaft and Friedrich-Alexander-University Erlangen-Nuremberg within the funding program Open Access Publishing. Verena Popp, Claudia Knoll, Nina Oks and Karina Dresel are thanked for excellent technical assistance.
Disclosure of conflict of interest
None.
References
- 1.Acevedo VD, Ittmann M, Spencer DM. Paths of FGFR-driven tumorigenesis. Cell Cycle. 2009 Feb 15;8:580–588. doi: 10.4161/cc.8.4.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Martino E, Tomlinson DC, Knowles MA. A decade of FGF receptor research in bladder cancer: past, present, and future challenges. Adv Urol. 2012 Jul 31;2012:429213. doi: 10.1155/2012/429213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsimafeyeu I, Demidov L, Stepanova E, Wynn N, Ta H. Overexpression of fibroblast growth factor receptors FGFR1 and FGFR2 in renal cell carcinoma. Scand J Urol Nephrol. 2011 Feb 18;45:190–195. doi: 10.3109/00365599.2011.552436. [DOI] [PubMed] [Google Scholar]
- 4.Koufou S, Lunz JC, Borchardt A, Keck B, Kneitz B, Gaisa NT, Hafner C, Giedl C, Rau TT, Rogler A, Wieland WF, Hartmann A, Stoehr R. Mutational activation of FGFR3 is not involved in the development of prostate cancer. Pathobiology. 2010 Nov 29;77:249–252. doi: 10.1159/000317055. [DOI] [PubMed] [Google Scholar]
- 5.Stoehr CG, Stoehr R, Hartmann A, Hofstaedter F, Junker K, Blaszyk H, Wieland WF, Otto W, Denzinger S, Walter B. Mutational activation of FGFR3: no involvement in the development of renal cell carcinoma. J Cancer Res Clin Oncol. 2012;138:359–361. doi: 10.1007/s00432-011-1130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourdin J, Sastre-Garau X, Chopin D, Thiery JP, Radvanyi F. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 7.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013 Feb 15;22:795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreou A, Lamy A, Layet V, Cailliez D, Gobet F, Pfister C, Menard M, Frebourg T. Ealry-onset low-grade papillary carcinoma of the bladder associated with apert syndrome and a germline FGFR2 mutation (Pro253Arg) Am J Med Genet A. 2006 Oct 15;140:2245–2247. doi: 10.1002/ajmg.a.31430. [DOI] [PubMed] [Google Scholar]
- 9.Ricol D, Capellen D, El Marjou A, Gil-Diez-di-Medina S, Girault JM, Yoshida T, Ferry G, Tucker G, Poupon MF, Chopin D, Thiery JP, Radvanyi F. Tumour suppressive properties of fibroblast growth factor receptor 2-IIIb in human bladder cancer. Oncogene. 1999;18:7234–7243. doi: 10.1038/sj.onc.1203186. [DOI] [PubMed] [Google Scholar]
- 10.Mostofi FK, Davis CJJ, Sesterhenn IA. World Health Organization International Histological Classification. New York: Springer; 1999. Histological typing of urinary bladder tumours. [Google Scholar]
- 11.Sobin LH, Wittekind C. TNM Classification of Malignant Tumours. Hoboken, New Jersey: John Wiley & Sons; 2002. [Google Scholar]
- 12.Stoehr R, Zietz S, Burger M, Filbeck T, Denzinger S, Obermann EC, Hammerschmied C, Wieland WF, Knuechel R, Hartmann A. Deletions of chromosomes 9 and 8p in histologically normal urothelium in patients with bladder cancer. Eur Urol. 2005;47:58–63. doi: 10.1016/j.eururo.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Krakstad C, Birkeland E, Seidel D, Kusonmano K, Petersen K, Mjos S, Hoivik EA, Wik E, Halle MK, Oyan AM, Kalland KH, Werner HMJ, Trovik J, Salvesen H. High-throughput mutation profiling or primary and metastatic endometrial cancers identifies KRAS, FGFR2 and PIK3CA to be frequently mutated. PLoS One. 2012;7:e52795. doi: 10.1371/journal.pone.0052795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petzoldt JL, Leigh IM, Duffy PG, Sexton C, Masters JR. Immortalisation of human urothelial cells. Urol Res. 1995;23:377–380. doi: 10.1007/BF00698738. [DOI] [PubMed] [Google Scholar]
- 15.Katoh M. FGFR abnormalities underlie a spectrum of bone, skin, and cancer pathologies. J Invest Dermatol. 2009 Apr 29;129:1861–1867. doi: 10.1038/jid.2009.97. [DOI] [PubMed] [Google Scholar]
- 16.Katoh M. Cancer genomics and genetics of FGFR2. Int J Oncol. 2008;33:233–237. [PubMed] [Google Scholar]
- 17.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, Dean M, Huang Y, Jia W, Zhou Q, Tang A, Yang Z, Li X, Song P, Zhao X, Ye R, Zhang S, Lin Z, Qi M, Wan S, Xie L, Fan F, Nickerson ML, Zou X, Hu X, Xing L, Lv Z, Mei H, Gao S, Liang C, Gao Z, Lu J, Yu Y, Liu C, Li L, Fang X, Jiang Z, Yang J, Li C, Zhao X, Chen J, Zhang F, Lai Y, Lin Z, Zhou F, Chen H, Chan HC, Tsang S, Theodorescu D, Li Y, Zhang X, Wang J, Yang H, Gui Y, Wang J, Cai Z. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45:1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balbás-Martínez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Márquez M, Vazquez M, Lapi E, Castro-Giner F, Beltran S, Bayés M, Carrato A, Cigudosa JC, Domínguez O, Gut M, Herranz J, Juanpere N, Kogevinas M, Langa X, López-Knowles E, Lorente JA, Lloreta J, Pisano DG, Richart L, Rico D, Salgado RN, Tardón A, Chanock S, Heath S, Valencia A, Losada A, Gut I, Malats N, Real FX. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45:1464–1469. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross JS, Wang K, Gay LM, Al-Rohil RN, Nazeer T, Sheehan CE, Jennings TA, Otto GA, Donahue A, He J, Palmer G, Ali S, Nahas M, Young G, Labrecque E, Frampton G, Erlich R, Curran JA, Brennan K, Downing SR, Yelensky R, Lipson D, Hawryluk M, Miller VA, Stephens PJ. A high frequency of activating extracellular domain ERBB2 (HER2) mutations in micropapillary urothelial carcinoma. Clin Cancer Res. 2014 Jan 1;20:68–75. doi: 10.1158/1078-0432.CCR-13-1992. DOI: 10.1158/1078-0432.CCR-13-1992. [DOI] [PubMed] [Google Scholar]
- 20.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, He M, Li Z, Sun X, Jia W, Chen J, Yang S, Zhou F, Zhao X, Wan S, Ye R, Liang C, Liu Z, Huang P, Liu C, Jiang H, Wang Y, Zheng H, Sun L, Liu X, Jiang Z, Feng D, Chen J, Wu S, Zou J, Zhang Z, Yang R, Zhao J, Xu C, Yin W, Guan Z, Ye J, Zhang H, Li J, Kristiansen K, Nickerson ML, Theodorescu D, Li Y, Zhang X, Li S, Wang J, Yang H, Wang J, Cai Z. Frequent mutations of chromatin remodelling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–879. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gil Diez de Medina S, Chopin D, El Marjou A, Delouvee A, LaRochelle WJ, Hoznek A, Abbou C, Aaronson SA, Thiery JP, Radnanyi F. Decreased expression of keratinocyte growth factor receptor in a subset of human transitional cell bladder carcinomas. Oncogene. 1997;14:323–330. doi: 10.1038/sj.onc.1200830. [DOI] [PubMed] [Google Scholar]


