Abstract
Usage of presumed 5′UTR or downstream in-frame AUG codons, next to non-AUG codons as translation start codons contributes to the diversity of a proteome as protein isoforms harboring different N-terminal extensions or truncations can serve different functions. Recent ribosome profiling data revealed a highly underestimated occurrence of database nonannotated, and thus alternative translation initiation sites (aTIS), at the mRNA level. N-terminomics data in addition showed that in higher eukaryotes around 20% of all identified protein N termini point to such aTIS, to incorrect assignments of the translation start codon, translation initiation at near-cognate start codons, or to alternative splicing. We here report on more than 1700 unique alternative protein N termini identified at the proteome level in human and murine cellular proteomes. Customized databases, created using the translation initiation mapping obtained from ribosome profiling data, additionally demonstrate the use of initiator methionine decoded near-cognate start codons besides the existence of N-terminal extended protein variants at the level of the proteome. Various newly identified aTIS were confirmed by mutagenesis, and meta-analyses demonstrated that aTIS reside in strong Kozak-like motifs and are conserved among eukaryotes, hinting to a possible biological impact. Finally, TargetP analysis predicted that the usage of aTIS often results in altered subcellular localization patterns, providing a mechanism for functional diversification.
Eukaryotic protein-coding genes can give rise to multiple translation products of which the expression is regulated at multiple levels. In contrast to transcriptional regulation, protein translational regulation permits for more immediate effects to take place. Initiation, elongation, termination, and ribosome recycling constitute the different phases of the eukaryotic translation process, with translation initiation acting as the gate-keeping step by the successive steps of ternary complex recruitment, scanning, AUG codon selection, and ribosomal subunit joining. Overall, this process requires over 30 different proteins including the eukaryotic initiation factors (eIFs)1 (1). In eukaryotes, the translation start codon is typically found by ribosome scanning, referred to as the canonical mechanism of translation initiation. Here, the 43S pre-initiation complex (PIC) composed of the initiator Met-tRNA (Met-tRNAi) pre-loaded onto the small (40S) ribosomal subunit, binds near the 5′ end of the mRNA molecule in a m7G-cap structure/eukaryotic initiation factors 4 (i.e. eIF4E, 4G, and 4A; jointly referred to as the eIF4F complex) mediated fashion. This complex then starts to scan successive triplets of the 5′ untranslated region (5′UTR) in the 3′ direction until an AUG start codon or, alternatively, a near-cognate start codon entered the P (peptidyl) decoding site of the ribosome. Start codon recognition requires base-pairing with the anticodon loop of Met-tRNAi and triggers a scanning arrest and GTP hydrolysis of the eIF2-GTP-Met-tRNAi ternary complex, ultimately leading to the formation of the 48S initiation complex. The latter is then followed by factor displacement, enabling the joining of the large (60S) subunit and assembly of the elongation-competent 80S initiation complex, which can now accommodate the second amino acid encoding aminoacyl-tRNA into the aminoacyl site (A-site) and thus formation of the first peptide bond in the process of translation elongation upon recruitment of translation elongation factors.
Secondary RNA structures might influence the processivity and efficiency of scanning and as such regulate translation initiation. mRNAs that contain secondary structures in their 5′UTR require ATP proportional to the degree of secondary structure (2) in addition to helicase activity to enhance 43S PIC binding and scanning.
Although the scanning mechanism of translation initiation is used by most mRNAs, an alternative manner of translation initiation of a specific subset of mRNAs is mediated by internal ribosomal entry sites (IRES).
Viruses use internal ribosomal entry as a mechanism of translation, engaging host cell ribosomes while bypassing the need for (a subset of) the limiting eIFs. Internal ribosomal entry sequences are typically long and highly structured elements that mimic the functions of eIFs while requiring trans-acting factors such as the polypyrimidine tract binding protein PTB or the La autoantigen (3). Several IRESes were also discovered in various cellular mRNAs expressed during apoptosis or mitosis or following cell stress, when cap-dependent translation is known to be impaired (4–5). Moreover, other specific mechanisms of translation initiation exist, such as the structural mRNA element driven, cap-dependent and IRES-like mechanism of histone H4 translation initiation, related to the fact that the noncanonical histone H4 mRNA features—such as its short 5′ UTR—prevent conventional scanning and translation initiation (6).
Besides IRES, a second common type of alternative translation is leaky ribosomal scanning. Here, the sequence context surrounding the first encountered AUG is suboptimal, leading to leaky scanning and translational initiation at both this first AUG codon and additional downstream AUG codons (7).
Further, translation re-initiation after a short upstream ORF (uORF) is another common regulatory control mechanism of translation initiation (8–9). In fact, up to 50% of all mammalian genes encode mRNAs that have at least one short uORF residing upstream of the main protein-encoding ORF and that consists of about 30 codons on average (10). Here, some translation factors remain associated with the ribosome, thereby enabling scanning after translating the uORF and thus enabling re-initiation of translation at downstream sites.
Finally, 5′ mRNA leader sequence recapping can also give rise to alternative translates (11–12), and thus contributes to the translational initiation landscape.
Cis-acting sequence elements steer recognition of the correct initiation codon to ensure the fidelity of translation initiation. Usually, this AUG triplet resides in an optimal context (i.e. gcc[A/G]ccAUGG(not T)), with a purine at position −3 and a guanine at position +4 relative to the A of the AUG codon which is designated as +1 (7). Control of translation initiation codon recognition and thereby translation initiation can additionally be exerted through various trans-acting factors such as eIFs, where the conserved eIF1 acts as a key determinant. eIF1 mutations resulting in premature eIF1 dissociation were shown to increase initiation rates at near-cognate start codons (13) and are thus key in maintaining the fidelity of initiation (14). Further, eIF1A thought to occupy the A-site, regulates start codon selection in a dual fashion as its N-terminal region decreases the initiation accuracy and promotes eIF1 dissociation at AUG codons, whereas its C-terminal region increases the stringency of start codon selection and promotes continued scanning at non-AUG codons (15). Further, eIF2 and eIF5 also help to ensure the fidelity of initiation codon selection. In general, phosphorylation of the alpha subunit of eIF2 (eIF2αP) is known to reduce translation initiation, contradictory however, translational induction of GCN4, a yeast transcriptional activator, has been observed by reducing translation initiation at four uORFs (16), thereby overcoming the inhibitory effect of these uORFs on re-initiation at the GCN4 ORF (17).
Ribosome profiling, a recently developed genomics-based strategy, enables systematic monitoring of protein translation events by deep sequencing of ribosome-protected mRNA fragments. To date, this methodology was applied to study the translatomes and the changes thereof in human (18–21), mouse (22), zebrafish (23), nematode (24), plants (25–26), yeast (17, 27–28), bacteria (Escherichia coli (29–30) and Bacillus subtilis (30)), human cytomegalovirus (31), and bacteriophage lambda (32).
When used in combination with initiation-specific translation inhibitors, this technique allows for the study of (alternative) translation (initiation) with subcodon or even single-nucleotide resolution, the latter referred to as Global Translation Initiation sequencing or GTI-seq (22, 33). As such, ribosome profiling provided a wealth of information on the mRNA engagement of (initiating) ribosomes and revealed the omnipresence of alternative translation initiation events in human and mice as nearly half the transcripts harbor multiple translation initiation sites or TIS in their sequence (22, 33). Besides a handful of cases for which alternative TIS selection leads to (functionally) distinct proteins isoforms because of their N-terminal heterogeneity (i.e. protein stability (34), localization (35–39), function (40), etc.), the overall functional outcome of alternative mRNA engagement, the factors and mechanisms involved in TIS selection, and the overall outcome of expanding the proteome diversity remain largely elusive.
Upon ribosome emergence, nascent protein chains (i.e. 30 to 50 amino acid long protein N termini) can be subjected to various cotranslational modification events, including proteolysis (removal of the initiator methionine (iMet) by the MetAPs (methionine aminopeptidases) (41–42)) and N-terminal (de)blocking modifications (N-terminal acetylation (Nt-acetylation) (43–46) or deformylation (47)); ubiquitous modifications in eukaryotes and prokaryotes respectively. 50% of all soluble yeast proteins and 80–90% of all soluble human proteins are modified by acetylation of the α-amino group of the amino-terminal residue (Nt-acetylation) (48–51). The utmost N-terminal amino acid is the major determining factor whether or not a given protein is Nt-acetylated and by which N-terminal acetyltransferase (NAT) this occurs (52), although some redundancy among the different NATs can be observed (50, 53). Because Nt-acetylation is considered to mainly occur cotranslationally (54), in vivo acetylated protein N termini can thus be considered as proxies of translation initiation, though read out at the proteome-wide level.
In this study, N-terminal proteomics was used to map the TIS landscape in human and mouse cells. Overall, more than 20% of all identified protein N termini point to aTIS and we report on more than 1700 unique alternative protein N termini next to the more than 4500 database annotated protein N termini identified in the proteomes of study, thereby linking about one-third of the uniquely identified protein N termini to alternative translation initiation events.
EXPERIMENTAL PROCEDURES
Cell Culture
Human epithelial colon (HCT116) and human epidermoid (A-431) cells were grown in Dulbecco's Modified Eagle's Medium (DMEM) medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. Human monocytic THP-1 cells and human primary B-cells were grown in HyClone Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal bovine serum and penicillin/streptomycin (all Invitrogen, Carlsbad, CA). Primary mouse dendritic cells (55), mouse macrophage Mf4/4 (56), mouse lymphoblast YAC-1, human lymphoblast K-562 and Jurkat (57) and human epithelial cervix HeLa (50) cells were cultured as described previously. All cell lines were purchased from American Type Culture Collection, Manassas, VA.
N-terminal COFRADIC and LC-MS/MS Analysis
N-terminal COFRADIC analyses were performed as described previously (58). To enable the assignment of in vivo Nt-acetylation events, all primary protein amines were blocked making use of a (stable isotopic encoded) N-hydroxysuccinimide ester at the protein level (i.e. NHS esters of 13C2D3 or D3 acetate).
LC-MS/MS analysis was performed as described previously ((50) and (48)). The generated MS/MS peak lists were searched with Mascot using the Mascot Daemon interface (version 2.2.0, Matrix Science, Boston, MA). Searches were performed in the Swiss-Prot database with taxonomy set to human or mouse (UniProtKB/Swiss-Prot database version 2012_03, containing 20,254 human and 16,513 mouse entries (535,248 sequence entries in total)) or using custom databases (combination of UniProtKB/Swiss-Prot and Ribo-seq derived translation sequences (59)).
13C2D3- or D3-acetylation at lysines, carbamidomethylation of cysteine and methionine oxidation to methionine-sulfoxide were set as fixed modifications for the N-terminal COFRADIC analyses. Variable modifications were 13C2D3-acetylation or D3-acetylation and acetylation of protein N termini. Pyroglutamate formation of N-terminal glutamine was additionally set as a variable modification. Endoproteinase Arg-C/P (Arg-C specificity with arginine-proline cleavage allowed) was set as enzyme allowing no missed cleavages. The mass tolerance on the precursor ion was set to 10 ppm, 0.2 Da and 0.5 Da, and on fragment ions to 0.5 Da, 0.1 Da, 0.5 Da for the Orbitrap, Q-TOF Premier and Ion Trap analyses respectively. The peptide charge was set to 1+, 2+, 3+ and instrument setting was put to ESI-TRAP for Orbitrap and Ion Trap analyses and to ESI-QUAD-TOF for Q-TOF Premier analyses. Only peptides that were ranked one and scored above the threshold score, set at 99% confidence, were withheld. The estimated false discovery rate by searching decoy databases (a shuffled version of the yeast Swiss-Prot database made by the DBToolkit algorithm (60)) was found to lie below 1.5% on the spectrum level. All annotated highest scoring spectra of the N-terminal peptides reported in supplemental Table S1 are provided as supplemental data.
Selection of N termini
From the mouse and human N-terminal data sets, N-terminally blocked peptides were selected and classified. The high confident TIS encompass: (1) all (partially) in vivo Nα-acetylated N termini and in vivo unmodified N termini of which the start position corresponded with a Swiss-Prot isoform, Ensembl and/or TrEMBL annotated TIS site; (2) iMet processed or iMet retaining counterparts of in vivo Nα-acetylated N termini; (3) N termini matching TIS previously identified by ribosome profiling; (4) N termini annotated as dbTIS in (a) prior Swiss-Prot release(s); (5) N termini for which the iMet processed and/or iMet retaining orthologous N-terminal peptide (HomoloGene) was identified as being (partially) Nα-acetylated in vivo.
Sequence Logo Analysis
All experimentally observed alternative N termini were aligned based on their translation start codon. The N-terminal peptides lacking the initiator methionine (iMet) were preceded with the iMet to rule out codon shifts in the sequence logo creation. Afterward, all peptides were mapped to their coding sequence (Perl scripting using the Ensembl API). Sequence logos were created based on the aligned transcript sequences (12 bp upstream and 9 bp downstream) using WebLogo 3 (http://weblogo.threeplusone.com, (61)). Sequence logos were plotted using both the residue probability and information content in bits as measure. Sequence logos were created for both the dTIS and corresponding dbTIS flanking regions. Also, an extra positive control to the dTIS sequence logos was generated based on 5000 randomly selected coding sequences corresponding to annotated translation initiation sites from CCDS (62) proteins were aligned for nucleotide context logo creation.
Ribosome Profiling and Genome-wide Visualization
Raw sequencing reads of the mESC ribosome profiling data (22) were downloaded from the Gene Expression Omnibus (data set GSE30839). All reads from the control (cycloheximide treated, also referred to as CHX treated, sample GSM765292) and harringtonine treated (sample GSM765295) were remapped using bowtie (v.0.12.7) on the mouse genome (assembly version 37) using the protocol described (63). All HEK293 cell line GTI-seq data (27) was downloaded from the NCBI Sequence Read Archive (accessions: SRX172392, SRX172361, SRX172360, SRX172315) and processed similarly.
Genome-wide visualization of the experimental data, in combination with publicly available data, was accomplished using an in-house developed genome browser (H2G2, http://h2g2.ugent.be/biobix.html). Information tracks containing the ribosome profile/GTI-seq mappings of the CHX treated samples (generating a translation profile all over the coding mRNA) and harringtonine/lactimidomycin treated samples (translation profile accumulation at the TIS) are available (see (22) for more information). Furthermore, an information track is constructed showing the predicted translation products, based on the TIS-predictions from Ingolia et al. (22) and Lee et al. (27) (for respectively the mESC and HEK293 cell line sample) and the UCSC transcript annotation. The genomic locations of the N-terminal peptides identified by means of N-terminal COFRADIC are also visualized in the H2G2 browser. Other visualization tracks include genomic information from a local Ensembl (64) instance NCBIM37.66, transcript tracks holding the annotated TIS within the UniProtKB and Ensembl database and a conservation track based on the phastCons (65–66) conservation scores among others. More information on how to use the H2G2 browser can be found in Supplemental File S1 and as indicated below.
Genomic Annotation of the Identified dbTIS and aTIS
All Swiss-Prot annotated N termini (dbTIS) and alternative N termini (aTIS) were mapped to their corresponding reference genome (GRCh37 for human and NCBIM37 for mouse) based on the UniProt-KB/Swiss-Prot accession number (or alternatively the Swiss-Prot gene name) and the N-terminal peptide sequence (PeptideMapper script based on the BioMart (67) and Ensembl (68) API, version 66). The genomic locations of the experimental aTIS and dbTIS locations were made available as a visualization track in theH2G2 genome browser (see above). Two projects are made available (named TIS Human and TIS Mouse) using a public login (see supplemental File S1 for more details). These projects hold several visualization tracks (see Fig 5 and 6 for examples): (A) an Ensembl gene track (B) an Ensembl transcript track (visible after mouse-click on the gene track), and tracks holding (C) the annotated TIS within the UniProtKB database split into Swiss-Prot and trEMBL (D) the annotated TIS within the Ensembl database (E) the reported aTIS and dbTIS locations (F) a conservation track based on the phastCons (65–66) conservation scores based on alignment of 45 and 29 vertebrate genomes respectively for human and mouse. Furthermore all genes where an aTIS or dbTIS has been identified by means of the N-terminal COFRADIC experiments are listed in “GeneDigest” reports within the H2G2 genome browser environment.
Fig. 5.
Representation of the single exon encoding mouse and human JUNB protein in the H2G2 genome browser. Several information tracks are presented. From top to bottom: (I) Ensembl Gene annotation, (II) Ensembl Transcript annotation, (III) ribosome profile data of control human HEK293t (A) and mESC (B) cell line sample showing profiles alongside the CDS, (IV) ribosome profile data of lactimidomycin (LTM) treated human HEK293t (A) and Harringtonine mESC (B) cell line sample showing profiles alongside the CDS, (V) Ribo-seq predicted TIS, (VI) the translation product prediction based on the two aforementioned tracks, (VII) TrEmbl annotated TIS, (VIII) Ensembl annotated TIS and (IX) dbTIS and (X) alternative TIS identified using N-terminal COFRADIC. Two zoomed figures (in black boxes) are also depicted, representing a more detailed view of the genomic region around the translation initiation site of the N-terminal truncated JUNB protein isoform in human (A) and mouse (B) identified using N-terminal COFRADIC, clearly demonstrating the use of an alternative initiation site (ATG) and accumulated Ribo-seq signals at this start site in human and mouse. Furthermore an alignment of the mouse and human N-terminal protein sequences of the transcription factor jun-B and d(b)TIS conservation is presented together with the (C) autoradiograph showing translation initiation of in vitro transcribed JUNB at the AUG start codons encoding M1 and M5.
Fig. 6.
Representation of the human UCHL1 protein in the H2G2 genome browser. Information tracks are presented as is Fig. 5. Furthermore an alignment of the N-terminal UCHL1 mouse and human protein sequences and d(b)TIS conservation (A) is presented together with the (B) autoradiograph showing translation initiation of in vitro transcribed ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1) at the AUG start codons encoding M1 and M6 and M12.
The aTIS “GeneDigest” report lists all genes wherefore an alternative start site is reported, whereas the dbTIS “GeneDigest” report lists all genes of which a Swiss-Prot database annotated TIS has been identified. A third “Genedigest” report lists extra translation start sites identified from the N-terminomics experiments searching a protein product database constructed based on ribosome profiling sequence information (22) (see above). Further, experimental Ribo-seq data (22) are also presented as custom tracks, allowing manual inspection of co-occurrence of N-terminal COFRADIC and Ribo-seq experimental evidence.
Conservation Analysis
To assess the evolutionary conservation potential of the identified d(b)TIS and their flanking sequences as compared with 5000 randomly chosen, BioMart (67) annotated CCDS translation initiation sites, their orthologous positions in various vertebrate genomes were extracted using phastCons (65–66) and scored in a multiple sequence alignment. The phastCons program computes conservation scores based on a phylo-HMM, a type of probabilistic model that describes both the process of DNA substitution at each site in a genome and the way this process changes from one site to the next. The value plotted at each site is the posterior probability that the corresponding alignment column was “generated” by the conserved state of the phylo-HMM.
TargetP Analysis
To categorize the subcellular location of the proteins translated from their annotated versus their alternative N termini, a targetP prediction (v1.1b, (69)) was performed. The location assignment is based on a predicted N-terminal presequence: a mitochondrial targeting peptide, or a secretory pathway signal peptide.
Generation of TIS Mutagenized CDS and in Vitro Translation Assays
pOTB7 vectors (RZPD Imagenes, Germany) encoding aminoacyl tRNA synthase complex-interacting multifunctional protein 2 (AIMP2), inhibitor of kappa light polypeptide gene enhancer in B-cells kinase gamma (NEMO), Zinc finger protein 296 (ZN296), splicing factor 3a subunit 3 (SF3A3), cytoplasmic aspartate-tRNA ligase (SYDC), ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1), transcription factor jun-B (JUNB), cytokine receptor-like factor 3 (CRLF3), Nucleosome assembly protein 1-like 1 (NP1L1), and pyridoxamine 5′-phosphate oxidase (PNPO) served as templates for site-directed PCR-mutagenesis (QuickChange, Stratagene, La Jolla, CA) according to the manufacturer's instructions using the primer pairs indicated in supplemental Table S2. The correctness of all (mutant) cDNA sequences generated was confirmed by DNA-sequencing.
The mutagenized constructs were used as templates for in vitro coupled transcription/translation in a rabbit reticulocyte lysate system according to the manufacturer's instructions (IVTT; Promega, Madison, WI) to generate [35S]methionine labeled protein products. 5 μl of the translate reaction was diluted 10-fold in 10 mm Tris pH 8.0. NuPAGE® LDS Sample Buffer (Invitrogen) was added and the samples heated for 10 min at 70 °C. Samples were separated on 4–12% NuPAGE® Bis-Tris gradient gels (1.0 mm x 12 well) (Invitrogen) using MOPS Buffer. Subsequently, proteins were transferred onto a PVDF membrane, air-dried and exposed to a film suitable for radiographic detection (ECL Hyperfilms, Amersham Biosciences, Buckinghamshire, UK). Radiolabeled proteins were visualized by radiography.
RESULTS
Mapping of the Translation Initiation Landscape in Human and Mice Using N-terminomics Reveals Numerous Alternative Translation Initiation Events
In this study, a proteome-wide map of the translation initiation landscape in human and mouse was created by mass spectrometry assisted analysis of protein N termini isolated by N-terminal COFRADIC (70). A TIS compilation was made from previously generated N-terminal proteomics data ((50, 55–57) and unpublished data) derived from the proteomes of the human HeLa, HCT116, A-431, THP-1, K-562, and Jurkat cell lines in addition to primary B-cells as well as the mouse cell lines Mf4/4 and YAC-1 next to primary dendritic mouse cells. Here, prior to tryptic digestion, all primary amines, and thus free protein N termini, were mass tagged by acetylation using nonnatural, stable isotope encoded groups such as trideutero-acetate. In this way, in vivo Nt-acetylated and in vivo free N termini can be distinguished and the degree of Nt-acetylation determined (71). After tryptic digestion, all protein N termini will thus be blocked, whereas all other internal peptides will have a newly generated primary α-amine. Subsequently, N-terminal peptides are enriched for by means of strong cation exchange (SCX) step at low pH and further segregated from remaining internal peptides using a diagonal chromatography strategy. Selected protein N termini are subsequently identified following LC-MS/MS analysis (72). Identified protein N termini were grouped by their TIS context. First, protein N termini with a Swiss-Prot database annotated protein start position (i.e. N termini starting at protein position one or two in the protein sequence) are referred to as database annotated TIS or dbTIS. Overall, 2879 human and 1771 mouse dbTIS-indicative N termini originating from 2723 and 1708 unique Swiss-Prot protein entries were identified (supplemental Table S1).
Second, based on the cotranslational nature of N-terminal acetylation of protein N termini (48) by the NATs, the near universal requirement of a Met-encoding initiator codon (iMet) and the cotranslational processing of iMet by methionine aminopeptidases (MetAPs), all in vivo free and/or Nt-acetylated peptides with start positions downstream the database annotated TIS were grouped. In this way, 1231 human (1060 proteins) and 465 mouse (418 proteins) N termini hinted to protein N termini originating from the usage of in-frame, downstream TIS (dTIS), thus giving rise to N-terminal truncated protein isoforms. The N termini hinting to dTIS were further subdivided into two subcategories (See also Experimental procedures); the high confident dTIS encompassing all (partially) in vivo Nt-acetylated peptides among others. In vivo unmodified dTIS compliant with the rules of iMet-processing and Nt-acetylation (the latter for example considering that, without exception, (X)-Pro-starting N termini are unmodified (73)) were withheld as low confident dTIS and this only when their protein start position did not overlap with any proteolytic cleavage event reported in public repositories (74–77) (i.e. protein signal processing sites or reported proteolytic cleavage sites after or before a Met residue). Finally, and whenever Ribo-seq or ortholog mapping hinted to translation initiation events, low confident dTIS were recataloged as high confident dTIS (see below and supplemental Table S1).
For dbTIS, the discrepancy between the numbers of identified protein N termini and the actual proteins is only because of the observed incompleteness of iMet-processing (i.e. cases where both the iMet processed and unprocessed N termini were identified) and thus heterogeneity of the N-terminal protein ends, whereas in the case of dTIS, multiple dTIS were observed for several proteins.
Further, the identified N termini were grouped by mapping them to the first or a subsequent exon, with the former category hinting to alternative translation events by leaky scanning or re-initiation, whereas the latter, besides representing putative dTIS, might point to TIS originating from alternative splicing (Table I and supplemental Table S1). Overall 1220 human dTIS (out of the 1231 Swiss-Prot N termini identified) and all 465 mouse dTIS N termini could be mapped onto their corresponding reference genome and their confidence level is given in Table I and supplemental Table S1. Meta data related to the d(b)TIS identifications are made available as visualization tracks in the H2G2 genome browser (http://h2g2.ugent.be/biobix.html, see also supplementary information).
Table I. All experimentally observed alternative translation initiation sites (aTIS: 1231 human, 465 mouse) were mapped onto their corresponding reference genomes (Ensembl human GRCh37 and mouse NCBIM37). The dTIS locations were detected throughout several mass spectrometry analyses and a compilation was extracted from the in-house ms-lims system (109) obtained from previously generated N-terminal proteomics data ((50, 55–57) and previously unpublished data) (supplemental Table S1). The overlap with annotated Ensembl, Swiss-Prot or TrEMBL annotated TIS is also provided. For the Ensembl mapping an extra subdivision was done based on TIS location in the first (Exon1) or the consecutive exons (Exon>1). Also, a comparison was made with the TIS identified within two ribosome profiling studies on HEK293 and mESC cell lines (18,19). See the “selection of N-termini” paragraph within the material and methods section for more explanation on the subdivision based on confidence level (either H or L). “(+ meta)” indicates that available isoform, transcript, Ribo-seq and/or orthologues dTIS metadata is available for dTIS originally assigned as low confidence (See also supplemental Table S1).
| Human dTIS |
Mouse dTIS |
|||||
|---|---|---|---|---|---|---|
| Confidence level |
Total | Confidence level |
Total | |||
| H | L | H | L | |||
| Identified peptides | 858 | 373 | 1231 | 274 | 191 | 465 |
| Mapped peptides | 850 | 370 | 1220 | |||
| Ensembl TIS | ||||||
| Exon1 | 41 | 4 | 45 | 25 | 4 | 29 |
| Exon>1 | 146 | 32 | 178 | 39 | 3 | 42 |
| Subtotal | 187 | 36 | 223 | 64 | 7 | 71 |
| No Ensembl TIS | ||||||
| Exon1 | 301 | 47 | 348 | 128 | 48 | 176 |
| Exon>1 | 362 | 287 | 649 | 82 | 136 | 218 |
| Subtotal | 663 | 334 | 997 | 210 | 184 | 394 |
| Swiss-Prot isoform TIS | 17 | 1 | 18 | 36 | 1 | 37 |
| No Swiss-Prot isoform TIS | 833 | 369 | 1202 | 238 | 190 | 428 |
| TrEMBL TIS | 159 | 32 | 191 | 43 | 6 | 49 |
| No TrEMBL TIS | 691 | 338 | 1029 | 213 | 185 | 416 |
| Ribo-seq TIS | 96 | 9 | 105 | 57 | 12 | 69 |
| No Ribo-seq TIS | 754 | 361 | 1115 | 217 | 179 | 396 |
| Identified peptides(+meta) | 900 | 331 | 1231 | 298 | 167 | 465 |
| Mapped peptides(+meta) | 892 | 328 | 1220 | |||
Of the dTIS N termini identified, 18% (n = 223) and 15% (n = 71) of the Swiss-Prot nonannotated human and mouse TIS mapped to Swiss-Prot isoform entries and/or indicative transcripts in TrEMBL and/or Ensembl, validating our selection strategy for identifying dTIS, as these have been experimentally proven to give rise to N-terminally truncated protein isoforms (supplemental Table S1). Overall, these numbers are indicative for the fact that our data set is of high quality and thus holds numerous hitherto unreported dTIS sites, here discovered at the level of the proteome.
TIS Sequence Context Analyses
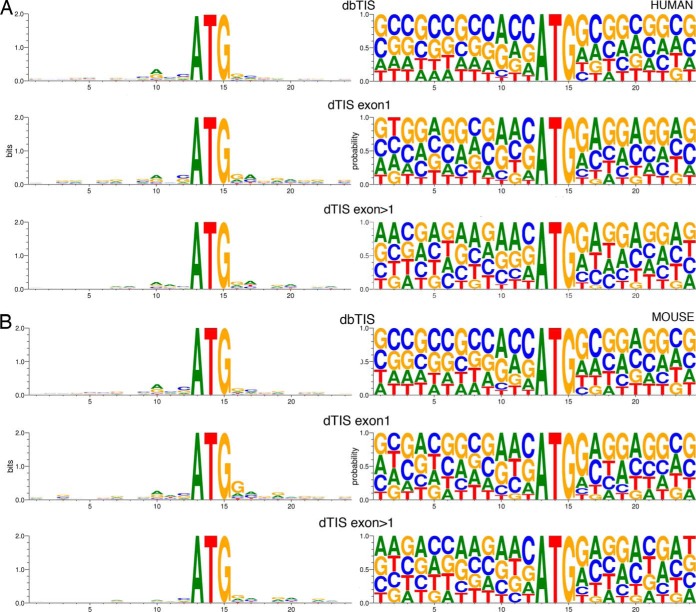
A survey of the sequence context flanking the dbTIS and dTIS of the Exon1 and Exon>1 categories using WebLogo (78) revealed a preference of the most crucial Kozak context elements being a purine at position −3 and guanine at position +4 (79) in the Kozak motif gcc[A/G]ccAUGG(not U) with the dbTIS and dTISexon1 equally well conserved, followed by the dTISexon>1 category. Only d(b)TIS locations were taken into account where the complete flanking region does not span any splice junction. For the dbTIS, additionally the higher GC content in the flanking nucleotide context becomes noticeable (80–81) (see Fig. 1A and 1B).
Fig. 1.
A, Homo sapiens TIS WebLogos. The flanking sequences (12 bases upstream, 9 bases downstream) of the corresponding dbTIS for which a dTIS has been identified, the experimentally observed dTIS, located in exon 1 (n = 374), and the experimentally observed dTIS, located in subsequent exons (n = 791) are used to create WebLogos. B, Mus musculus TIS WebLogos. The flanking sequences (12 bases upstream, 9 bases downstream) of the corresponding dbTIS for which a dTIS has been identified, the experimentally observed dTIS, located in exon 1 (n = 197), and the experimentally observed dTIS, located in subsequent exons (n = 251) are used to create WebLogos. Both the probability and bits values are plotted. The discrepancy between the numbers given above and the numbers in Table I are because splice site spanning flanking sequences were not used to create the WebLogos.
A detailed analysis of the human Swiss-Prot dbTIS for which we report an alternative start site located in the first exon (i.e. the dTISexon1 category) additionally revealed an increased occurrence of suboptimal start codon contexts (with a pyrimidine in position −3 upstream of AUG instead of purine (82)). As compared with the start codon contexts of all identified dbTIS, an increased suboptimal versus optimal measure of 35.2% versus 19.5% is observed (as deduced from input data used in Fig. 1 and S1). According to the leaky scanning model (83), the 40S ribosomal subunits can miss an AUG codon in a suboptimal context and initiate translation at downstream AUG(s), which is in corroboration with the data obtained from GTI-seq data, showing that the strongest Kozak consensus sequence was observed in the gene group with no detectable dTIS but with dbTIS initiation, whereas this context was largely absent in the group of genes lacking a detectable translation initiation at dbTIS (33). The downstream flanking sites of these downstream start sites in both the dTISexon1 and dTISexon>1 categories were further inspected using the AUG_Hairpin software, enabling the prediction of downstream secondary structure influencing translation start site recognition (82, 84–86). Following the strategy of Kochetov et al. only those dTIS that show a stable stem-loop structure (Etot < −20 kcal/mol) located between 13 and 19 nucleotides downstream from the start site were retained. Average energies of eligible stem-loop structures (Etot) were −32.2 kcal/mol and −32.6 kcal/mol for the dTIS with suboptimal and optimal start codon contexts respectively (also the distributions of Etot values proved not to be significantly different according to a Kolmogorov-Smirnov two-sample test). Overall, the presence of the Kozak sequence context in all categories is further indicative for real TIS events.
Conservation Analysis
To assess the possibility of evolutionary conservation of the identified dTIS and their flanking sequences as compared with their corresponding dbTIS, the orthologous positions in various vertebrate genomes were extracted using phastCons (65–66) and scored in a multiple sequence alignment, thereby generating a metagenic conservation plot (Fig. 2). Also, an analysis was made between the identified dTIS and a set of 5000 randomly chosen, BioMart (67) annotated complete CDS (CCDS) translation initiation sites (serving as a proxy for the global dbTIS landscape, supplemental Fig. S2). Only d(b)TIS locations were taken into account where the complete flanking region does not span any splice junction. In general, the phastCons score (between 0 and 1) gives a probability that each nucleotide belongs to a conserved element (see Material and Methods for detailed explanation). Overall, the human and mouse conservation plot indicated that the dTIS are highly conserved, with a mean conservation score of 0.97 (± 0.002, 95% confidence interval) and 0.97 (± 0.005) for respectively the Exon1 and Exon>1 groups compared with the dbTIS with a mean conservation score of 0.96 (± 0.01) and are thus indicative for the fact that the dTIS translation start sites are very well conserved within eukaryotic genomes in analogy to what was previously reported by Bazykin et al. (87) using in silico predictive analyses and using in vivo GTI-seq experiments (33).
Fig. 2.
Conservation plots for the flanking exonic regions of human and mouse TIS. Conservation plots for the flanking (12 bp upstream, 9 bp downstream) exonic regions of human (left pane) and mouse d(b)TIS (right pane) are plotted. The conservation measure averages the phastCons score at every position of all flanking sequences within the subgroups (corresponding dbTIS, dTIS located in exon 1 (Exon1 subgroup), and dTIS located in subsequent exons (Exon>1 subgroup)) after alignment based on their translation start site. For all flanking positions, the mean is provided together with its 95% confidence interval. The upper panels show the conservation plot of the flanking region of corresponding dbTIS for which a dTIS has been identified. The middle and lower panel are respectively based on flanking regions of dTIS of the Exon1 (n = 374 for human, n = 197 for mouse) and Exon>1 (n = 791 for human, n = 251 for mouse) subgroups. Only d(b)TIS locations were taken into account where the complete flanking region does not span any splice junction.
Further, the conservation scores of the dTIS flanking regions of both the Exon1 and Exon>1 groups are high, ranging from 0.9 to 1. Here, next to the translation start codon, other Kozak hallmarks such as the guanine at position +4 and purine at position −3 are well conserved. Also notable in the dTIS conservation plots—and expected given the higher coding potential of the first two nucleotides—is the slightly higher conservation of the first two nucleotides of the coding triplets in the translated sequence (88). This feature is most pronounced in the human dTISexon>1 plot. As opposed to the dTIS plots, the flanking 5′ upstream sequences in the dbTIS plots score significantly lower as these presumably contain untranslated sequence (UTR) in contrast to the 5′ upstream region of the dTIS that contain translated sequence encoding for N-terminal protein extensions. No significant differences are obvious between the dTIS conservation plots of the Exon1 and Exon>1 groups.
Statistical testing was performed to assess the sequence conservation surrounding the Kozak motif and to increase confidence that the identified sites (dTIS) are genuine translation initiation sites. For that purpose we compiled a data set of decoy sites meeting the following criteria: (I) Consensus Coding Sequences (CCDS) were scanned for downstream Kozak sequence motifs [A/G]ccAUGG, (II) the identified Kozak sequence motif sites that overlap with dTIS identified in the N-terminal COFRADIC datasets reported in this study were discarded, (III) the ones showing an overlap with dTIS identified within the ribosome profiling experiments re-analyzed in this study (human (33) and mouse (22)) were also discarded. This group of decoy sites was compared with the different categories of TIS described in the study: database annotated TIS (dbTIS), downstream TIS located in exon1 (dTISexon1) and downstream TIS located in further downstream exons (dTISexon>1). The PhastCons conservation scores at positions (-3,+1,+2,+3,+4), the most crucial Kozak context positions, were averaged for further calculation. A low p value (4.701e−12 and <2.2e−16 for respectively the human and mouse data sets) in a Welsh one-way ANOVA is indicative for a difference among the four TIS groups (after testing for heteroscedasticity using the Levene's test). In order to determine which particular group of TIS deviates the most, a Tukey's Honestly Significant Difference (Tukey-HSD) post-hoc test (accounting for heteroscedasticity by using a heteroscedastic consistent covariance estimation) was performed showing a clear difference in the sequence conservation surrounding the Kozak motif between the decoy group and the three other sets (dbTis, dTISexon1 and dTISexon>1, p value < 0.001) at a 95% confidence level.
Finally, and to further analyze the degree of conservation of dTIS between our human and mouse data sets, the experimentally identified mouse and human dTIS were compared. In total, of 200 orthologous dTIS pairs, both the human and mouse N termini could be identified (i.e. for 43% of all mouse dTIS identified the human orthologous N termini could be identified).
Of these, 29 human and 31 mouse dTIS were originally classified as low confident dTIS. Based on the MS/MS-based evidence of Nt-acetylation of these orthologous N termini, three human and nine dTIS could now be re-cataloged under the reliability class 1 (supplemental Table S1).
TargetP Analysis
To have a first approximation of the functional impact of alternative TIS usage, a TargetP analysis was performed predicting the subcellular location of the full-length proteins (i.e. proteins translated starting from their dbTIS) versus their N-terminally truncated counterparts identified (dTISexon1 and dTISexon>1) (89). Fig. 3 (upper and lower pane for respectively human and mouse) show that although only a small percentage of the dbTIS protein products is predicted to contain a mitochondrial targeting or signal peptide (i.e. most likely an underrepresentation, because in most cases signal or transit peptide maturation has occurred), a noticeable decrease of secreted or mitochondrial targeted proteins can be observed when assessing their N-terminally truncated counterparts (see Fig. 3 pie charts, Chi-squared test of Independency, p value < 2.2e−16 for both the human and mouse data sets). The bar plots within Fig. 3 give a more detailed view, making an extra subdivision based on (I) the reliability of the TargetP prediction (class 1 to 5) and (II) whether the dTIS is localized in exon1 or in an exon downstream exon1 (exon>1). The more detailed bar plots also show a significant drop for the mitochondrial and secretory pathway localization categories (“M” and “S”) independent of the reliability classes (1–5) in both the exon1 and exon>1 groups (green versus blue bars).
Fig. 3.
TargetP analysis of the protein products generated by dbTIS versus dTIS usage. TargetP predicts both N-terminal mitochondrial targeting peptide (mTP) and signal peptides (SP) processing, respectively reflecting mitochondrial and secretory pathway localization. Both human (upper pane) and mouse (lower pane) are plotted. The pie charts depict the overall localization patterns of the dTIS and dbTIS translation products. The more detailed bar charts make an extra subdivision based on (I) the reliability of the TargetP prediction (class 1 to 5, where 1 indicates the strongest prediction) and (II) whether the dTIS is localized in exon1 or in an exon downstream exon1 (exon>1). The green and blue bars respectively correspond to N-terminal isoforms raised upon dbTIS and dTIS usage, dark and clear bars represent the Exon1 and Exon>1 group respectively. The x axis shows the predicted localizations (“M” stands for mitochondrial, “S” for secretory pathway) and the reliability of that prediction (class 1 to 5). The rightmost bars depict the combination of all reliability classes (“All M” and “All S”). The y axis corresponds to the total number of TIS events falling within the groups depicted in the x axis.
Overall, the TargetP output strengthens the idea that dTIS usage has an impact on protein subcellular localization (35–36, 90), which was also hypothesized and computationally investigated by Cai et al. (91) and in fact proven for a variety of N-terminal protein isoforms generated by means of alternative translation initiation.
Ribosome Profiling Data Provide Independent Experimental Support for N-terminomics Data
Interestingly, of the here identified TIS, complementary Ribo-seq TIS profiling data are available for 861 of the 1755 transcript-matching mouse dbTIS (49%), 69 of the 465 mouse dTIS (15%), 1150 of the 2841 human dbTIS (40%), and 105 of the 1220 human dTIS (9%) (supplemental Table S1), thereby providing evidence that these represent genuine translation initiation sites in mouse and human transcripts (22), and thus that these N termini are representative for N-terminal protein variants.
As such, the experimental evidence obtained by ribosome profiling-assisted TIS identification categorizes 47 extra mouse dTIS and 66 extra human dTIS to reliability class 1, thereby increasing the percentage of validated dTIS to 27% (n = 125) and 22% (n = 272) in mouse and human respectively (supplemental Table S1).
Overall, and when taking into account the available isoform, transcript, Ribo-seq and orthologous dTIS metadata, 24 extra mouse dTIS, and 42 extra human dTIS originally assigned as low confidence, are now classified as highly likely genuine dTIS, respectively summing up to 73% (n = 900) and 64% (n = 298) of all identified human and mouse dTIS having data that support their translation initiation potency (supplemental Table S1).
Because the translatome (i.e. the ORF delineation and the translation initiation landscape) can be specifically delineated using ribosome profiling data, usage of this type of data does not necessitate translation into its three reading frames, hence decreasing the search space tremendously. Alongside, noncanonical codons serving as alternate initiation codons are depicted. These near cognate translation initiation codons can either be decoded as the expected initiator methionine residue or alternatively to their coding-matching amino acid as for example leucine-decoded CUG starts of translation initiations have been reported (92–93). For these reasons, customized sample-oriented and ribosome profiling derived protein databases were created which served as custom-made reference data sets for the proteomes of study (59).
In the mouse proteomes, besides the additional full-length protein N termini identified (i.e. N termini not enclosed in the Swiss-Prot database), 17 N termini indicating N-terminal protein extensions were identified (supplemental Table S1). Four N-terminal extensions were generated upon translation initiation at an AUG codon, in addition, 13 were produced by translation initiation at near cognate start codons; being GUG (6 N termini), CUG (4), and ACG (3) normally encoding for respectively Val, Leu, and Thr, but decoded to Met as evident from the iMet-retaining N termini identified. Besides, the N termini of an uORF and a dTIS protein product could be identified using respectively GUG and CUG as start codon.
In human, 17, 4, and 2 N termini respectively hinted to N-terminal protein extensions, N-terminally truncated and overlapping uORF protein products, 22 of which were generated upon translation initiation at a near-cognate start codon (supplemental Table S1). Besides, these database searches led to the identification of some additional dbTIS, not contained in the Swiss-Prot database (supplemental Table S1).
TIS Mutagenesis Analyses Reveal that N-terminal Protein Isoforms of the Class dTISexon1 are Generated by Means of Leaky Ribosomal Scanning
To further verify whether some of the alternative TIS products identified are raised by alternative translation initiation, in vitro translation studies of (mutagenized) dTIS holding coding sequences (CDS) flanked by (a part of) their presumed 5′UTR were performed using coupled in vitro transcription/translation assays. In general, mutation of AUG to CUG typically abolished or greatly diminished translation initiation at these sites. In all cases distinct protein bands corresponding to the short N-terminal protein isoform(s) (i.e. dTIS products), identified by proteomics means, and the database annotated variant could be determined (Fig. 4). Further, because the mutation of the canonical initiation site affected the production of the truncated isoform(s) and vice versa (i.e. some truncated N-terminal isoforms were only detected when the dbTIS were mutagenized), our results strongly indicate that the dTIS products are produced by alternative translation initiation via leaky ribosomal scanning at internal translation start sites. Besides, in some cases a deviation from the 5′ polarity of scanning could be observed for closely spaced AUG codons (up to 16–19 nt). As a result of the proposed reverse directionality of scanning (3′ to 5′), a lower initiation frequency at a 5′ proximal AUG could be observed in the presence of (a) nearby downstream AUG(s), suggestive of downstream nucleotides inferring a restricted relaxation to the forward directionality of scanning of the proximal AUG (94).
Fig. 4.
In vitro translation of TIS-mutagenized constructs reveal the existence of the by proteomics identified N-terminal protein variants. Wild type and d(b)TIS mutagenized pOTB7 constructs encoding N-terminal variants of aminoacyl tRNA synthase complex-interacting multifunctional protein 2 (AIMP2), inhibitor of kappa light polypeptide gene enhancer in B-cells kinase gamma (NEMO), Zinc finger protein 296 (ZN296), splicing factor 3a subunit 3 (SF3A3), cytoplasmic aspartate-tRNA ligase (SYDC), cytokine receptor-like factor 3 (CRLF3), Nucleosome assembly protein 1-like 1 (NP1L1), and pyridoxamine 5′-phosphate oxidase (PNPO) were in vitro transcribed and translated. Following SDS-PAGE and electroblotting, radiolabeled proteins were visualized by radiography. Assignments of the precursor band corresponding to the database annotated protein sequences (black arrowhead) and protein products raised upon dTIS usage (orange and dashed arrowheads) were verified by mutating their respective initiator methionines. A gray arrow points to translation initiation events at dTIS which were not identified by proteomics means. An asterisk is indicative of an unspecific protein band produced in the control in vitro transcription and translation reaction (i.e. a reaction without input DNA). In each case the theoretical molecular weights of the identified N-terminal protein variants are indicated.
DISCUSSION
The most acknowledged mechanism of protein diversification in mammalian genomes is alternative splicing, where different mRNAs are derived from the same nascent transcript. Only recently, alternative translation initiation from a single mature transcript was recognized as an important and wide-spread mechanism of protein diversification, further highlighting the importance of gene functionality (22, 33).
As previously postulated, targeted analysis of protein N termini is ideally suited to study N-terminal protein isoform diversity (35).
In this study, we report on more than 1700 alternative translation initiation events in mouse and human cell lines by applying stringent rules for mass spectrometric based identifications of N-terminal peptides. Besides, our detailed understanding of the specificity of the Nt-acetyltransferases and cotranslational processes in general assists in judging if such peptides indeed report protein translation events and thereby allow for the functional (re-)annotation of genomes. For a significant fraction of the here reported TIS, available metadata from transcripts, ribosome profiling, TIS sequence context and conservation analyses served as evidence that our N-terminal selection provides a very powerful strategy to map the TIS landscape in higher eukaryotes. In addition, and as is the case for the high mobility group proteins HMGB1, HMGB2, and HMGB3, next to the orthologs matching dTIS sites, corresponding dTIS sites in all three homologs could be identified (supplemental Table S1). These observations are further strengthened by the fact that previously reported dTIS products of the Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) (95), Glucocorticoid receptor (GCR) (96), Insulin-degrading enzyme (97), Regulator of G-protein signaling 2 (98), and the BAG family molecular chaperone regulator 1 (99)—of which the N-terminal isoform expression was shown to display a stage- and site-specific expression profile during mouse development—were also identified in this study (supplemental Table S1).
Ribo-seq and GTI-seq in mammalian cells revealed that only half of the TIS codons made use of AUG as the translation initiation codon. However, in the study of Lee et al. (33), TIS codon usage was shown to be distinct when residing in the presumed 5′UTR (uTIS) as opposed to the annotated CDS. When outside the dbTIS/dTIS reading frame, uTIS are mostly associated with short ORFs and were mostly non-AUG codons, whereas dTIS, typically encoding for N-terminal truncated protein variants predominantly made use of AUG codons, an observation which is in line with our data and the fact that only 37 protein products were raised upon translation initiation at near-cognate start codons, typically giving rise to N-terminal protein extensions, were identified in our mouse and human data sets based on available Ribo-seq data sets (22, 33). Further, a recent computational analyses of ribosome profiling data calculating the efficiencies of individual translation initiation sites, revealed that despite the high frequency of non-AUG translation initiation sites identified by means of ribosome profiling, the probability of initiation at non-AUG codons was found to be considerably lower than at AUG codons (data presented by Pavel Baranov at the EMBO Conference Series “Protein Synthesis and Translational Control,” Heidelberg, Germany 2013 (100)), likely explaining their general underrepresentation in the N-terminomics data sets here presented.
In addition to translation re-initiation for alternative translation initiation, GTI-seq demonstrated that leaky scanning was the major contributing mechanism leading to TIS selection because the strongest Kozak consensus sequences were observed in the gene group with dbTIS selection but no detectable dTIS, whereas dTIS selection was observed when a weak or no consensus sequence context of the dbTIS was present, enabling for an estimation of the leakiness of the first AUG codon and again confirming our proteome data.
Our TargetP analyses as well as other multiple lines of evidence indicate that alternative translation initiation can give rise to iso-functional though localization-specific N-terminal protein variants making translation initiation a very attractive mechanism of regulating protein localization as previously for example reported for the p43 Component of the Multisynthetase Complex (37) where a mitochondrial targeting sequence is lost when translation initiation proceeds via a dTIS, whereas in contrast, translation initiation at a dTIS in Flap endonuclease 1 (FEN-1) exposes a cryptic mitochondrial targeting signal (38), two cases contributing to the increased complexity of the mitochondrial proteome (39).
Although translation initiation at dTIS found in close proximity to dbTIS are more likely to yield isofunctional and localization nondistinct N-terminal isoforms, thereby providing a potential fail-safe mechanism for translation initiation to occur, it is noteworthy that the N-terminal identity of a protein is critically important in determining protein stability. More specifically, the N-end rule relates the regulation of the in vivo half-life of a protein to the identity of (the N-terminal modification of) its N-terminal residue. Therefore, the loss of even a single N-terminal amino acid or its modification can significantly impact protein stability (101), influence protein localization (102) and protein complex formation (103) among others. Finally, our TargetP analyses shows a clearly noticeable change in localization in both exon1 and exon>1 groups for both the human and mouse data sets (Fig. 3) indicative that the findings of altered subcellular localization/functional diversification also applies to the broader set of alternative translation events identified in this study.
Besides steering protein localization and protein stability, TIS selection can also regulate protein expression levels. For example, hypo- or hypermorphic point mutations introducing premature translation termination codons in the first exon can result in the production of truncated protein variants through translation initiation at in-frame methionines downstream the nonsense mutation (104–106). Two such examples for which various dTIS sites were identified in this study include the nuclear factor kappa B (NF-κB) essential modulator (IKKγ/NEMO) and the NF-κB inhibitor IκBα (104–106) (Fig. 4 and supplemental Table S1). Premature translation termination codons in these genes—although leading to the residual production of a truncated variant sufficient for the nonlethality observed during development—have been shown to underlie specific cases of the genetic disorder anhydrotic ectodermal dysplasia with immune deficiency. The (presumed) dTIS of both reported truncated protein variants have here been identified as being in vivo Nt-acetylated (Ac-M38LHLPSEQGAPETLQR (NEMO) and Ac-M37KDEEYEQMVKELQEIR (IκBα)), indicative for the fact that, as demonstrated to be the case for wild type NEMO transfected cells, a (limited) translation initiation of IκBα at these alternative methionines can also occur under normal cellular conditions. The dTIS product of IκBα lacks the two amino-terminal IκB kinase (IKK) phosphorylation sites known to be essential for targeting IκBα for proteasomal degradation, and as a result the degradation-resistant variant acts as a dominant negative regulator of NF-κB activity. The mutation in IKKγ/NEMO, the scaffolding subunit of the IKK complex, gives rise to a truncated but functional variant that is produced in limited, insufficient amounts for the development of protective immune responses. In vitro transcription/translation assays independently confirmed that the 44 kDa isoform of NEMO is the product of alternative translation initiation at the internal Met38. Here, close to the canonical start codon, other start codons are located downstream of the database annotated iMet1 (i.e. GxxAUGG and GxxAUGC), corresponding to the (surrounding) nucleotide motifs of methionines 13 and 38. The likelihoods of these AUG codons to act as translation initiation sites were estimated 0.29 and 0.27 versus 0.54 for the first AUG codon (TxxAUGA) (translation initiation prediction at http://atgpr.dbcls.jp). As such, various mutagenized constructs were made (Fig. 4) in which the presumed initiator Met1, the internal Met13, and/or Met38 were mutagenized to monitor translation initiation at these sites. Expression from the coding sequence alone resulted in the production of two clearly distinct protein forms (Fig. 4, WT sample), of which the expression of shorter isoform was lower as compared with its full-length counterpart, indicative for leaky ribosome scanning. Further, mutagenesis of Met13 also resulted in two distinct bands of which the higher MW band runs ∼1 kDa lower as compared the highest MW band of the WT construct and thus probably indicative for the fact that the higher MW band observed encompasses the products of translation at Met1 and Met13, an observation which is confirmed when Met 13 or alternatively Met 38 is mutated. The fact that translation initiation may occur at Met 1 and Met 13 is further supported by the observation that cellular expression of the WT and M38A mutant resulted in a somewhat smeared out precursor band (106) and that the TrEMBL and Ensembl databases hold preliminary entries of this Met13 initiated protein variant. The combined Met 13/38 mutant seems to express the 44 kDa form besides residual translation initiation at the first noncanonical mutant CUG codon. Although, in vitro translation from the triple AUG to CUG mutant construct is significantly impaired, residual translation initiation (mainly at the near-cognate start codon decoded to Met38) can still be observed. Overall, we observed translation at three different AUG codons in NEMO, of which the translation product starting with Met38 was identified using N-terminal proteomics in the proteomes of K562, THP-1, and B-cells.
Further, ribosome profiling data where the utmost 5′ AUG triplet resides in a suboptimal context (absence of a purine at −3 and/or G at +4) suggested that more than one quarter of the human transcripts showed clear evidence of downstream translation initiation and thus could display a bi- or multicistronic behavior (33), meaning that these mRNAs could produce more than one polypeptide through leaky scanning. Many of these hypothetical cases would be expected to involve the production of small peptides or N-terminal truncated protein isoforms that have routinely been excluded from database sequence annotations.
Despite the multitude of alternative TIS here identified, linking over 30% of the protein N termini to alternative translation initiation, for the majority of them their spectral counts (supplemental Table S1) hint to a general lower translation efficiency and thus likely lower concentration of protein products expressed from such secondary initiation codons. Nonetheless, potent functions of such possibly lower abundance dTIS protein products have been demonstrated as in the case of the mitogenic osteogenic growth peptide (OGP) translated by leaky scanning from mammalian histone H4 mRNA (107). Strikingly however, when TIS are relatively closely spaced (i.e. resulting in N-terminal protein isoforms differing less than about six amino acids), spectral counts and our in vitro translation results hint to a more balanced expression, likely linking this to a proximity effect previously shown to modify the strict sequential constraint of regular leaky scanning into a more competitive feature (94). In this respect, it is important to note that such protein products can easily be overlooked by conventional detection technologies such as Western blotting despite their potent expression, as here shown to be the case by mutagenesis studies enabling visualization of the N-terminal AIMP2, JUNB, UCHL1 and CRLF3 isoforms generated by translation initiation at closely spaced AUG start codons (Fig. 4–6).
Further, the bioinformatics-assisted integration of positional proteomics and available ribosome profiling data enabled for a more sensitive and comprehensive protein discovery, thereby enabling a global (re-)annotation of the translation initiation landscape (31, 59). Interestingly, the high overlap of alternative translation products identified in this comprehensive study with those from a previous positional proteomics study that focused on the TIS-landscape in mouse embryonic stem (mESC) cells, further hints to their functional importance and conservation. In contrast however to the majority of translation products linked to alternative translation initiation as observed in ribosome profiling, the repertoire reported in this study is mainly confined to translation products raised upon downstream AUG besides some upstream noncognate codon usage, meaning that the products of uORF and out-of-frame translation generally remain undetected. Although the sensitivity of ribosome profiling to detect TIS sites and translation products remains unprecedented, the bioinformatics-oriented approaches used to assign TIS in ribosome profiling studies in some cases appear ineffective (e.g. the dTIS in JUNB here identified, clearly provided a discrete LTM and Harringtonin signal in the human and mouse Ribo-seq data sets (22, 33), though remained undetected by the strict training algorithm used in the original Ingolia et al. study (Fig. 5)). This, as well as the occurrence of cotranslational protein modification events and the recent finding that mRNA ribosome occupancy does not necessarily hints to effective translation (108), necessitate the need for proteomics endeavors to identify the (mature) translation products. As such, we foresee that the complementary use of proteomics approaches and ribosome profiling will further assist in the comprehensive cataloguing of TIS ultimately leading to detectable and functional translation products.
Supplementary Material
Acknowledgments
We thank Prof. Kris Gevaert for critical reading of the manuscript, A.V. Kochetov for the batch analysis of the AUG_Hairpin software and Kimberly Demeyer for technical assistance.
Footnotes
Author contributions: P.V.D. and G.M. designed research; P.V., D.G., and G.M. performed research; P.V. and G.M. analyzed data; P.V. wrote the paper; W.V.C. supervised research.
* P.V.D. and G.M. are Postdoctoral Fellows of the Research Foundation - Flanders (FWO-Vlaanderen). D.G. is supported by a Ph.D. grant of the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen). P.V.D. acknowledges support from the Research Foundation - Flanders (FWO-Vlaanderen), project number G.0269.13N. G.M. and W.V.C. also acknowledge the Nucleotide 2 Networks Multidisciplinary Research Partnership (Special Research Fund Ghent University).
 This article contains supplemental Figs. S1 and S2, Tables S1 and S2, and Text file S1.
This article contains supplemental Figs. S1 and S2, Tables S1 and S2, and Text file S1.
1 The abbreviations used are:
- eIF
- eukaryotic Initiation Factor
- IRES
- Internal Ribosomal Entry Site
- aTIS
- alternative Translation Initiation Site
- CHX
- Cycloheximide
- COFRADIC
- COmbined FRActional DIagonal Chromatography
- dbTIS
- database annotated Translation Initiation Site
- dTIS
- downstream Translation Initiation Site
- GTI-seq
- Global Translation Initiation sequencing
- H2G2
- HitchHikers Guide to the Genome
- iMet
- initiator methionine
- LTM
- Lactimidomycin
- mESC
- mouse Embryonic Stem Cell
- NAT
- N-terminal acetyltransferase
- PIC
- Pre-Initiation Complex
- Ribo-seq
- Ribosome profiling sequencing
- RNA-seq
- RNA sequencing
- SCX
- Strong Cation Exchange
- uORF
- upstream Open Reading Frame
- uTIS
- upstream Translation Initiation Site
- UTR
- Untranslated Region
- CCDS
- Consensus Coding Sequence
- MetAP
- methionine aminopeptidase.
REFERENCES
- 1. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jackson R. J. (1991) The ATP requirement for initiation of eukaryotic translation varies according to the mRNA species. Eur. J. Biochem. 200, 285–294 [DOI] [PubMed] [Google Scholar]
- 3. Thompson S. R. (2012) Tricks an IRES uses to enslave ribosomes. Trends Microbiol. 20, 558–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Holcik M., Sonenberg N., Korneluk R. G. (2000) Internal ribosome initiation of translation and the control of cell death. Trends Genet. 16, 469–473 [DOI] [PubMed] [Google Scholar]
- 5. Spriggs K. A., Stoneley M., Bushell M., Willis A. E. (2008) Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell 100, 27–38 [DOI] [PubMed] [Google Scholar]
- 6. Martin F., Barends S., Jaeger S., Schaeffer L., Prongidi-Fix L., Eriani G. (2011) Cap-assisted internal initiation of translation of histone H4. Mol. Cell 41, 197–209 [DOI] [PubMed] [Google Scholar]
- 7. Kozak M. (1991) Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266, 19867–19870 [PubMed] [Google Scholar]
- 8. Gaba A., Wang Z., Krishnamoorthy T., Hinnebusch A. G., Sachs M. S. (2001) Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 20, 6453–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kozak M. (1987) Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 7, 3438–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calvo S. E., Pagliarini D. J., Mootha V. K. (2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 106, 7507–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Plessy C., Bertin N., Takahashi H., Simone R., Salimullah M., Lassmann T., Vitezic M., Severin J., Olivarius S., Lazarevic D., Hornig N., Orlando V., Bell I., Gao H., Dumais J., Kapranov P., Wang H., Davis C. A., Gingeras T. R., Kawai J., Daub C. O., Hayashizaki Y., Gustincich S., Carninci P. (2010) Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat. Methods 7, 528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schoenberg D. R., Maquat L. E. (2009) Re-capping the message. Trends Biochem. Sci. 34, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheung Y. N., Maag D., Mitchell S. F., Fekete C. A., Algire M. A., Takacs J. E., Shirokikh N., Pestova T., Lorsch J. R., Hinnebusch A. G. (2007) Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 21, 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pisarev A. V., Kolupaeva V. G., Pisareva V. P., Merrick W. C., Hellen C. U., Pestova T. V. (2006) Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 20, 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fekete C. A., Mitchell S. F., Cherkasova V. A., Applefield D., Algire M. A., Maag D., Saini A. K., Lorsch J. R., Hinnebusch A. G. (2007) N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 26, 1602–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 17. Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsieh A. C., Liu Y., Edlind M. P., Ingolia N. T., Janes M. R., Sher A., Shi E. Y., Stumpf C. R., Christensen C., Bonham M. J., Wang S., Ren P., Martin M., Jessen K., Feldman M. E., Weissman J. S., Shokat K. M., Rommel C., Ruggero D. (2012) The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reid D. W., Nicchitta C. V. (2012) Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J. Biol. Chem. 287, 5518–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fritsch C., Herrmann A., Nothnagel M., Szafranski K., Huse K., Schumann F., Schreiber S., Platzer M., Krawczak M., Hampe J., Brosch M. (2012) Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 22, 2208–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ingolia N. T., Lareau L. F., Weissman J. S. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of Mammalian proteomes. Cell 147, 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bazzini A. A., Lee M. T., Giraldez A. J. (2012) Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stadler M., Artiles K., Pak J., Fire A. (2012) Contributions of mRNA abundance, ribosome loading, and post- or peri-translational effects to temporal repression of C. elegans heterochronic miRNA targets. Genome Res. 22, 2418–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zoschke R., Watkins K. P., Barkan A. (2013) A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25, 2265–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu M. J., Wu S. H., Wu J. F., Lin W. D., Wu Y. C., Tsai T. Y., Tsai H. L. (2013) Translational Landscape of Photomorphogenic Arabidopsis. Plant Cell 25, 3699–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brar G. A., Yassour M., Friedman N., Regev A., Ingolia N. T., Weissman J. S. (2012) High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerashchenko M. V., Lobanov A. V., Gladyshev V. N. (2012) Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 109, 17394–17399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oh E., Becker A. H., Sandikci A., Huber D., Chaba R., Gloge F., Nichols R. J., Typas A., Gross C. A., Kramer G., Weissman J. S., Bukau B. (2011) Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li G. W., Oh E., Weissman J. S. (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stern-Ginossar N., Weisburd B., Michalski A., Le V. T., Hein M. Y., Huang S. X., Ma M., Shen B., Qian S. B., Hengel H., Mann M., Ingolia N. T., Weissman J. S. (2012) Decoding human cytomegalovirus. Science 338, 1088–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu X., Jiang H., Gu Z., Roberts J. W. (2013) High-resolution view of bacteriophage lambda gene expression by ribosome profiling. Proc. Natl. Acad. Sci. U.S.A. 110, 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee S., Liu B., Huang S. X., Shen B., Qian S. B. (2012) Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 109, E2424–E2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song K. Y., Choi H. S., Hwang C. K., Kim C. S., Law P. Y., Wei L. N., Loh H. H. (2009) Differential use of an in-frame translation initiation codon regulates human mu opioid receptor (OPRM1). Cell. Mol. Life Sci. 66, 2933–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kobayashi R., Patenia R., Ashizawa S., Vykoukal J. (2009) Targeted mass spectrometric analysis of N-terminally truncated isoforms generated via alternative translation initiation. FEBS Lett. 583, 2441–2445 [DOI] [PubMed] [Google Scholar]
- 36. Rossmanith W. (2011) Localization of human RNase Z isoforms: dual nuclear/mitochondrial targeting of the ELAC2 gene product by alternative translation initiation. PLoS One 6, e19152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shalak V., Kaminska M., Mirande M. (2009) Translation initiation from two in-frame AUGs generates mitochondrial and cytoplasmic forms of the p43 component of the multisynthetase complex. Biochemistry 48, 9959–9968 [DOI] [PubMed] [Google Scholar]
- 38. Kazak L., Reyes A., He J., Wood S. R., Brea-Calvo G., Holen T. T., Holt I. J. (2013) A cryptic targeting signal creates a mitochondrial FEN1 isoform with tailed R-Loop binding properties. PLoS One 8, e62340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kazak L., Reyes A., Duncan A. L., Rorbach J., Wood S. R., Brea-Calvo G., Gammage P. A., Robinson A. J., Minczuk M., Holt I. J. (2013) Alternative translation initiation augments the human mitochondrial proteome. Nucleic Acids Res. 41, 2354–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomas D., Plant L. D., Wilkens C. M., McCrossan Z. A., Goldstein S. A. (2008) Alternative translation initiation in rat brain yields K2P2.1 potassium channels permeable to sodium. Neuron 58, 859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kischkel F. C., Hellbardt S., Behrmann I., Germer M., Pawlita M., Krammer P. H., Peter M. E. (1995) Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14, 5579–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Giglione C., Boularot A., Meinnel T. (2004) Protein N-terminal methionine excision. Cell. Mol. Life Sci. 61, 1455–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pestana A., Pitot H. C. (1974) N-terminal acetylation of histone-like nascent peptides on rat-liver polyribosomes in-vitro. Nature 247, 200–202 [DOI] [PubMed] [Google Scholar]
- 44. Pestana A., Pitot H. C. (1975) Acetylation of nascent polypeptide-chains on rat-liver polyribosomes in vivo and in vitro. Biochemistry 14, 1404–1412 [DOI] [PubMed] [Google Scholar]
- 45. Strous G. J., Berns A. J., Bloemendal H. (1974) N-terminal acetylation of the nascent chains of alpha-crystallin. Biochem. Biophys. Res. Commun. 58, 876–884 [DOI] [PubMed] [Google Scholar]
- 46. Strous G. J., van Westreenen H., Bloemendal H. (1973) Synthesis of lens protein in vitro. N-terminal acetylation of alpha-crystallin. Eur. J.Biochem. / FEBS 38, 79–85 [DOI] [PubMed] [Google Scholar]
- 47. Giglione C., Fieulaine S., Meinnel T. (2009) Cotranslational processing mechanisms: towards a dynamic 3D model. Trends Biochem. Sci. 34, 417–426 [DOI] [PubMed] [Google Scholar]
- 48. Arnesen T., Van Damme P., Polevoda B., Helsens K., Evjenth R., Colaert N., Varhaug J. E., Vandekerckhove J., Lillehaug J. R., Sherman F., Gevaert K. (2009) Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. U.S.A. 106, 8157–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brown J. L., Roberts W. K. (1976) Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nalpha-acetylated. J. Biol. Chem. 251, 1009–1014 [PubMed] [Google Scholar]
- 50. Van Damme P., Hole K., Pimenta-Marques A., Helsens K., Vandekerckhove J., Martinho R. G., Gevaert K., Arnesen T. (2011) NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS Genet 7, e1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Starheim K. K., Gevaert K., Arnesen T. (2012) Protein N-terminal acetyltransferases: when the start matters. Trends Biochem. Sci 37, 152–161 [DOI] [PubMed] [Google Scholar]
- 52. Polevoda B., Sherman F. (2003) N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 325, 595–622 [DOI] [PubMed] [Google Scholar]
- 53. Van Damme P., Lasa M., Polevoda B., Gazquez C., Elosegui-Artola A., Kim D. S., De Juan-Pardo E., Demeyer K., Hole K., Larrea E., Timmerman E., Prieto J., Arnesen T., Sherman F., Gevaert K., Aldabe R. (2012) N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc. Natl. Acad. Sci. U.S.A. 109, 12449–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kendall R. L., Yamada R., Bradshaw R. A. (1990) Cotranslational amino-terminal processing. Method Enzymol. 185, 398–407 [DOI] [PubMed] [Google Scholar]
- 55. Impens F., Colaert N., Helsens K., Ghesquiere B., Timmerman E., De Bock P. J., Chain B. M., Vandekerckhove J., Gevaert K. (2010) A quantitative proteomics design for systematic identification of protease cleavage events. Mol. Cell. Proteomics 9, 2327–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lamkanfi M., Kanneganti T. D., Van Damme P., Vanden Berghe T., Vanoverberghe I., Vandekerckhove J., Vandenabeele P., Gevaert K., Nunez G. (2008) Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteomics 7, 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Van Damme P., Maurer-Stroh S., Plasman K., Van Durme J., Colaert N., Timmerman E., De Bock P. J., Goethals M., Rousseau F., Schymkowitz J., Vandekerckhove J., Gevaert K. (2009) Analysis of protein processing by N-terminal proteomics reveals novel species-specific substrate determinants of granzyme B orthologs. Mol. Cell. Proteomics 8, 258–272 [DOI] [PubMed] [Google Scholar]
- 58. Staes A., Van Damme P., Helsens K., Demol H., Vandekerckhove J., Gevaert K. (2008) Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC). Proteomics 8, 1362–1370 [DOI] [PubMed] [Google Scholar]
- 59. Menschaert G., Van Criekinge W., Notelaers T., Koch A., Crappe J., Gevaert K., Van Damme P. (2013) Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics 12, 1780–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Martens L., Vandekerckhove J., Gevaert K. (2005) DBToolkit: processing protein databases for peptide-centric proteomics. Bioinformatics 21, 3584–3585 [DOI] [PubMed] [Google Scholar]
- 61. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pruitt K. D., Harrow J., Harte R. A., Wallin C., Diekhans M., Maglott D. R., Searle S., Farrell C. M., Loveland J. E., Ruef B. J., Hart E., Suner M. M., Landrum M. J., Aken B., Ayling S., Baertsch R., Fernandez-Banet J., Cherry J. L., Curwen V., Dicuccio M., Kellis M., Lee J., Lin M. F., Schuster M., Shkeda A., Amid C., Brown G., Dukhanina O., Frankish A., Hart J., Maidak B. L., Mudge J., Murphy M. R., Murphy T., Rajan J., Rajput B., Riddick L. D., Snow C., Steward C., Webb D., Weber J. A., Wilming L., Wu W., Birney E., Haussler D., Hubbard T., Ostell J., Durbin R., Lipman D. (2009) The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 19, 1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ingolia N. T., Brar G. A., Rouskin S., McGeachy A. M., Weissman J. S. (2012) The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Flicek P., Amode M. R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., Fitzgerald S., Gil L., Gordon L., Hendrix M., Hourlier T., Johnson N., Kahari A. K., Keefe D., Keenan S., Kinsella R., Komorowska M., Koscielny G., Kulesha E., Larsson P., Longden I., McLaren W., Muffato M., Overduin B., Pignatelli M., Pritchard B., Riat H. S., Ritchie G. R., Ruffier M., Schuster M., Sobral D., Tang Y. A., Taylor K., Trevanion S., Vandrovcova J., White S., Wilson M., Wilder S. P., Aken B. L., Birney E., Cunningham F., Dunham I., Durbin R., Fernandez-Suarez X. M., Harrow J., Herrero J., Hubbard T. J., Parker A., Proctor G., Spudich G., Vogel J., Yates A., Zadissa A., Searle S. M. (2012) Ensembl 2012. Nucleic Acids Res. 40, D84–D90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L. W., Richards S., Weinstock G. M., Wilson R. K., Gibbs R. A., Kent W. J., Miller W., Haussler D. (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fan X., Zhu J., Schadt E. E., Liu J. S. (2007) Statistical power of phylo-HMM for evolutionarily conserved element detection. BMC Bioinformatics 8, 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Haider S., Ballester B., Smedley D., Zhang J., Rice P., Kasprzyk A. (2009) BioMart Central Portal–unified access to biological data. Nucleic Acids Res. 37, W23–W27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Flicek P., Amode M. R., Barrell D., Beal K., Brent S., Chen Y., Clapham P., Coates G., Fairley S., Fitzgerald S., Gordon L., Hendrix M., Hourlier T., Johnson N., Kahari A., Keefe D., Keenan S., Kinsella R., Kokocinski F., Kulesha E., Larsson P., Longden I., McLaren W., Overduin B., Pritchard B., Riat H. S., Rios D., Ritchie G. R., Ruffier M., Schuster M., Sobral D., Spudich G., Tang Y. A., Trevanion S., Vandrovcova J., Vilella A. J., White S., Wilder S. P., Zadissa A., Zamora J., Aken B. L., Birney E., Cunningham F., Dunham I., Durbin R., Fernandez-Suarez X. M., Herrero J., Hubbard T. J., Parker A., Proctor G., Vogel J., Searle S. M. (2011) Ensembl 2011. Nucleic Acids Res. 39, D800–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 70. Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., Vandekerckhove J. (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 21, 566–569 [DOI] [PubMed] [Google Scholar]
- 71. Van Damme P., Arnesen T., Gevaert K. (2011) Protein alpha-N-acetylation studied by N-terminomics. FEBS J 278, 3822–3834 [DOI] [PubMed] [Google Scholar]
- 72. Staes A., Impens F., Van Damme P., Ruttens B., Goethals M., Demol H., Timmerman E., Vandekerckhove J., Gevaert K. (2011) Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat. Protoc. 6, 1130–1141 [DOI] [PubMed] [Google Scholar]
- 73. Goetze S., Qeli E., Mosimann C., Staes A., Gerrits B., Roschitzki B., Mohanty S., Niederer E. M., Laczko E., Timmerman E., Lange V., Hafen E., Aebersold R., Vandekerckhove J., Basler K., Ahrens C. H., Gevaert K., Brunner E. (2009) Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol 7, e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lange P. F., Huesgen P. F., Overall C. M. (2012) TopFIND 2.0–linking protein termini with proteolytic processing and modifications altering protein function. Nucleic Acids Res. 40, D351–D361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Crawford E. D., Seaman J. E., Agard N., Hsu G. W., Julien O., Mahrus S., Nguyen H., Shimbo K., Yoshihara H. A., Zhuang M., Chalkley R. J., Wells J. A. (2013) The DegraBase: a database of proteolysis in healthy and apoptotic human cells. Mol. Cell. Proteomics 12, 813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rawlings N. D., Barrett A. J., Bateman A. (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40, D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Colaert N., Maddelein D., Impens F., Van Damme P., Plasman K., Helsens K., Hulstaert N., Vandekerckhove J., Gevaert K., Martens L. (2013) The Online Protein Processing Resource (TOPPR): a database and analysis platform for protein processing events. Nucleic Acids Res. 41, D333–D337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15, 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gu W., Zhou T., Wilke C. O. (2010) A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput. Biol. 6, e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mizuno M., Kanehisa M. (1994) Distribution profiles of GC content around the translation initiation site in different species. FEBS Lett. 352, 7–10 [DOI] [PubMed] [Google Scholar]
- 82. Kochetov A. V., Palyanov A., Titov II, Grigorovich D., Sarai A., Kolchanov N. A. (2007) AUG_hairpin: prediction of a downstream secondary structure influencing the recognition of a translation start site. BMC Bioinformatics 8, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jackson R. J. (2005) Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 33, 1231–1241 [DOI] [PubMed] [Google Scholar]
- 84. Kochetov A. V. (2008) Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays 30, 683–691 [DOI] [PubMed] [Google Scholar]
- 85. Kochetov A. V., Ahmad S., Ivanisenko V., Volkova O. A., Kolchanov N. A., Sarai A. (2008) uORFs, reinitiation and alternative translation start sites in human mRNAs. FEBS Lett. 582, 1293–1297 [DOI] [PubMed] [Google Scholar]
- 86. Volkova O. A., Kochetov A. V. (2010) Interrelations between the nucleotide context of human start AUG codon, N-end amino acids of the encoded protein and initiation of translation. J. Biomol. Struct. Dyn. 27, 611–618 [DOI] [PubMed] [Google Scholar]
- 87. Bazykin G. A., Kochetov A. V. (2011) Alternative translation start sites are conserved in eukaryotic genomes. Nucleic Acids Res. 39, 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Taylor F. J., Coates D. (1989) The code within the codons. Biosystems 22, 177–187 [DOI] [PubMed] [Google Scholar]
- 89. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 90. Adilakshmi T., Ness-Myers J., Madrid-Aliste C., Fiser A., Tapinos N. (2011) A nuclear variant of ErbB3 receptor tyrosine kinase regulates ezrin distribution and Schwann cell myelination. J. Neurosci. 31, 5106–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cai J., Huang Y., Li F., Li Y. (2006) Alteration of protein subcellular location and domain formation by alternative translational initiation. Proteins 62, 793–799 [DOI] [PubMed] [Google Scholar]
- 92. Nemeth A. L., Medveczky P., Toth J., Siklodi E., Schlett K., Patthy A., Palkovits M., Ovadi J., Tokesi N., Nemeth P., Szilagyi L., Graf L. (2007) Unconventional translation initiation of human trypsinogen 4 at a CUG codon with an N-terminal leucine. A possible means to regulate gene expression. FEBS J 274, 1610–1620 [DOI] [PubMed] [Google Scholar]
- 93. Schwab S. R., Shugart J. A., Horng T., Malarkannan S., Shastri N. (2004) Unanticipated antigens: translation initiation at CUG with leucine. PLoS Biol 2, e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Matsuda D., Dreher T. W. (2006) Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA 12, 1338–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Le H. T., Sorrell A. M., Siddle K. (2012) Two isoforms of the mRNA binding protein IGF2BP2 are generated by alternative translational initiation. PLoS One 7, e33140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lu N. Z., Cidlowski J. A. (2006) Glucocorticoid receptor isoforms generate transcription specificity. Trends Cell Biol. 16, 301–307 [DOI] [PubMed] [Google Scholar]
- 97. Leissring M. A., Farris W., Wu X., Christodoulou D. C., Haigis M. C., Guarente L., Selkoe D. J. (2004) Alternative translation initiation generates a novel isoform of insulin-degrading enzyme targeted to mitochondria. Biochem. J. 383, 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gu S., Anton A., Salim S., Blumer K. J., Dessauer C. W., Heximer S. P. (2008) Alternative translation initiation of human regulators of G-protein signaling-2 yields a set of functionally distinct proteins. Mol. Pharmacol. 73, 1–11 [DOI] [PubMed] [Google Scholar]
- 99. Crocoll A., Blum M., Cato A. C. (2000) Isoform-specific expression of BAG-1 in mouse development. Mech. Dev. 91, 355–359 [DOI] [PubMed] [Google Scholar]
- 100. Michel A., O'connor P., Choudhury K. R., Firth A., Li G. W., Ingolia N. T., Weissman J. S., Atkins J., Baranov P. (2013) Elucidating mechanisms of translation with computational analysis of ribo-seq data. Abstract EMBO Conference Series entitled ‘Protein Synthesis and Translational Control’, Heidelberg, Germany 2013 [Google Scholar]
- 101. Hwang C. S., Shemorry A., Varshavsky A. (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Behnia R., Panic B., Whyte J. R., Munro S. (2004) Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat. Cell Biol. 6, 405–413 [DOI] [PubMed] [Google Scholar]
- 103. Shemorry A., Hwang C. S., Varshavsky A. (2013) Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell 50, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lopez-Granados E., Keenan J. E., Kinney M. C., Leo H., Jain N., Ma C. A., Quinones R., Gelfand E. W., Jain A. (2008) A novel mutation in NFKBIA/IKBA results in a degradation-resistant N-truncated protein and is associated with ectodermal dysplasia with immunodeficiency. Hum. Mutat. 29, 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. McDonald D. R., Mooster J. L., Reddy M., Bawle E., Secord E., Geha R. S. (2007) Heterozygous N-terminal deletion of IkappaBalpha results in functional nuclear factor kappaB haploinsufficiency, ectodermal dysplasia, and immune deficiency. J. Allergy Clin. Immunol. 120, 900–907 [DOI] [PubMed] [Google Scholar]
- 106. Puel A., Reichenbach J., Bustamante J., Ku C. L., Feinberg J., Doffinger R., Bonnet M., Filipe-Santos O., de Beaucoudrey L., Durandy A., Horneff G., Novelli F., Wahn V., Smahi A., Israel A., Niehues T., Casanova J. L. (2006) The NEMO mutation creating the most-upstream premature stop codon is hypomorphic because of a reinitiation of translation. Am. J. Hum. Genet. 78, 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bab I., Smith E., Gavish H., Attar-Namdar M., Chorev M., Chen Y. C., Muhlrad A., Birnbaum M. J., Stein G., Frenkel B. (1999) Biosynthesis of osteogenic growth peptide via alternative translational initiation at AUG85 of histone H4 mRNA. J. Biol. Chem. 274, 14474–14481 [DOI] [PubMed] [Google Scholar]
- 108. Guttman M., Russell P., Ingolia N. T., Weissman J. S., Lander E. S. (2013) Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Helsens K., Colaert N., Barsnes H., Muth T., Flikka K., Staes A., Timmerman E., Wortelkamp S., Sickmann A., Vandekerckhove J., Gevaert K., Martens L. (2010) ms_lims, a simple yet powerful open source laboratory information management system for MS-driven proteomics. Proteomics 10, 1261–1264 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.