Abstract
Bacterial secreted proteins constitute a biologically important subset of proteins involved in key processes related to infection such as adhesion, colonization, and dissemination. Bacterial extracellular proteases, in particular, have attracted considerable attention, as they have been shown to be indispensable for bacterial virulence.
Here, we analyzed the extracellular subproteome of Clostridium difficile and identified a hypothetical protein, CD2830, as a novel secreted metalloprotease. Following the identification of a CD2830 cleavage site in human HSP90β, a series of synthetic peptide substrates was used to identify the favorable CD2830 cleavage motif. This motif was characterized by a high prevalence of proline residues. Intriguingly, CD2830 has a preference for cleaving Pro–Pro bonds, unique among all hitherto described proteases. Strikingly, within the C. difficile proteome two putative adhesion molecules, CD2831 and CD3246, were identified that contain multiple CD2830 cleavage sites (13 in total). We subsequently found that CD2830 efficiently cleaves CD2831 between two prolines at all predicted cleavage sites. Moreover, native CD2830, secreted by live cells, cleaves endogenous CD2831 and CD3246.
These findings highlight CD2830 as a highly specific endoproteinase with a preference for proline residues surrounding the scissile bond. Moreover, the efficient cleavage of two putative surface adhesion proteins points to a possible role of CD2830 in the regulation of C. difficile adhesion.
Clostridium difficile is an anaerobic, Gram-positive, spore-forming bacterium. Human intake of spores occurs through the fecal-oral route. Upon leaving the stomach and becoming exposed to bile acids in the small intestine, the C. difficile spores germinate. Once germinated, the vegetative cells in the colon encounter an unreceptive environment (1). Competition with the normal flora, immune responses, gastric fluids (2), and specialized antimicrobial peptides all act against a developing infection. Moreover, the physical barrier formed by a layer of glycoproteins (mucins) covering the underlying epithelial cells forms a major hurdle for firm adhesion.
Many enteric pathogens express factors that reduce competition, allow evasion of host immune responses, and promote adhesion and/or invasion of tissues. These virulence factors are located in the cell membrane or wall (controlling adhesion and protection) or are secreted (modifying the surrounding environment). The best studied virulence factors in C. difficile are the toxins TcdA and TcdB (3, 4), which cause destruction of the intestinal barrier by disrupting the epithelial actin cytoskeleton. It is speculated that the subsequent increased permeability of the intestinal epithelium leads to increased exudation of fluids, including nutritional substances. Damage to the intestinal mucosa causes the main symptoms of C. difficile infection, including pseudomembranous colitis (1).
Data illustrating how C. difficile circumvents host defense mechanisms are limited. As an example, in response to attack by antimicrobial peptides, C. difficile expresses a set of genes that change the surface charge, thereby diminishing the interaction of cationic antimicrobial peptides on the bacterial surface (5). C. difficile cell membrane and cell wall proteins are obvious candidate molecules for direct interaction with the host and are most likely involved in processes such as adhesion and colonization. Footholds on the host cell surface proteins include the extracellular matrix components fibronectin, laminin, collagen, and fibrinogen, which all have been implicated in C. difficile adhesion (6–9). In addition, C. difficile secretory proteins are released into the surrounding environment, where they can exert their function. However, besides the toxins, little is known about extracellular factors that contribute to C. difficile infection (8, 10).
In this study we analyzed the secreted proteins of C. difficile and characterized a very specific, highly active secreted metalloprotease, CD2830. We demonstrate that it has a unique preference for hydrolyzing Pro–Pro bonds in an overall proline-rich cleavage motif. The identification of two C. difficile LPXTG surface proteins as highly efficient substrates for CD2830 indicates a role for this enzyme in bacterial motility.
EXPERIMENTAL PROCEDURES
Chemicals Used
IgA was purchased from VWR International (Amsterdam, the Netherlands) (401098), and recombinant HSP90β (SPR-102A) and recombinant HSP90α (SPR-101A) were obtained from Sanbio (Uden, the Netherlands). Sequencing-grade modified trypsin was acquired from Promega (Madison, WI). Peptides were synthesized at the Leiden University Medical Center facility as described previously (11). Synthetic FRET peptides contained an N-terminal Lys-Dabcyl and a C-terminal Glu-Edans. 1,10-Phenantroline was purchased from Sigma-Aldrich.
C. difficile Strains and Growth Conditions
The C. difficile strains were grown anaerobically in a microaerobic cabinet (Don Whitley DG 250) at 37°C in pre-reduced 3% Bacto Tryptose, 2% yeast extract (Difco), and 0.1% thioglycolate (pH 7.4) medium (TY); brain heart infusion broth (Oxoid, Basingstoke, MA) supplemented with 0.5% yeast extract and 0.01% l-cysteine (Sigma) (12); or minimal medium broth (13). When required, the broths were supplemented with appropriate antibiotics. Mid-logarithmic growth phase pre-cultures (A600 0.4–0.8) were used to inoculate pre-reduced TY broth to a starting A600 of 0.05. Optical density readings were taken hourly in the exponential growth phase and at 12 and 24 h post-inoculation.
Proteomic Analysis of the C. difficile Exoproteome
For analysis of the exoproteome, 12 ml of minimal medium from C. difficile strain 630 grown to early stationary phase was collected. Intact bacterial cells were removed via centrifugation for 10 min at 7000 × g. The resulting supernatant was filtered through a 0.45-μm filter followed by a 0.2-μm filter (Whatman FP 30). The filtered supernatant was subsequently concentrated to 0.5 ml using an Amicon Ultra-15 Centrifugal Filter Unit NMWL 3 (Millipore, Billerica, MA). The concentrated sample (20 μl) was separated on a SDS-PAGE gel (NuPAGE 4–12% gradient Bis-Tris gels, Invitrogen, Carlsbad, CA) and Coomassie stained (Simply Blue Safe Stain, Invitrogen). In-gel digestion and LC–ion trap MS/MS analysis were performed as described previously (14). Peak lists were generated using Data Analysis 4.0 (Bruker Daltonics, Bremen, Germany) with default settings and exported as Mascot generic files. Peptides were identified in the C. difficile database (strain 630, 3712 sequences, 1,164,103 residues, released October 5, 2010) using the Mascot algorithm (Mascot 2.4.1, Matrix Science, London, UK), applying a combined search in Mascot Deamon 2.2.2. An MS tolerance of 0.6 Da (with 13C = 1) and an MS/MS tolerance of 0.5 Da were used. Trypsin was designated as the enzyme, and up to one missed cleavage site was allowed. Carbamidomethylcysteine was selected as a fixed modification, and oxidation of methionine as a variable modification. The false discovery rate of this analysis (peptide matches above the identity threshold) was less than 1% using a decoy database. For further analysis, protein hits with at least two unique peptides with a score above 30 were selected.
Bioinformatics Analysis
Predictions of signal sequences were carried out using the SignalP 4.1 Server. Prediction of cell wall binding motifs, anchors, and subcellular localization were performed through an NCBI conserved domain search, the CBS LipoP server, and PSORTB.
Sequence alignments were performed with the use of the Clustal Omega Multiple Sequence Alignment tool. All predictions were performed using standard settings. Structural models of CD2830 were generated by the Phyre2 protein fold recognition server. All predictions were done using the standard parameters. WebLogo generation was performed at the Computational Genomics Research Group lab at the University of California, Berkeley.
PCR Detection to Determine the Genetic Prevalence of CD2830
The PCR reactions used to determine the genetic prevalence of the CD2830 gene were performed with GoTaq polymerase, according to the manufacturer's instructions, with the forward primer AGGGATGGGAAGGTACTGGA and the reverse primer GTTTGTGGACAAGCTGATTTTAACT.
DNA Constructs
To construct the his10-tagged CD2830 expression plasmid, the CD2830 sequence was amplified via PCR from C. difficile strain 630 genomic DNA, using primers AGGGAATCATATGGATAGTACTACTATACAACAAAATAAAGACAC and TATTGGATCCCTATTTAGCTAAATTTTGCAAAAAGC. The PCR product was digested with NdeI and BamHI and ligated into vector pET16b (Novagen, Darmstadt, Germany), similarly digested with NdeI and BamHI. This resulted in the construction of a CD2830 expression vector containing 10 consecutive histidines at its N-terminus, replacing the signal sequence (supplemental Fig. S1B). To construct his-tagged CD2831, a similar approach was used, using primers AGTTCCATATGAAGCAAGGTTATGCTTTTGAAGC and TCTGGCTCGAGCTATGTAGTACTATCCCCTGTTTTTGG, which were NdeI and XhoI cloned in pET16b, resulting in an expression construct containing amino acids 732 to 947 of CD2831.
The pET28a 6xHis-HSP90B plasmid was kindly provided by Dr. Daumantas Matulis.
Purification of rCD2830, rCD2831, and rHSP90β
The bacterial expression constructs were transformed into the Escherichia coli DE3 C43 strain (Lucigen, Middleton, WI), and a single colony was cultivated on a rotary shaker (200 rpm) in 100 ml of Luria broth at 37¦°C until an A600 of 0.5 was reached, after which the cultures were induced with 1 mm isopropylthio-β-galactoside for 5 h, using ampicillin (50 μg/ml) as a selection marker. We followed the protocol for the preparation of cleared E. coli lysates under native conditions from Qiagen (Venlo, the Netherlands) as described in the fifth edition of the QIAexpressionist, with the following modifications: The E. coli lysates (50 mm sodium phosphate buffer, pH 7.4, 5 mm 2-mercaptoethanol, 0.1% Nonidet P-40, 300 mm NaCl) containing his-tagged proteins were loaded on a 1-ml HisTrap HP column (GE Healthcare). The column was washed with 20 ml of lysis buffer. The his-tagged proteins were eluted at 150 mm imidazole using a 25-ml linear gradient ranging from 20 to 250 mm imidazole.
Proteolytic Assays with CD2830
CD2830 proteolytic assays were performed at 37°C in phosphate-buffered saline (PBS) (pH 7.4), 0.5 mm ZnCl2 for a duration of 16 h, unless otherwise stated. Reactions were stopped by the addition of Laemmli loading buffer and by heating the samples at 95°C. Proteolytic assays with the single protein targets were performed with 0.5 μg of rCD2830 and 3 μg of HSP90α, 3 μg of HSP90β, 5 μg of IgA1 and A2 and 2 or 5 μg of rCD2831. IgA1 and IgA2 were preincubated with 50 μm 2-mercaptoethanol in PBS for 30 min at 50°C prior to digestion. For the proteolytic assay with a Caco-2 lysate, the lysate was prepared as described in the manual for Pierce IP lysis buffer (Thermo Scientific). 10 μg of lysate was incubated with 1 μg of rCD2830. Protease activity was visualized via SDS-PAGE of the samples followed by Coomassie staining. The peptide cleavage assays were performed with 20 pmol peptides and 1 μg of rCD2830. After overnight incubation at 37°C, samples were analyzed with MALDI-TOF-MS (Ultraflex II and UltrafleXtreme, Bruker) as described previously (15). Cleavage was scored as positive when both product peptides were present and one of them had an intensity of at least 25% of the substrate ion.
The CD2830-mediated cleavage of FRET peptides was followed by fluorescence detection at an excitation wavelength of 355 nm and an emission wavelength of 485 nm in a 96-well plate reader (Mithras LB940, Berthold Technologies, Bad Wildbad, Germany). For determination of the steady-state kinetics, incubations were stopped at 30-s time intervals by mixing 10 μl of reaction mix with 10 μl of 0.25% TFA. These samples were further diluted to 100 μl using 80 μl of PBS and measured as described above. Initial velocities were determined based on the slope of the reaction curve during the first 2 min. A calibration curve plotting the fluorescence intensity versus the cleaved peptide concentration after complete cleavage of the peptide was used to convert the initial velocity from fluorescence units per second to nanomoles per second. Data were then fitted to the Michaelis–Menten equation, from which the Km, Vmax, and Kcat values were derived.
Identification of CD2830 Cleavage Products of HSP90β
In-gel digestion of untreated and CD2830-treated HSP90β was performed as described previously (14). Peptides were injected onto a trap column equilibrated with 0.1% formic acid (Acclaim PepMap100, 100 μm × 2 cm, nanoViper C18, 5 μm, 100 Å, Thermo Scientific) and separated on an analytical reverse phase column (Acclaim PepMap RSLC, 75 μm × 50 cm, nanoViper C18, 2 μm, 100 Å, Thermo Scientific) coupled to an ion trap mass spectrometer (amaZon Speed, Bruker) at a flow rate of 300 nl/min. The liquid chromatography was performed with a piece-linear gradient (0–10 min at 2% B, 10–25 min to 5% B, 25–165 min to 25% B, 165–175 min to 30% B, 175–190 min to 35% B, 190–195 min to 100% B, and 195–210 min at 100% B, where B is 95% acetonitrile/0.1% formic acid). For mass spectrometric analysis, ions were generated using a Captive Spray (Bruker) at a spray voltage of 1.4 kV. The temperature of the heated capillary was 180°C.
For the analysis of tryptic digests, eluting peptides were analyzed using the data-dependent MS/MS mode over an m/z 300–1400 range. The 10 most abundant ions in an MS spectrum were selected for MS/MS analysis via collision-induced dissociation using helium as the collision gas.
For direct identification of the CD2830 cleavage products from HSP90β and recombinant CD2831, proteolytic digestions were performed as described above. After desalting on a reverse phase cartridge as described above, peptides were eluted using 50% acetonitrile, 0.1% formic acid and then directly infused into the Q-TOF mass spectrometer (maXis, Bruker) with a syringe pump (flow rate = 30 μl/h). For mass spectrometric analysis, ions were generated using the Captive Spray (Bruker) with the same parameters as described above. Data were recorded for 5 min (mass range: m/z 300–1400).
Identification of CD2830 Cleavage Products of CD2831 and CD3246 in Conditioned Medium
The conditioned minimal growth medium of C. difficile cells (10 ml of the supernatant after 30 min at 30,000g) was desalted on a reverse phase cartridge (C-18 Oasis HLB, 1 cc, 30 mg, Waters, Etten-Leur, the Netherlands), and peptides were eluted stepwise using 150 μl of 20% and then 50% acetonitrile in 0.1% formic acid. The samples were dried in a vacuum concentrator until the volume was approximately 25 μl. Peptides were analyzed via LC–ion trap MS/MS using the same LC parameters as described above. During the whole run, MS/MS data acquisition was continuously performed on m/z values corresponding to the expected peptides (m/z 680.8 (CD2831 peptide) and m/z 638.8 (CD3246 peptide)).
RESULTS
Proteomics Analysis of the Extracellular Medium of C. difficile
In order to identify proteins secreted by C. difficile strain 630, we analyzed the culture medium at early stationary growth phase using SDS-PAGE followed by in-gel tryptic digestion and reverse phase LC–ion trap MS/MS (supplemental Fig. S1A). This approach identified 115 proteins in total (supplemental Tables S1 and S2). It is well known that such a proteomic subfraction often contains not only truly secreted proteins, but also, for example, major cytoplasmic proteins (16). Bioinformatic analysis (SignalP (17)) showed that 39 proteins (34%) contained a signal sequence (Table I) and only a few proteins seemed to be truly cleaved and released (no lipid anchors or cell wall binding motifs). One of the predicted genuinely secreted proteins, the uncharacterized protein CD2830 (Q183R7), appeared to be highly expressed, based on the number of identified unique peptides (10) and overall sequence coverage (47%). This finding prompted us to analyze CD2830 in more detail.
Table I. Proteomic analysis of the C. difficile exoproteome. C. difficile cells were grown in minimal medium until the beginning of the stationary phase. Conditioned medium was collected, filtered, concentrated, and analyzed with SDS-PAGE (supplemental Fig. S1A). Following in-gel digestion with trypsin, proteins were identified via LC–ion trap MS/MS and database searching (supplemental Tables S1 and S2). From this list, only proteins containing a signal peptide were selected and sorted based on the sequence coverage. Prediction of cell wall binding motifs, anchors, and subcellular localization was performed using NCBI Conserved Domain Search, the LipoP server, and PSORTB.
| Locus | Protein name | Protein I.D. | SignalP cleavage | LPP lipid attachment | Cell wall binding motif | Localization (PSORTB) | Sequence coverage | Unique peptides |
|---|---|---|---|---|---|---|---|---|
| CD2793 | Precursor of the S-layer proteins | Q183M8 | Yes | CW binding 2 | Cell wall | 80 | 63 | |
| CD2768 | Putative cell-wall hydrolase | Q183K6 | Yes | SH3_3 | Extracellular | 61 | 11 | |
| CD2767 | Putative cell surface protein | Q183K2 | Yes | CW binding 2 | Cell wall | 48 | 28 | |
| CD2830 | Uncharacterized protein | Q183R7 | Yes | Unknown | 47 | 10 | ||
| CD0873 | ABC-type transport system, sugar-family extracellular solute-binding protein | Q18A65 | Yes | Yes | Unknown | 45 | 11 | |
| CD2791 | Cell wall binding protein | Q183M5 | Yes | CW binding 2 | Cell wall | 44 | 25 | |
| CD1469 | Putative cell surface protein putative penicillin-binding protein | Q18BY9 | Yes | CW binding 2 | Unknown | 43 | 54 | |
| CD1522 | Putative polysaccharide deacetylase | Q18C42 | No | Extracellular | 43 | 12 | ||
| CD2784 | Putative N-acetylmuramoyl-l-alanine amidase,autolysin (EC 3.5.1.28) | Q183L9 | Yes | CW binding 2 | Cell wall | 42 | 29 | |
| CD2193 | Fragment of putative cell wall-binding protein (N-terminal region) pseudogene | Q185Y1 | Yes | CW binding 2 | Cell wall | 39 | 18 | |
| CD1156 | Uncharacterized protein | Q18B20 | No | Yes | Unknown | 37 | 16 | |
| CD0183 | Putative cell wall hydrolase | Q18CS3 | Yes | SH3_3 | Extracellular | 36 | 10 | |
| CD2672 | ABC-type transport system, oligopeptide-family solute-binding protein | Q183A6 | Yes | Yes | Cell wall | 34 | 17 | |
| CD1135 | Putative SH3-domain protein | Q18AZ7 | Yes | SH3_3 | Cell wall | 33 | 12 | |
| CD1035 | Putative N-acetylmuramoyl-l-alanine amidase,autolysin (EC 3.5.1.28) | Q18AN5 | Yes | CW binding 2 | Cytoplasmic/membrane | 31 | 19 | |
| CD2388 | Putative peptidoglycan-binding/hydrolysing protein | Q181X4 | Yes | PG_binding_1 | Extracellular | 31 | 8 | |
| CD2789 | Cell surface protein | Q183M7 | Yes | CW binding 2 | Cell wall | 30 | 17 | |
| CD2798 | Cell wall binding protein | Q183N5 | Yes | CW binding 2 | Cell wall | 29 | 10 | |
| CD2127 | Putative exported protein | Q185R5 | No | Cytoplasmic/membrane | 29 | 6 | ||
| CD2797 | Putative calcium-binding adhesion protein | Q183N3 | Yes | FA58C (discoin) | Unknown | 28 | 51 | |
| CD1304 | Acd (mannosyl-glycoprotein endo-β-N-acetylglucosamidase) (EC 3.2.1.96) | Q5S4P9 | Yes | SH3_3 | Extracellular | 28 | 15 | |
| CD2408 | Putative multidrug efflux pump | Q181Z3 | Yes | Unknown | 27 | 9 | ||
| CD3043 | Uncharacterized protein | Q184N0 | No | Unknown | 25 | 6 | ||
| CD0844 | Putative cell wall–binding protein | Q18A40 | Yes | CW binding 2 | Cell wall | 25 | 8 | |
| CD2794 | Cell wall binding protein | Q183N1 | No | CW binding 2 | Cell wall | 24 | 14 | |
| CD2177 | ABC-type transport system,cystine/aminoacid-family extracellular solute-binding protein | Q185W5 | No | Yes | Unknown | 23 | 4 | |
| CD2787 | Cell surface protein (EC 3.4.22.15) | Q183M1 | Yes | CW binding 2 | Cell wall | 22 | 12 | |
| CD2402 | Putative cell wall hydrolase phosphatase-associated protein (EC 3.4.-.-) | Q181Y8 | Yes | SH3_3 | Extracellular | 21 | 8 | |
| CD3669 | Putative exported protein | Q181S4 | Yes | Yes | Unknown | 18 | 4 | |
| CD2795 | Cell wall binding protein | Q183N0 | Yes | CW binding 2 | Cell wall | 16 | 7 | |
| CD3192 | Putative cell surface peptidase, M4 family | Q181V6 | Yes | CW binding 2 | Cell wall | 15 | 6 | |
| CD2796 | Cell surface protein | Q183N2 | Yes | CW binding 2 | Cell wall | 10 | 3 | |
| CD2518 | Cell surface protein | Q182K4 | Yes | CW binding 2 | Cell wall | 9 | 3 | |
| CD3366 | Putative exported protein | Q180L0 | Yes | Unknown | 9 | 2 | ||
| CD0545 | Uncharacterized protein | Q188Y7 | Yes | Yes | Unknown | 8 | 2 | |
| CD0514 | Putative hemagglutinin/adhesin | Q188W0 | Yes | CW binding 2 | Cell wall | 7 | 10 | |
| CD0855 | ABC-type transport system, oligopeptide-family extracellular solute-binding protein | Q18A51 | No | Yes | Cell wall | 6 | 2 | |
| CD3145 | Putative serine-aspartate repeat-containing protein SdrF | Q184Y2 | Yes | Extracellular | 3 | 2 | ||
| CD2831 | Putative adhesin | Q183R6 | Yes | Cell wall | 2 | 2 |
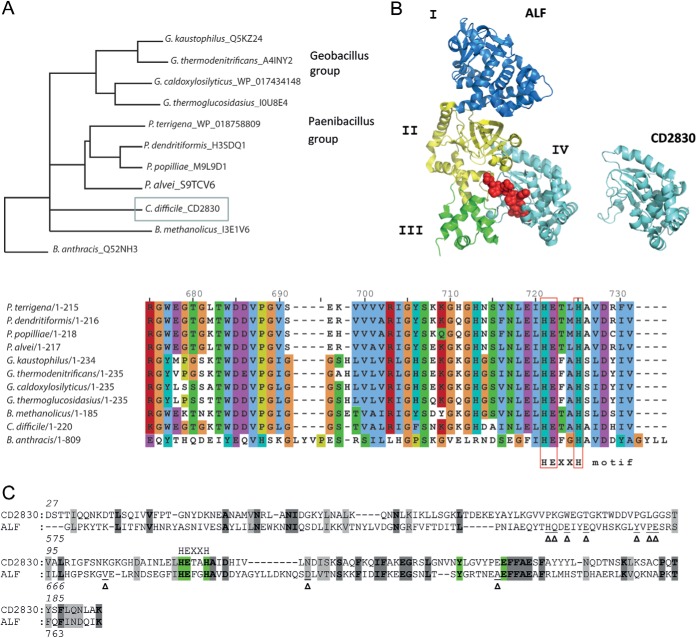
Based on a BLAST search of the nonredundant protein sequence database, C. difficile CD2830 showed the greatest homology to several proteins within the group of Geobacilli and Paenibacilli, represented by the phylogenetic tree in Fig. 1A. Surprisingly, no other close homologues were found in species belonging to the Clostridia class. Although the function of these homologues has not been experimentally determined, they have several common structural features. Based on the presence of a signal peptide, all are predicted to be secreted proteins (data not shown). In the UniProt database, several of them are annotated to contain a signature motif for metalloprotease activity, and all have a conserved HExxH motif in which the two histidine residues coordinate the zinc ion in the active site, common to zinc metalloproteases (Fig. 1A, sequence alignment in lower panel).
Fig. 1.
CD2830 from Clostridium difficile is a predicted metalloprotease with a fold similar to the anthrax lethal factor catalytic domain. A, neighbor-joining phylogenetic tree relating the C. difficile CD2830 protein with other closest related protein sequences. The closest related protein sequences were selected based on a CD2830 BLAST search of the database of non-redundant protein sequences, with the statistical significance (expected) threshold for reporting matches set at 1e-50. Species names are indicated, followed by UniProtKB accession numbers. The phylogenetic analysis was performed using Clustal Omega-Multiple Sequence Alignment, including Bacillus anthracis anthrax lethal factor protein. Protein alignment surrounding the conserved HExxH motif is shown in the lower panel. B, Three-dimensional stereo ribbon model of anthrax lethal factor (ALF) (left) and CD2830 predicted structure (right). ALF coloring and domain annotation are according to Pannifer et al. (19). The MAPKK target peptide is shown as a red ball-and-stick model. The ALF consists of four domains (I, II, III, and IV). The CD2830 consists of only a proteolytic domain with a fold similar to that of domain IV of ALF. C, amino acid sequence alignment of C. difficile CD2830 with ALF according to the Phyre2 protein structure prediction. Identical amino acids are shaded in dark gray, and similar residues shaded in gray. Zinc coordinating residues are highlighted in green. Arrowheads (Δ) point to ALF amino acids involved in peptide substrate recognition. The HEXXH metal binding site is indicated.
Moreover, analysis of CD2830 using the Phyre2 three-dimensional structure prediction server (18) demonstrated, with 100% confidence, over 97% alignment coverage, and 18% amino acid identity (Fig. 1B), similarity to the catalytic domain of anthrax toxin lethal factor (ALF)1 (residues 575–771) (19). Among the identical residues was the abovementioned HExxH motif (Fig. 1C). Two observations can be made with respect to the CD2830 fold in comparison to ALF. First, the structure of ALF comprises four structural domains (I, II, III, and IV) including an N-terminal domain that binds the membrane-translocating component (domain I, Fig. 1B). Structural similarity with CD2830 extends only to the C-terminal region containing the metalloprotease domain (domain IV, Fig. 1B). Secondly, identical residues between CD2830 and ALF mostly correspond to internal, structural fold–determining residues, and not to the specific contacts (19, 20) between ALF and its target peptide (Fig. 1C, arrowheads). In summary, this analysis indicates that CD2830 is a secreted, functionally active zinc metalloprotease with a fold similar to that of ALF, but with different targets.
Genetic Prevalence of CD2830
To determine the distribution of the cd2830 gene among the genomes of C. difficile, we evaluated the genomic DNA of 30 strains, encompassing the six major lineages (21), by means of PCR using specific primers for cd2830. We found that in all strains tested the cd2830 gene was present, emphasizing the importance of this gene for the species.
Recombinant CD2830 Is a Functional Protease Cleaving HSP90β
To investigate the predicted metalloprotease activity of CD2830, we cloned the cd2830 gene into a bacterial expression vector and overexpressed recombinant CD2830 protein (rCD2830) in E. coli and purified the protein (supplemental Fig. S1B). rCD2830 was measured via electrospray ionization Q-TOF MS under neutral conditions and after acidification, and we confirmed the presence of a bound Zn2+ (supplemental Figs. S1C and S1D).
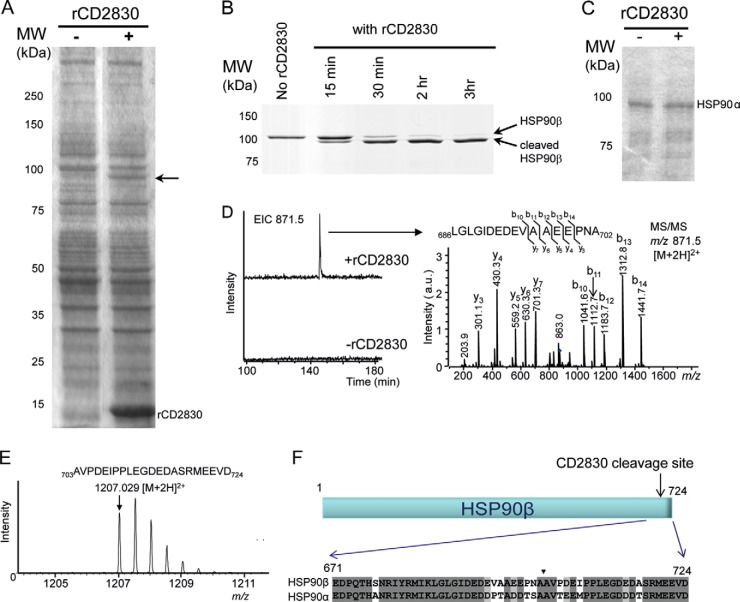
We then focused on the identification of substrates of CD2830. For this purpose, we incubated total C. difficile and Caco-2 cell lysates with and without rCD2830 and analyzed these samples via SDS-PAGE. No difference between the rCD2830 treated and untreated C. difficile samples was apparent (data not shown). However, when the human Caco-2 lysate was incubated with rCD2830, a clear cleavage product of ∼85 kDa was noticeable (arrow in Fig. 2A). We subsequently analyzed this 85-kDa cleavage product using reverse phase LC–ion trap MS/MS after in-gel tryptic digestion and identified the major protein as heat shock protein (HSP) 90β (data not shown).
Fig. 2.
Proteolytic assays reveal that human HSP90β is a substrate of CD2830. A, Caco-2 whole cell lysates analyzed via SDS-PAGE before (−) and after (+) incubation with rCD2830. Arrow points at cleavage product. B, recombinant human HSP90β analyzed via SDS-PAGE after incubation with rCD2830 for different time periods. C, SDS-PAGE analysis of human HSP90α after 20 h of incubation with (+) or without (−) rCD2830. D, LC–ion trap MS(/MS) analysis of tryptic digests of HSP90β before and after cleavage with rCD2830. A specific semi-tryptic peptide of HSP90β was identified only in the run from the CD2830-cleaved HSP90β. MS/MS analysis of this peptide demonstrated that HSP90β was cleaved by CD2830 after Ala-702 (see F). E, Electrospray ionization Q-TOF-MS analysis of the C-terminally released HSP90β peptide after cleavage by rCD2830. F, identification of the rCD2830 cleavage site in human HSP90β and alignment of the C-terminal amino acid sequences of human HSP90β and HSP90α. Arrowhead indicates the CD2830 cleavage site between Ala-702 and Ala-703 of human HSP90β. MW, molecular weight in kilodaltons.
To confirm that HSP90β was cleaved by rCD2830, we performed a proteolytic assay with purified HSP90β and also included the HSP90α isoform, which has 93% sequence identity with HSP90β. As can be seen in Fig. 2B, purified HSP90β is cleaved by rCD2830. Surprisingly, purified HSP90α was not cleaved (Fig. 2C). In order to determine the precise cleavage site, we employed two strategies. First, we compared the LC-MS/MS analyses of the in-gel tryptic digests of the rCD2830-cleaved and uncleaved HSP90β. A clear signal at m/z 871.5 [M+2H]2+ was noticeable in the digest of CD2830-treated HSP90β but was absent in the untreated sample (Fig. 2D, left-hand panel). On the basis of MS/MS data, this species was identified as the semi-tryptic HSP90β peptide Leu686-Ala702 (Fig. 2D, right-hand panel). Second, we measured the small HSP90β fragment originating after rCD2830 cleavage by means of direct-infusion electrospray ionization Q-TOF mass spectrometry (Fig. 2E). The mass of this peptide corresponds to the C-terminal part of HSP90β (Ala703-Asp724, theoretical m/z 1207.039 [M+2H]2+). Therefore, both approaches clearly demonstrated that HSP90β is cleaved by rCD2830 between alanine 702 and 703 (Fig. 2F). Because both homologous alanines are also present in HSP90α (Fig. 2F) but are insusceptible to CD2830 cleavage, this indicates that the surrounding amino acid residues are important for the specificity of CD2830.
Identification of the CD2830 Cleavage Site Motif
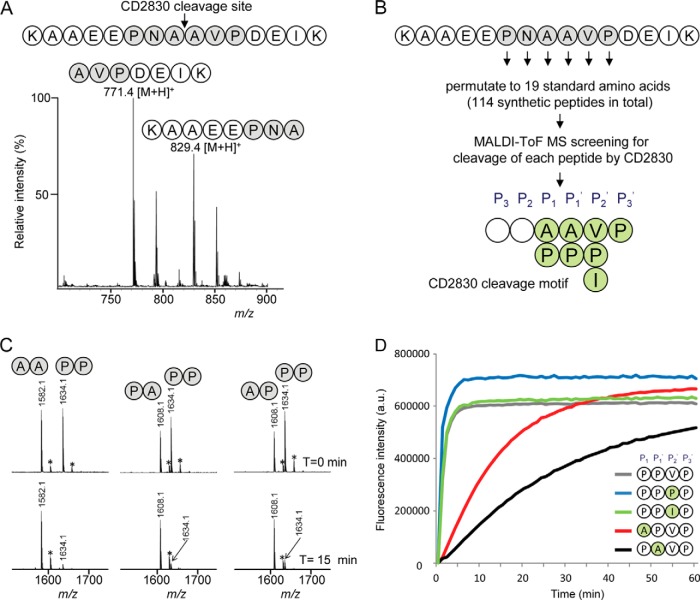
To determine whether CD2830 is also capable of cleaving small peptides, we generated a synthetic peptide (KAAEEPNAAVPDEIK) based on the identified cleavage site of HSP90β and incubated this with rCD2830. Using MALDI-TOF-MS analysis, we observed cleavage of this peptide, and the products at m/z 829.40 [M+H]+ (KAAEEPNA) and m/z 771.41 [M+H]+ (AVPDEIK) confirmed the CD2830 cleavage between the two alanine residues (Fig. 3A). When we assayed an HSP90α peptide containing the homologous sequence surrounding the cleavage site, DTSAAVTEE (Fig. 2F), no cleavage was observed, again demonstrating the specificity of CD2830 for the C-terminal region of HSP90β (data not shown).
Fig. 3.
Identification of the CD2830 cleavage motif based on a peptide library screen reveals a preference for cleavage between proline residues. A, MALDI-TOF-MS spectrum of a synthetic peptide containing the HSP90β sequence AAEEPNAAVPDEI (amino acids 696–708) after 16 h of incubation with rCD2830. B, a synthetic peptide library was constructed in which all six positions surrounding the CD2830 scissile bond were permutated to the 19 standard amino acids within a core synthetic peptide (KAAEEPNAAVPDEIK), resulting in a total set of 114 peptides. Peptides were individually incubated with rCD2830, and cleavage was measured via MALDI-TOF-MS. The resulting CD2830 cleavage motif based on this peptide library screen is shown. C, binary mixtures of synthetic peptides with either a proline or an alanine at the P1 and P1′ positions in the core peptide (see above) were incubated with rCD2830 and analyzed via MALDI-TOF-MS after 15 min of incubation. After this short incubation, cleavage of the peptide containing a proline at both P1 and P1′ was more efficient than cleavage of the peptides containing one or two alanines. *+Na+. D, within a core synthetic FRET peptide (DabcylLys-EVNPPVPD-EdansGlu), permutations were introduced at the P1, P1′, and P2′ (PPV) positions. All peptides (50 μm) were incubated with rCD2830, and the formation of cleavage products was followed in time using fluorescence detection (see “Experimental Procedures” for details).
Next we aimed at determining the CD2830 cleavage motif in order to screen databases for additional putative CD2830 substrate proteins. For this purpose, we generated a synthetic peptide library in which the six positions flanking the cleavage site (P, N, ǍA, V, P) within the peptide described above (KAAEEPNAAVPDEIK) were permutated to each of the 19 standard amino acids (Fig. 3B). Each of the resulting 114 peptides was individually incubated for 16 h with rCD2830 and then measured using MALDI-TOF-MS to determine whether cleavage had occurred. Amino acid residues in a substrate undergoing cleavage are commonly designated P3, P2, P1, P1′, P2′, and P3′. In brief (Fig. 3B), the screening of the 114 synthetic peptides demonstrated that the most stringent position was P3′ (i.e. only peptides with a proline at this position were cleaved). The requirement of a proline at position P3′ explains why HSP90α (threonine at P3′; Fig. 2F) was not cleaved. The two positions flanking the scissile bond, ǍA, only tolerated replacement by a proline. The P2′ position could, in addition to valine, also accommodate a proline or an isoleucine. Less stringent requirements were observed for positions P3 and P2, where many amino acids were tolerated (P3: P, F, V, C, I, Y, W, E, M; P2: N, F, L, M, H, Y, W, R).
Overall, a striking prevalence of multiple proline residues surrounding the scissile bond was observed, and the possibility of a proline at P1′ is especially remarkable because most proteolytic enzymes, including trypsin, do not allow for this. To gain more insight about the relative preference for proline residues around the scissile bond, we incubated binary mixtures of synthetic peptides with rCD2830. These peptides varied in the presence of either a proline or an alanine at the P1 and P1′ positions within our reference peptide (KAAEEPNAAVPDEIK, KAAEEPNPAVPDEIK, KAAEEPNAPVPDEIK, KAAEEPNPPVPDEIK). Assaying the cleavage of mixtures of two of these peptides after 15 min of incubation with rCD2830 showed that the peptide containing a proline at both P1 and P1′ was more efficiently cleaved than peptides with either one or two alanines at these positions (Fig. 3C). This was corroborated by the appearance of the product peptides (data not shown). Subsequently, we synthesized peptides in which we increased the number of proline residues and tested these for cleavage by CD2830. Intriguingly, even a peptide with six consecutive proline residues within our reference peptide was cleaved (supplemental Fig. S2).
We subsequently developed a fluorescent assay using FRET peptides that allowed fast, real-time quantitative analysis of CD2830 activity. First of all, we demonstrated that CD2830 is inhibited by the chelating agent o-phenanthroline (supplemental Fig. S3), confirming that CD2830 is a metalloprotease. We also used these FRET peptides to gain more insight into the parameters of the kinetics of CD2830 activity. This again showed that peptides having two prolines around the scissile bond were cleaved at the highest rate (Fig. 3D and supplemental Fig. S4A), confirming the data shown above (Fig. 3C). Moreover, of the three different amino acid options at the P2′ position, proline also appeared to be the most efficient, although real-time kinetics were very similar to having either a Val or an Ile at that position (Fig. 3D). The Km, Vmax, and Kcat values with the peptide having four consecutive prolines (spanning the P1–P3′ positions) were 77 ± 9 μm (n = 4), 0.08 ± 0.009 nmol/s (n = 4), and 19 ± 2 s−1 (n = 4), respectively (supplemental Fig. S4B).
Next, we wanted to test whether such a proline-rich sequence would also be cleaved in the context of an intact protein. One putative protein target of CD2830 containing a proline-rich motif is IgA2, because the flexible hinge region of the heavy chain contains the sequence PVPPPPPC. Several bacterial species produce an IgA protease suggested to be important for immune evasion. We decided to investigate the cleavage of IgA2 by CD2830 and indeed observed cleavage of IgA2 within the hinge region PVP̌PPPPC (supplemental Fig. S5).
CD2830 Substrate Cleavage Sites Are Found in C. difficile LPXTG Proteins
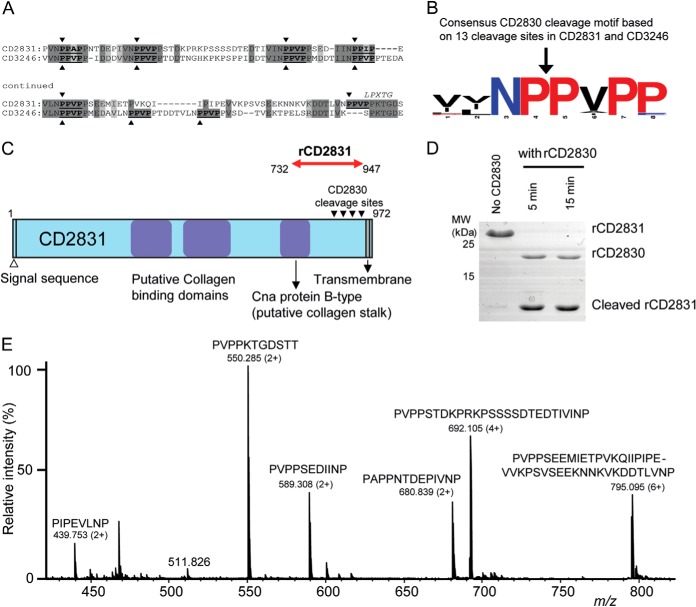
To find potential targets of CD2830 in the C. difficile proteome, we performed a ScanProsite search (22) using the above-described cleavage motif [PA]PA][VPI]P. We found only four potential signal sequences containing proteins harboring a CD2830 cleavage motif within the C. difficile strain 630 proteome: CD0515 (carboxypeptidase), CD2831 (putative adhesin, collagen binding), CD3043 (putative transglutaminase), and CD3246 (putative collagen binding). Remarkably, CD2831, located next to CD2830 in the genome, and CD3246 contained six and seven consecutive potential cleavage sites, respectively (Fig. 4A). The region containing these sites seemed conserved between CD2831 and CD3246 (shaded areas in Fig. 4A). Moreover, CD2831 and CD3246 had features common to LPXTG cell wall anchored proteins, and the putative protease cleavage sites were directly adjacent to this peptidoglycan anchor motif. The multitude of potential target sites suggests effective cleavage of these two cell surface proteins, and based on the 13 potential cleavage sites in these two proteins a consensus motif was constructed (Fig. 4B). In agreement with the results of our peptide screen described above, a striking overall proline-rich motif is apparent, and preferences for a proline at P4′ and asparagine at P2 seem plausible.
Fig. 4.
Identification of multiple CD2830 cleavage sites in putative C. difficile surface proteins. A, identification of multiple CD2830 cleavage sites (indicated by arrowheads) in the C-terminal region of the C. difficile putative adhesion proteins CD2831 and CD3246. Both putative substrates contain an LPXTG motif (PPXTG and SPXTG, respectively) and show considerable sequence similarity around the CD2830 cleavage sites. B, prevalence of amino acids around the scissile bond in the 13 CD2830 cleavage sites identified in CD2831 and CD3246. The cleavage sites were aligned using WebLogo and visualized using the following coloring: red, proline; black, nonpolar; blue, polar (not proline). C, schematic representation of C. difficile CD2831 showing the putative collagen binding domains and transmembrane domain. Recombinant CD2831 (rCD2831) protein corresponding to amino acids 732–947 (red arrow) was produced containing all the CD2830 cleavage sites. D, SDS-PAGE analysis of rCD2831 treated with rCD2830 for different time periods. E, mass spectrometric analysis (Q-TOF-MS) of the small peptide cleavage products derived from rCD2831 after incubation with rCD2830 for 30 min.
Based on the multitude of possible cleavage sites, we decided to study CD2830-mediated cleavage of these C. difficile putative adhesion molecules in more detail. Noteworthy, CD3246, but not CD2831, has been lost in the genomes of C. difficile ribotype 027, including the virulent R20291 strain (23). Therefore, we initially focused our research on CD2831. To confirm cleavage of CD2831 (putative adhesion protein; Fig. 4C) by CD2830, we produced part of CD2831 as a recombinant protein and tested it for cleavage. As shown in Fig. 4D, within 5 min of incubation with rCD2830, all recombinant CD2831 was cleaved. Therefore, this cleavage was much more efficient than that observed for HSP90β (Fig. 2B) and IgAs (supplemental Fig. S5). Because the gel analysis did not allow us to discriminate which of the six putative target sites were cleaved, we also analyzed the CD2831 cleavage products via direct-infusion electrospray ionization Q-TOF-MS. This showed that each predicted CD2830 cleavage site within CD2831 was efficiently cleaved (Fig. 4E).
CD2830 Is Functionally Secreted from C. difficile Cells and Cleaves Endogenous CD2831 and CD3246
In order to test whether secreted native CD2830 is active within the medium of C. difficile growing cells, we incubated a synthetic peptide containing a CD2830 cleavage site identified in CD2831 (KDTIVINP̌PVPPSEK) with conditioned minimal medium. Subsequent MALDI-TOF-MS analysis (Fig. 5A) revealed cleavage of the peptide, thereby demonstrating that CD2830 is secreted as a functional protease from C. difficile cells. Moreover, using our fluorescent assay, we demonstrated that more than 90% of the proteolytic activity was present in the medium and only less than 10% in the cell pellet, supporting the evidence that CD2830 was actively secreted in the medium (data not shown).
Fig. 5.
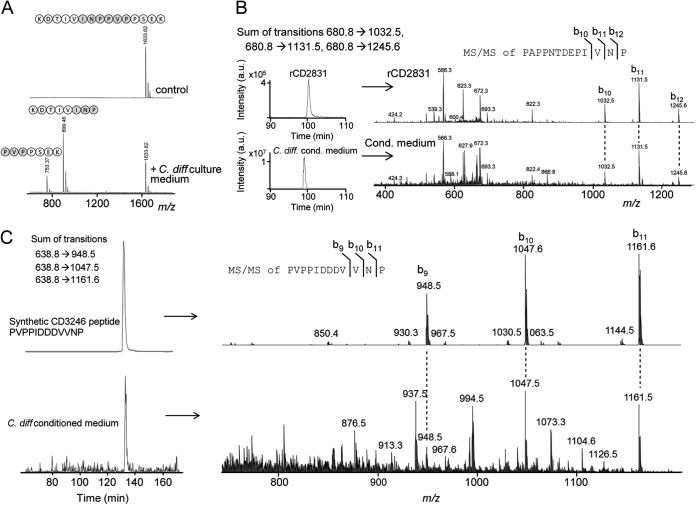
Detection of native CD2830 protease activity in the secretome of C. difficile and cleavage of endogenous CD2831 and CD3246. A, a synthetic peptide containing one of the CD2830 cleavage sites in CD2831 (KDTIVINPPVPPSEK) was incubated with the conditioned medium collected from cultures of C. difficile cells. After incubation for 16 h, samples were analyzed via MALDI-TOF-MS to determine substrate peptide cleavage. B, upper panel: LC–ion trap MS/MS analysis of recombinant CD2831 after treatment with rCD2830 demonstrating the elution profile (left) and MS/MS identification (right) of the CD2831 cleavage product PAPPNTDEPIVNP. Lower panel: LC-MS/MS analysis of C. difficile conditioned medium. Shown are the sum of the specific transitions 680.8 → 1032.5, 680.8 → 1131.5, and 680.8 → 1245.6 in the LC-MS/MS elution profile (left) and MS/MS identification (right), which are both equivalent to that observed with rCD2831 (upper panel), thereby unambiguously demonstrating the presence of the CD2831 cleavage product in C. difficile conditioned medium. C, upper panel: a synthetic peptide corresponding to one of the putative cleavage products of CD3246 (PVPPIDDDVVNP) was analyzed via LC–ion trap MS/MS to determine its elution time and MS/MS spectrum. Lower graph: LC-MS/MS analysis of C. difficile conditioned medium. Shown are the sum of the specific transitions 638.8 → 948.5, 638.8 → 1047.6, and 638.8 → 1161.6 (left) and MS/MS identification (right), which are both equivalent to that observed with the synthetic peptide (upper panel), thereby clearly demonstrating the presence of the CD3246 cleavage product in C. difficile conditioned medium.
Having established that CD2830 is actively secreted, we investigated whether this activity is sufficient to cleave endogenous CD2831. For this purpose, we set up a method for the targeted measurement of CD2831 cleavage products via LC–ion trap MS/MS analysis. As a reference for retention time and fragmentation pattern, we first analyzed the rCD2831 sample treated with rCD2830 protease. We focused on m/z 680.8 [M+2H]2+, corresponding to one of the CD2831 product peptides (PAPPNTDEPIVNP; Fig. 4E). This peptide eluted around 100 min, as shown by the sum of the intensities of the transitions 680.8 → 1032.5, 680.8 → 1131.5, and 680.8 → 1245.6, characteristic for this peptide (Fig. 5B, upper part). Analysis of the conditioned medium of C. difficile unambiguously identified the presence of the same CD2831 cleavage product, based on both retention time and MS/MS fragmentation (Fig. 5B, lower part). This clearly shows that native CD2830 activity cleaved endogenous CD2831. This could also explain the identification of CD2831 in our secretome analysis (Table I).
Similarly, we tested whether a CD3246 cleavage product could be identified in the medium. As a reference, we used a synthetic putative CD2830 product peptide from CD3246 (PVPPIDDDVVNP; Fig. 4A). We first analyzed this peptide via LC–ion trap MS/MS to determine the retention time and MS/MS spectrum (Fig. 5C, upper panels). Then, we analyzed the conditioned cell culture medium and observed a clear signal corresponding to the same peptide (Fig. 5C, lower part), demonstrating the presence of the CD3246 cleavage product in the conditioned medium.
Overall, the above data clearly demonstrate that C. difficile cells secrete active CD2830 that can cleave the endogenous putative cell surface molecules CD2831 and CD3246.
DISCUSSION
Bacterial secreted proteins constitute a biologically important subset of proteins that are involved in multiple processes to aid proliferation and infection. Bacterial extracellular proteases, in particular, have attracted considerable attention, as these have often been shown to be indispensable for bacterial virulence. Here, we studied a novel secreted metalloprotease, CD2830, which we identified as a genuinely secreted protein from C. difficile.
After the initial identification of HSP90β as a substrate for CD2830, we identified a CD2830 cleavage motif using a synthetic peptide library screen. The identified CD2830 cleavage motif is unique and has hitherto not been described for any other protease (24, 25). The strong preference for proline residues at the P1, P1′, and P3′ positions is especially intriguing. The distinctive cyclic structural characteristics of proline create conformational constraints to proline-containing peptides. Peptide bonds containing a proline residue are in general less sensitive to proteolytic cleavage, especially with a proline at the P1′ position. However, some proline-specific enzymes have evolved, most of which target either N- or C-terminally located residues (26, 27). Even though proteolytic cleavage at a Pro–Pro bond has been observed before (28, 29), to the best of our knowledge, CD2830 is the first enzyme with a high preference for hydrolysis of such a peptide bond. Our peptide library screen indicated less stringency for amino acid residues at the P2 and P3 positions. Because in the current study we permutated the amino acid positions based on the initial cleavage site (PNǍAVP), additional screens and kinetic experiments should elucidate a more detailed picture of the optimal CD2830-cleavage motif with respect to these two positions. In addition, structural analysis of the catalytic domain of CD2830 in complex with the target peptide might reveal specific contacts important for proteolytic activity.
Using the CD2830 protease cleavage motif from our peptide library screen, we found many potential CD2830 substrates (∼600) in the human subset of signal-peptide-containing proteins (membrane-associated or secreted proteins). Given the preference for prolines surrounding the scissile bond, this list would be restricted to 238 possible candidate substrates. Several of these proteins might play a role in defense against bacterial infections. Clearly, the presence of a putative CD2830 cleavage site does not make such a protein an assured substrate, as the cleavage site might be inaccessible or the protease and substrate might never co-localize or be synthesized at the same time. Based on the cleavage motif and the preference for multiple prolines, we studied the cleavage of IgA2 by CD2830. Except for Clostridium ramosum, whose IgA protease cleaves one isoform of human IgA2 (IgA2(1)) (30), the IgA2 subclass is resistant to other bacterial IgA proteases. We have now shown that IgA2 heavy chain can be cleaved by CD2830 in vitro within the hinge region. Notably, numerous bacterial species produce proteases that cleave at Pro–Ser and Pro–Thr sites within the hinge region of IgA1 (31–33). This IgA1 region contains two consecutive Pro-Pro-Thr-Pro motifs. In accordance with our identified motif, the Thr presumably prevents cleavage by CD2830 within this region, although glycosylation and accessibility of the cleavage site within the hinge region might also play a role. Whether CD2830 can cleave IgA in vivo and is relevant for immune evasion remains to be determined. Although several studies have provided a link between virulence and the expression of IgA proteases, it has been difficult to prove that such proteases are virulence factors in vivo. This is related to the fact that only human, gorilla, chimpanzee, and orangutan IgA are susceptible to the hitherto described IgA proteases (32, 34, 35).
Several observations within our study demonstrated that the C. difficile proteins CD2831 and CD3246 are candid CD2830 substrates. First, both proteins contain multiple CD2830 cleavage sites. Second, recombinant CD2831 was very efficiently cleaved by rCD2830 at all predicted cleavage sites. Third, cleavage of endogenous CD2831 and CD3246 by native CD2830 was observed in cultures of C. difficile cells.
The two C. difficile substrates identified are putative adhesin proteins and contain C-terminal PPXTG (CD2831) and SPXTG (CD3246) motifs, respectively. In Gram-positive bacteria, surface proteins harboring such a signal are covalently linked to the cell wall by the transpeptidase sortase, and several studies have shown the importance of these cell surface proteins for bacterial virulence (36–39). In C. difficile strain 630, only one putative sortase has been identified (8), and the number of putative sortase substrates appears to be limited (8, 9, 40, 41), but as yet no study has revealed genuine sortase substrates in C. difficile.
Cleavage of adhesion proteins has been shown to be an important mechanism for the control of their surface levels. In Staphylococcus aureus, a secreted protease (aureolysin) cleaves the clumping factor ClfB (42), a protein involved in adherence to blood clots. The aureolysin homologue in Enterococcus faecalis, gelatinase GelE, cleaves a surface adhesion protein, thereby controlling the ability of cells to adhere to collagens (43). In Bacillus anthracis, adhesion to human endothelial cells is regulated by cleavage of one of the major surface layer proteins (BslA) by the abundantly secreted protease InhA (39). Clear examples also are found in Gram-negative bacteria. In Bordetella pertussis, the SphB1 protease releases a major adhesin from the cell surface. Interestingly, an SphB1 knockout shows greater adhesion in vitro but attenuated colonization in vivo (44).
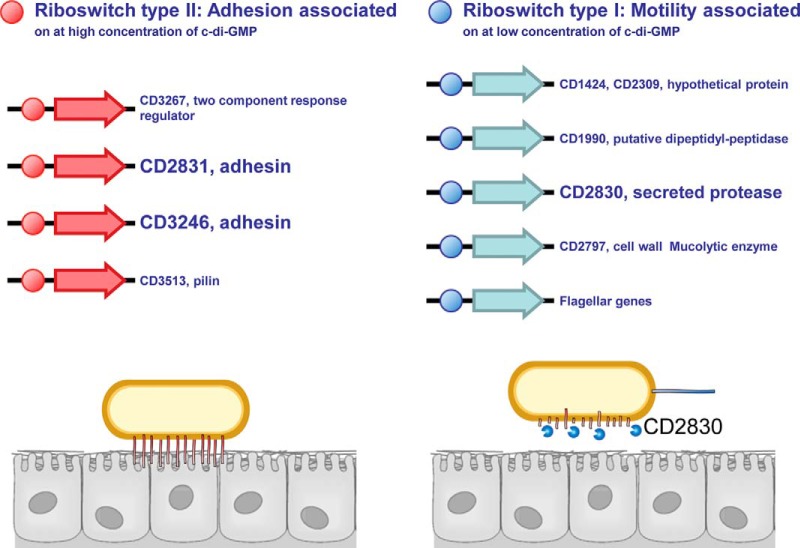
These examples all point to a role for a specific protease in the control of adhesion versus motility. We propose a similar role for CD2830 through cleavage of CD2831 and CD3246. Clearly, in such a scenario, protease and adhesion proteins have opposite effects, requiring tight control in vivo. In the past decade, the bacterial second messenger c-di-GMP has emerged as a central mediator in regulating prokaryotic adhesion/biofilm formation versus motility (45, 46). In Pseudomonas fluorescens, for example, a c-di-GMP binding protein sequesters a protease responsible for cleavage of an adhesion protein at low levels of c-di-GMP but releases it at higher concentrations, thereby regulating bacterial motility (47, 48). Besides c-di-GMP binding proteins, other c-di-GMP-sensing regulatory structures are riboswitches, RNA structures controlling gene expression through binding of specific ligands. In C. difficile, two types of c-di-GMP riboswitches have been described (49–51) that are oppositely regulated by c-di-GMP; the class I riboswitch is activated at low levels of c-di-GMP, whereas the class II c-di-GMP riboswitch is activated at higher levels of c-di-GMP (Fig. 6).
Fig. 6.
Model for the proposed role of CD2830 in regulation of adhesion versus motility in C. difficile. The C. difficile secreted protease CD2830 cleaves adhesin proteins CD2831 and CD3246, thereby releasing these cell surface anchors. Both adhesin mRNAs contain a riboswitch type II (red balls), which is turned on by elevated levels of c-di-GMP, a central mediator of motility and adhesion in bacteria (top panel). In total, four genes in the C. difficile strain 630 genome contain the type II riboswitch. The cd2830 gene contains a riboswitch type I (blue balls), which is turned on at lower levels of c-di-GMP, as was also shown for the flagellar operon. We postulate that this opposite regulation of expression of adhesins and CD2830 by c-di-GMP plays an important role in the regulation of motility and adhesion: low c-di-GMP concentrations result in the down-regulation of surface levels of adhesins both by repressing their gene expression and by the concomitant cell surface release mediated by CD2830 protease cleavage.
Interestingly, the 5′UTR of cd2830 contains a class I c-di-GMP riboswitch that, for example, was also found to control the expression of flagellar genes (52). Strikingly, three of the four loci containing a class II riboswitch encode putative adhesion molecules, CD2831, CD3246, and CD3513 (putative pilin). Two of these three proteins were identified as CD2830 substrates in this study (Fig. 6). The inverse regulation of cd2830 and cd2831 expression by c-di-GMP was very recently confirmed in cells with inducible expression of one of the many (53) c-di-GMP synthases (54). Consequently, the remarkable picture emerges that in C. difficile, c-di-GMP inversely regulates gene expression of a protease-encoding gene (CD2830) and its adjacent gene encoding its substrate (CD2831). The control of adhesion versus motility in C. difficile and the role of c-di-GMP and specific surface proteins are only starting to be elucidated, but our data suggest that CD2830 plays a pivotal role in this process. Testing this model (Fig. 6) will be the main focus of our future studies.
In conclusion, we have characterized CD2830 as a highly active secreted metalloprotease from C. difficile with a unique preference for multiple proline residues surrounding the scissile bond. The identification of two C. difficile adhesion proteins as efficient CD2830 substrates suggests that CD2830 plays a role in C. difficile adhesion and motility.
Supplementary Material
Acknowledgments
We thank Dr. Ron Hokke and Dr. Manfred Wuhrer for critical reading of the manuscript and Dr. Rico Derks for valuable suggestions for the data analysis.
Footnotes
Author contributions: P.J.H., O.I.K., A.M.D., E.J.K., J.C., J.D., and H.C.v. designed research; P.J.H., O.I.K., D.B., V.J.v., N.R., A.C.K., R.A.C., I.D., and H.C.v. performed research; P.J.H., O.I.K., J.D., and H.C.v. contributed new reagents or analytic tools; P.J.H., O.I.K., I.D., J.D., and H.C.v. analyzed data; P.J.H. and H.C.v. wrote the paper.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ALF
- anthrax toxin lethal factor
- HSP
- heat shock protein.
REFERENCES
- 1. Rupnik M., Wilcox M. H., Gerding D. N. (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7, 526–536 [DOI] [PubMed] [Google Scholar]
- 2. Muniz L. R., Knosp C., Yeretssian G. (2012) Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front. Immunol. 3, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuehne S. A., Cartman S. T., Heap J. T., Kelly M. L., Cockayne A., Minton N. P. (2010) The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713 [DOI] [PubMed] [Google Scholar]
- 4. Lyras D., O'Connor J. R., Howarth P. M., Sambol S. P., Carter G. P., Phumoonna T., Poon R., Adams V., Vedantam G., Johnson S., Gerding D. N., Rood J. I. (2009) Toxin B is essential for virulence of Clostridium difficile. Nature 458, 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McBride S. M., Sonenshein A. L. (2011) The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 157, 1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin Y. P., Kuo C. J., Koleci X., McDonough S. P., Chang Y. F. (2011) Manganese binds to Clostridium difficile Fbp68 and is essential for fibronectin binding. J. Biol. Chem. 286, 3957–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hennequin C., Janoir C., Barc M. C., Collignon A., Karjalainen T. (2003) Identification and characterization of a fibronectin-binding protein from Clostridium difficile. Microbiology 149, 2779–2787 [DOI] [PubMed] [Google Scholar]
- 8. Sebaihia M., Wren B. W., Mullany P., Fairweather N. F., Minton N., Stabler R., Thomson N. R., Roberts A. P., Cerdeno-Tarraga A. M., Wang H., Holden M. T., Wright A., Churcher C., Quail M. A., Baker S., Bason N., Brooks K., Chillingworth T., Cronin A., Davis P., Dowd L., Fraser A., Feltwell T., Hance Z., Holroyd S., Jagels K., Moule S., Mungall K., Price C., Rabbinowitsch E., Sharp S., Simmonds M., Stevens K., Unwin L., Whithead S., Dupuy B., Dougan G., Barrell B., Parkhill J. (2006) The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38, 779–786 [DOI] [PubMed] [Google Scholar]
- 9. Tulli L., Marchi S., Petracca R., Shaw H. A., Fairweather N. F., Scarselli M., Soriani M., Leuzzi R. (2013) CbpA: a novel surface exposed adhesin of Clostridium difficile targeting human collagen. Cell Microbiol. 15, 1674–1687 [DOI] [PubMed] [Google Scholar]
- 10. Vedantam G., Clark A., Chu M., McQuade R., Mallozzi M., Viswanathan V. K. (2012) Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes 3, 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hiemstra H. S., Duinkerken G., Benckhuijsen W. E., Amons R., De Vries R. R., Roep B. O., Drijfhout J. W. (1997) The identification of CD4+ T cell epitopes with dedicated synthetic peptide libraries. Proc. Natl. Acad. Sci. U.S.A. 94, 10313–10318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bakker D., Smits W. K., Kuijper E. J., Corver J. (2012) TcdC does not significantly repress toxin expression in Clostridium difficile 630DeltaErm. PLoS One 7, e43247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cartman S. T., Minton N. P. (2010) A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 76, 1103–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zauner G., Hoffmann M., Rapp E., Koeleman C. A., Dragan I., Deelder A. M., Wuhrer M., Hensbergen P. J. (2012) Glycoproteomic analysis of human fibrinogen reveals novel regions of O-glycosylation. J. Proteome Res. 11, 5804–5814 [DOI] [PubMed] [Google Scholar]
- 15. Balog C. I., Mayboroda O. A., Wuhrer M., Hokke C. H., Deelder A. M., Hensbergen P. J. (2010) Mass spectrometric identification of aberrantly glycosylated human apolipoprotein C-III peptides in urine from Schistosoma mansoni-infected individuals. Mol. Cell. Proteomics 9, 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boetzkes A., Felkel K. W., Zeiser J., Jochim N., Just I., Pich A. (2012) Secretome analysis of Clostridium difficile strains. Arch. Microbiol. 194, 675–687 [DOI] [PubMed] [Google Scholar]
- 17. Petersen T. N., Brunak S., von H. G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 18. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 19. Pannifer A. D., Wong T. Y., Schwarzenbacher R., Renatus M., Petosa C., Bienkowska J., Lacy D. B., Collier R. J., Park S., Leppla S. H., Hanna P., Liddington R. C. (2001) Crystal structure of the anthrax lethal factor. Nature 414, 229–233 [DOI] [PubMed] [Google Scholar]
- 20. Turk B. E., Wong T. Y., Schwarzenbacher R., Jarrell E. T., Leppla S. H., Collier R. J., Liddington R. C., Cantley L. C. (2004) The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor. Nat. Struct. Mol. Biol. 11, 60–66 [DOI] [PubMed] [Google Scholar]
- 21. Knetsch C. W., Terveer E. M., Lauber C., Gorbalenya A. E., Harmanus C., Kuijper E. J., Corver J., van Leeuwen H. C. (2012) Comparative analysis of an expanded Clostridium difficile reference strain collection reveals genetic diversity and evolution through six lineages. Infect. Genet. Evol. 12, 1577–1585 [DOI] [PubMed] [Google Scholar]
- 22. Castro E., Sigrist C. J., Gattiker A., Bulliard V., Langendijk-Genevaux P. S., Gasteiger E., Bairoch A., Hulo N. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He M., Sebaihia M., Lawley T. D., Stabler R. A., Dawson L. F., Martin M. J., Holt K. E., Seth-Smith H. M., Quail M. A., Rance R., Brooks K., Churcher C., Harris D., Bentley S. D., Burrows C., Clark L., Corton C., Murray V., Rose G., Thurston S., van T. A., Walker D., Wren B. W., Dougan G., Parkhill J. (2010) Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. U.S.A. 107, 7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Igarashi Y., Eroshkin A., Gramatikova S., Gramatikoff K., Zhang Y., Smith J. W., Osterman A. L., Godzik A. (2007) CutDB: a proteolytic event database. Nucleic Acids Res. 35, D546–D549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rawlings N. D., Barrett A. J., Bateman A. (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40, D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vanhoof G., Goossens F., De M. I., Hendriks D., Scharpe S. (1995) Proline motifs in peptides and their biological processing. FASEB J. 9, 736–744 [PubMed] [Google Scholar]
- 27. Cunningham D. F., O'Connor B. (1997) Proline specific peptidases. Biochim. Biophys. Acta 1343, 160–186 [DOI] [PubMed] [Google Scholar]
- 28. Patterson B. C., Sang Q. A. (1997) Angiostatin-converting enzyme activities of human matrilysin (MMP-7) and gelatinase B/type IV collagenase (MMP-9). J. Biol. Chem. 272, 28823–28825 [DOI] [PubMed] [Google Scholar]
- 29. Nolte W. M., Tagore D. M., Lane W. S., Saghatelian A. (2009) Peptidomics of prolyl endopeptidase in the central nervous system. Biochemistry 48, 11971–11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fujiyama Y., Iwaki M., Hodohara K., Hosoda S., Kobayashi K. (1986) The site of cleavage in human alpha chains of IgA1 and IgA2:A2m(1) allotype paraproteins by the clostridial IGA protease. Mol. Immunol. 23, 147–150 [DOI] [PubMed] [Google Scholar]
- 31. Qiu J., Brackee G. P., Plaut A. G. (1996) Analysis of the specificity of bacterial immunoglobulin A (IgA) proteases by a comparative study of ape serum IgAs as substrates. Infect. Immun. 64, 933–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woof J. M., Kerr M. A. (2006) The function of immunoglobulin A in immunity. J. Pathol. 208, 270–282 [DOI] [PubMed] [Google Scholar]
- 33. Gass J., Khosla C. (2007) Prolyl endopeptidases. Cell Mol. Life Sci. 64, 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Senior B. W., Woof J. M. (2005) The influences of hinge length and composition on the susceptibility of human IgA to cleavage by diverse bacterial IgA1 proteases. J. Immunol. 174, 7792–7799 [DOI] [PubMed] [Google Scholar]
- 35. Woof J. M., Russell M. W. (2011) Structure and function relationships in IgA. Mucosal Immunol. 4, 590–597 [DOI] [PubMed] [Google Scholar]
- 36. Budzik J. M., Marraffini L. A., Schneewind O. (2007) Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 66, 495–510 [DOI] [PubMed] [Google Scholar]
- 37. Hendrickx A. P., Budzik J. M., Oh S. Y., Schneewind O. (2011) Architects at the bacterial surface—sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 9, 166–176 [DOI] [PubMed] [Google Scholar]
- 38. Maresso A. W., Schneewind O. (2008) Sortase as a target of anti-infective therapy. Pharmacol. Rev. 60, 128–141 [DOI] [PubMed] [Google Scholar]
- 39. Tonry J. H., McNichol B. A., Ramarao N., Chertow D. S., Kim K. S., Stibitz S., Schneewind O., Kashanchi F., Bailey C. L., Popov S., Chung M. C. (2012) Bacillus anthracis protease InhA regulates BslA-mediated adhesion in human endothelial cells. Cell Microbiol. 14, 1219–1230 [DOI] [PubMed] [Google Scholar]
- 40. Pallen M. J., Lam A. C., Antonio M., Dunbar K. (2001) An embarrassment of sortases—a richness of substrates? Trends Microbiol. 9, 97–102 [DOI] [PubMed] [Google Scholar]
- 41. Comfort D., Clubb R. T. (2004) A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 72, 2710–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McAleese F. M., Walsh E. J., Sieprawska M., Potempa J., Foster T. J. (2001) Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. J. Biol. Chem. 276, 29969–29978 [DOI] [PubMed] [Google Scholar]
- 43. Pinkston K. L., Gao P., Diaz-Garcia D., Sillanpaa J., Nallapareddy S. R., Murray B. E., Harvey B. R. (2011) The Fsr quorum-sensing system of Enterococcus faecalis modulates surface display of the collagen-binding MSCRAMM Ace through regulation of gelE. J. Bacteriol. 193, 4317–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Coutte L., Willery E., Antoine R., Drobecq H., Locht C., Jacob-Dubuisson F. (2003) Surface anchoring of bacterial subtilisin important for maturation function. Mol. Microbiol. 49, 529–539 [DOI] [PubMed] [Google Scholar]
- 45. Mills E., Pultz I. S., Kulasekara H. D., Miller S. I. (2011) The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell Microbiol. 13, 1122–1129 [DOI] [PubMed] [Google Scholar]
- 46. Hengge R. (2009) Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273 [DOI] [PubMed] [Google Scholar]
- 47. Newell P. D., Boyd C. D., Sondermann H., O'Toole G. A. (2011) A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 9, e1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Newell P. D., Yoshioka S., Hvorecny K. L., Monds R. D., O'Toole G. A. (2011) Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0–1. J. Bacteriol. 193, 4685–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee E. R., Baker J. L., Weinberg Z., Sudarsan N., Breaker R. R. (2010) An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329, 845–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sudarsan N., Lee E. R., Weinberg Z., Moy R. H., Kim J. N., Link K. H., Breaker R. R. (2008) Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321, 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen A. G., Sudarsan N., Breaker R. R. (2011) Mechanism for gene control by a natural allosteric group I ribozyme. RNA 17, 1967–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Purcell E. B., McKee R. W., McBride S. M., Waters C. M., Tamayo R. (2012) Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 194, 3307–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bordeleau E., Fortier L. C., Malouin F., Burrus V. (2011) c-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 7, e1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soutourina O. A., Monot M., Boudry P., Saujet L., Pichon C., Sismeiro O., Semenova E., Severinov K., Le B. C., Coppee J. Y., Dupuy B., Martin-Verstraete I. (2013) Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 9, e1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.