Abstract
Normalization is an important step in the analysis of quantitative proteomics data. If this step is ignored, systematic biases can lead to incorrect assumptions about regulation. Most statistical procedures for normalizing proteomics data have been borrowed from genomics where their development has focused on the removal of so-called ‘batch effects.’ In general, a typical normalization step in proteomics works under the assumption that most peptides/proteins do not change; scaling is then used to give a median log-ratio of 0. The focus of this work was to identify other factors, derived from knowledge of the variables in proteomics, which might be used to improve normalization. Here we have examined the multi-laboratory data sets from Phase I of the NCI's CPTAC program. Surprisingly, the most important bias variables affecting peptide intensities within labs were retention time and charge state. The magnitude of these observations was exaggerated in samples of unequal concentrations or “spike-in” levels, presumably because the average precursor charge for peptides with higher charge state potentials is lower at higher relative sample concentrations. These effects are consistent with reduced protonation during electrospray and demonstrate that the physical properties of the peptides themselves can serve as good reporters of systematic biases. Between labs, retention time, precursor m/z, and peptide length were most commonly the top-ranked bias variables, over the standardly used average intensity (A). A larger set of variables was then used to develop a stepwise normalization procedure. This statistical model was found to perform as well or better on the CPTAC mock biomarker data than other commonly used methods. Furthermore, the method described here does not require a priori knowledge of the systematic biases in a given data set. These improvements can be attributed to the inclusion of variables other than average intensity during normalization.
The number of laboratories using MS as a quantitative tool for protein profiling continues to grow, propelling the field forward past simple qualitative measurements (i.e. cataloging), with the aim of establishing itself as a robust method for detecting proteomic differences. By analogy, semiquantitative proteomic profiling by MS can be compared with measurement of relative gene expression by genomics technologies such as microarrays or, newer, RNAseq measurements. While proteomics is disadvantaged by the lack of a molecular amplification system for proteins, successful reports from discovery experiments are numerous in the literature and are increasing with advances in instrument resolution and sensitivity.
In general, methods for performing relative quantitation can be broadly divided into two categories: those employing labels (e.g. iTRAQ, TMT, and SILAC (1)) and so-called “label-free” techniques. Labeling methods involve adding some form of isobaric or isotopic label(s) to the proteins or peptides prior to liquid chromatography-tandem MS (LC-MS/MS) analysis. Chemical labels are typically applied during sample processing, and isotopic labels are commonly added during cell culture (i.e. metabolic labeling). One advantage of label-based methods is that the two (or more) differently-labeled samples can be mixed and run in single LC-MS analyses. This is in contrast to label-free methods which require the samples to be run independently and the data aligned post-acquisition.
Many labs employ label-free methods because they are applicable to a wider range of samples and require fewer sample processing steps. Moreover, data from qualitative experiments can sometimes be re-analyzed using label-free software tools to provide semiquantitative data. Advances in these software tools have been extensively reviewed (2). While analysis of label-based data primarily uses full MS scan (MS1)1 or tandem MS scan (MS2) ion current measurements, analysis of label-free data can employ simple counts of confidently identified tandem mass spectra (3). So-called spectral counting makes the assumption that the number of times a peptide is identified is proportional to its concentration. These values are sometimes summed across all peptides for a given protein and scaled by protein length. Relative abundance can then be calculated for any peptide or protein of interest. While this approach may be easy to perform, its usefulness is particularly limited in smaller data sets and/or when counts are low.
This report focuses only on the use of ion current measurements in label-free data sets, specifically those calculated from extracted MS1 ion chromatograms (XICs). In general terms, raw intensity values (i.e. ion counts in arbitrary units) cannot be used for quantitation in the absence of cognate internal standards because individual ion intensities depend on a response factor, related to the chemical properties of the molecule. Intensities are instead almost always reserved for relative determinations. Furthermore, retention times are sometimes used to align the chromatograms between runs to ensure higher confidence prior to calculating relative intensities. This step is crucial for methods without corresponding identity information, particularly for experiments performed on low-resolution instruments. To support a label-free workflow, peptide identifications are commonly made from tandem mass spectra (MS/MS) acquired along with direct electrospray signal (MS1). Or, in alternative workflows seeking deeper coverage, interesting MS1 components can be targeted for identification by MS/MS in follow-up runs (4).
“Rolling up” the peptide ion information to the peptide and protein level is also done in different ways in different labs. In most cases, “peptide intensity” or “peptide abundance” is the summed or averaged value of the identified peptide ions. How the peptide information is transferred to the protein level differs between methods but typically involves summing one or more peptide intensities, following parsimony analysis. One such solution is the “Top 3” method developed by Silva and co-workers (5).
Because peptides in label-free methods lack labeled analogs and require separate runs, they are more susceptible to analytical noise and systematic variations. Sources of these obscuring variations can come from many sources, including sample preparation, operator error, chromatography, electrospray, and even from the data analysis itself. While analytical noise (e.g. chemical interference) is difficult to selectively reject, systematic biases can often be removed by statistical preprocessing. The goal of these procedures is to normalize the data prior to calculations of relative abundance. Failure to resolve these issues is the common origin of batch effects, previously described for genomics data, which can severely limit meaningful interpretation of experimental data (6, 7).
These effects have also been recently explored in proteomics data (8). Methods used to normalize proteomics data have been largely borrowed from the microarray community, or are based on a simple mean/median intensity ratio correction. Methods applied on microarray and/or gene chip and used on proteomics data include scaling, linear regression, nonlinear regression, and quantile normalizations (9). Moreover, work has also been done to improve normalization by subselecting a peptide basis (10). Other work suggests that linear regression, followed by run order analysis, works better than other methods tested (11). Key to this last method is the incorporation of a variable other than intensity during normalization. It is also important to note that little work has been done towards identifying the underlying sources of these variations in proteomics data. Although cause-and-effect is often difficult to determine, understanding these relationships will undoubtedly help remove and avoid the major underlying sources of systematic variations.
In this report, we have attempted to combine our efforts focused on understanding variability with the work initiated by others for normalizing ion current-based label-free proteomics data. We have identified several major variables commonly affecting peptide ion intensities both within and between labs. As test data, we used a subset of raw data acquired during Phase I of the National Cancer Institute's (NCI) Clinical Proteomics Technology Assessment for Cancer (CPTAC) program. With these data, we were able to develop a statistical model to rank bias variables and normalize the intensities using stepwise, semiparametric regression. The data analysis methods have been implemented within the National Institute of Standards and Technology (NIST) MS quality control (MSQC) pipeline. Finally, we have developed R code for removing systematic biases and have tested it using a reference standard spiked into a complex biological matrix (i.e. yeast cell lysate).
EXPERIMENTAL PROCEDURES
Data Sets
All of the CPTAC Phase I data sets used in this work are publicly available at https://cptac-data-portal.georgetown.edu/cptacPublic/. This site provides direct links to download the study data. Hereafter, data are referenced according to designated CPTAC Study number. Descriptions of the CPTAC yeast reference material, and SOPs used to acquire the data, can be found elsewhere (12). Briefly, the data for Study 6 were collected in the following order with a blank in between each study sample: (1) Sample 1B-NCI-20, (2) Sample 6-QC2 yeast only, (3) Sample 6A yeast + UPS1 at 0.25 fmol/μl, (4) Sample 6B, yeast + UPS1 at 0.74 fmol/μl, (5) Sample 6C, yeast + UPS1 at 2.2 fmol/μl, (6) Sample 6D, yeast + UPS1 at 6.7 fmol/μl, (7) Sample 6E, yeast + UPS1 at 20 fmol/μl, (8) Sample 6-QC1, UPS1 only at 20 fmol/μl. All yeast samples (prior to spike-ins) contained trypsin-digested yeast lysate at 60 ng/μl. A standard injection volume of 2 μl was used for all samples. All platforms used the same prepacked columns and a nano-ESI source. The Study 8 data were acquired without the use of the CPTAC SOP, according to individual lab protocol. The yeast sample was run in two amounts, 120 ng and 600 ng, which are referred to hereafter as ‘low’ and ‘high’, respectively.
Data Processing
All calculations on MS data files presented in this work were made by the NIST MSQC pipeline (v. 1.2.0) (13). All of the intensity values were extracted directly from output .msqc files using a parser written in Perl (available by request). The major changes from the previously published version of the software include the following: (1) the replacement of ReAdW4Mascot2.exe with a more refined program for processing MS1 data (ProMS), (2) the addition of MSPepSearch (v. 0.9.0) as the default search engine, (3) the introduction of a ‘full’ mode for providing peptide/protein relative quantitation information in the report. Several new metrics were also added that were used in this report. These include a calculation for the median intensity deviation.
The NIST MSQC pipeline is driven by a Perl program, which controls component programs in the following order: (1) ReAdW4Mascot2 for extracting spectra and retention times from RAW files to mzXML and MGF, (2) a search engine (MSPepSearch, SpectraST or OMSSA), (3) ProMS (optional but recommended), (4) nistms_metrics for calculating metrics and statistics within and between series, (5) merge_pep_results for final formatting of the result files. All command-line arguments and options for running the pipeline can be found at http://peptide.nist.gov/metrics or by running the pipeline without arguments. The newest version of the pipeline also comes with a GUI and WindowsTM installer. Additionally, MSFileReader from Thermo Fisher (http://sjsupport.thermofinnigan.com/public/detail.asp?id=703) can be used, allowing users without an installation of XCaliburTM to operate the pipeline. As with earlier versions, the pipeline requires an installation of Perl and runs only on the WindowsTM operating system. Thermo Fisher LTQ, LTQ-Orbitrap, LTQ-FT and QExactiveTM data files can be analyzed. A version utilizing ProteoWizard libraries for use with other vendor data files is under development. And at the time of writing, the pipeline was compatible with Agilent QTOF files.
Normalization and Variable Ranking
Normalization is the process of removing systematic bias in order to make runs more comparable. In this work, the data are ‘normalized’ based on the assumption that, on average, the peptide ion intensities do not change between runs. ‘Runs’ in this work denote technical replicates, replicates of the same sample between laboratories, or replicates of similarly engineered samples with known differences (as in the case of CPTAC Study 6.) Although the CPTAC data do not represent true biological experiments, they do represent best-case-scenario and a starting point from which further development of the aforementioned computational methods can be initiated.
Ideally, log2 ratios of peptide ion intensities (M) between any two technical replicates should either equal a constant value of 0, or log2(x) if runs have a known x-fold concentration difference. M values should then distribute randomly around the reference level without any significantly discernible pattern. However, in practice, many factors cause systematic bias in experimental data acquisition. An example of this is pen location during production of oligonucleotide arrays. Often, this extraneous variability is confounded with the biological variability of interest in the sample. It is thus necessary to remove systematic bias before proceeding to any relative quantification.
A collection of normalization methods borrowed from microarray analysis has been tested on proteomics data (9). These methods include mean/median removing, linear regression, locally-weighted regression and quantile techniques. However, in all but one case, only abundance (the average of peptide intensity between the two technical replicates in comparison) is used for normalization. In the current work, it was observed that abundance is not the only source of systematic bias. Therefore a scheme was developed to include a set of identified bias variables for effective data normalization. A detailed description of model development follows.
Let Mj(1) be the initial log ratio of intensities for peptide ion j in a given pair of runs, j = 1, …, N and N is the number of detected peptide ions. The normalization procedure starts with modeling the log ratio of intensities by
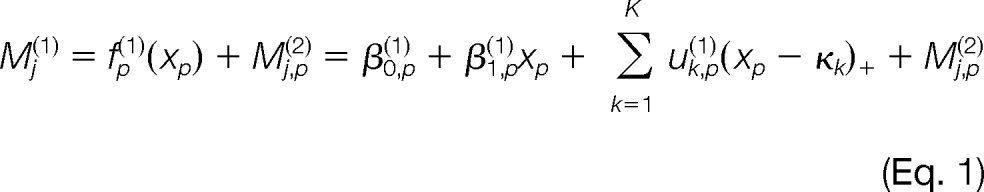 |
In Equation 1, xp is the pth variable in the P variables of interest, P = 1, K, P. The function fp(1)(·) is the regression function with variable xp. Both the simple linear and p-spline models are considered for fp(1)(·). The parameters β0,p(1), β1,p(1), and uk,p(1)'s are the regression coefficients in the model fp(1)(·). The knots (κk's (k = 1, K, K) are selected according to (14). The function (x − κk)+ = 0 if x ≤ κk and x − κk if x >κk. The regression residuals Mj,p(2) are then the normalized log ratios after adjusting for variable xp's impacts.
Each variable is fitted with individual fp(1)(xp) using the appropriate semiparametric (the p-spline) or simple linear regression models according to Equation 1. A deviance measure is designed to select the best normalization variable at each step. The goal is to select the variable that is most significantly related to systematic variation in the log ratio of intensities. The deviance for variable p at step 1 with function fp(1)(·) is defined as
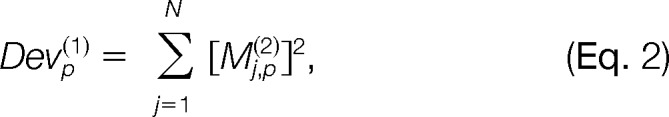 |
where Devp(1) is the deviance for variable p at step 1 and Mj,p(2) is the residuals from the regression model. The best variable selected at step 1 is the variable p* minimizing Devp(1). The normalized log ratio of intensities is then defined as Mj,p*(2) and the optimal deviance is Devp*(1).
The above procedure is repeated P times (the number of variables included), with the selected variable at each step removed from the following steps. Generally, at step s (s = 1, 2, L, P), the remaining variables xp's are fit into
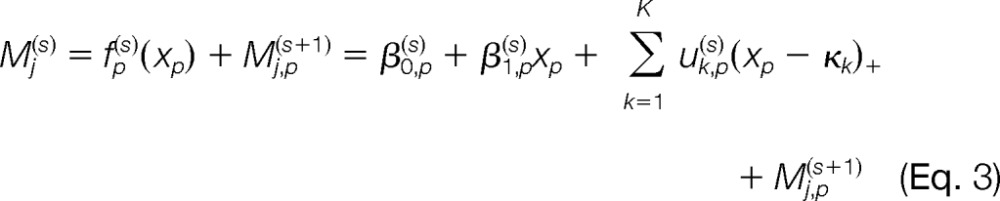 |
The best variable selected at step s is the one minimizing the deviance
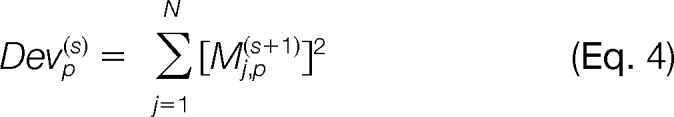 |
The ranking of the variables is based on the order they were selected in the above iterative procedure, and their impacts are measured by the reduction in deviance ΔDev(s) = Devp(s) − Devp*(s−1), where Devp*(s−1) is the sample variance of the original M when s = 1 and the deviance obtained at the selected variable p* when 1 < s ≤ P. The final normalized log ratio of peptide ion intensities is Mj(P+1), j = 1, L, N. R code for performing these calculations is available at http://homepages.uc.edu/∼wang2x7/.
Median Deviation
Median deviation is defined as the square root of the ratio of the 25th percentile to the median peptide intensity ratio. By using this statistic based on the center of the absolute deviations, the method is not sensitive to one observation deviating from an expected ratio of 1 and therefore provides a robust measure of the distribution of the deviations.
RESULTS
Previous work on the reproducibility of proteomic analyses by LC-MS formed the basis for this study (13). In that work, it was noted that peptide ion intensities are valuable reporters of overall reproducibility. Here, we describe their use as central indicators of systematic bias. As with any data analysis project in proteomics, software must be chosen that is capable of extracting the required data from the instrument-generated binary files. And for ion-current based label-free quantitative studies, routines for performing the necessary calculations on extracted ion chromatograms are also needed. While there are many software programs available, we chose to continue developing the tools within the NIST MSQC pipeline. As such, ProMS replaces ReAdW4Mascot2.exe for calculating intensity (Experimental Procedures). These values can now be written to the NIST MSQC output using the command-line option “–mode full” during processing.
Systematic Bias in the Peptide Ion Intensities
To begin our studies, we chose test data from CPTAC Study 8 (no SOP) and a subset of runs from Study 6 (SOP-controlled). The data were all processed through the NIST MSQC pipeline (v.1.2.0) as described under “Experimental Procedures.” In Study 8, three replicates of two amounts, 120 ng (low) and 600 ng (high), of the yeast material (Sample QC2) were analyzed in triplicate. The data from 6 different instruments in five different laboratories were analyzed. Additionally, data from Study 6 (yeast + UPS1) from two Orbitraps were also analyzed. To visualize the raw intensities, and the need for normalization, the distributions were plotted in Fig. 1.
Fig. 1.
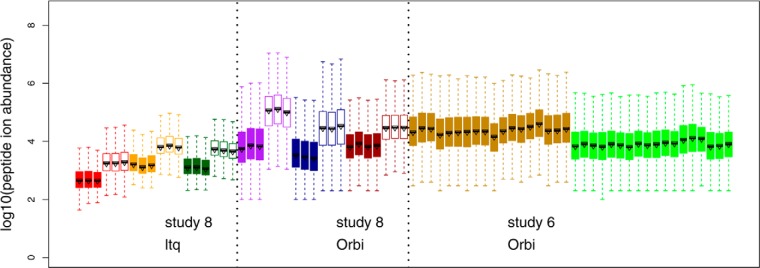
Raw peptide ion intensities from CPTAC Study 8 (no SOP) and a subset of runs from Study 6 (with SOP). Data in Study 8 include 2 LTQs and 3 Orbitrap instruments. Three experimental runs of two samples (2 μl injections), 60 ng/μl (“low,” filled boxplots) and 300 ng/μl (“high,” unfilled boxplots) of the yeast material (Sample QC2) were included in the analysis of Study 8. For Study 6, a subset of runs on two Orbitraps were also included. The runs were plotted in the order specified as below, each with three replicates: (1) Sample 6A yeast + UPS1 at 0.25 fmol/μl, (2) Sample 6B, yeast + UPS1 at 0.74 fmol/μl, (3) Sample 6C, yeast + UPS1 at 2.2 fmol/μl, (4) Sample 6D, yeast + UPS1 at 6.7 fmol/μl, (5) Sample 6E, yeast + UPS1 at 20 fmol/μl, (6) Sample 6-QC2 yeast only.
Several observations can be made from Fig. 1. First, the low versus high runs from Study 8 can be easily distinguished because of the five-fold difference in loading. The medians of the boxplots for high runs range from 3.2–2.9 in LTQs and 4.4–5.1 in Orbitraps, whereas the medians of low runs range from 2.6–3.2 in LTQs and 3.4–3.9 in Orbitraps. Second, the interquartile range (IQR) of peptide ion intensities measured on an Orbritrap is between 0.8–1.2, which is wider than IQR of 0.5–0.62 measured on an LTQ. Furthermore, even within the same type of instrument, obvious differences in median intensities and IQR exist between runs and between instruments. For example, the boxplots for Lab 1 in Study 6 have an average median around 4.3 with an IQR of ∼1. The boxplots for Lab 2 in Study 6 have a lower average median around 3.8 with IQR between 0.8–0.9. These results suggested the need for a data normalization step prior to any interpretation of differences within an experiment, and for normalization if data are to be compared between labs for both LTQ and Orbitrap instruments. Results on Orbitrap instruments are presented in detail in the main text, whereas results on LTQ instruments are included in Supplemental Data.
Systematic Bias as Identified by Relative Intensities
Next, we used the relative intensities and a group of variables to investigate systematic bias in the above data. Relative intensity is defined as M = log2 (IR1/IR2), where IR1 and IR2 are the MS1 intensities for the run R1 and R2, respectively. The underlying assumption is that the majority of relative intensities do not change between runs, allowing the user to examine dispersion in the data. Fig. 2 shows the systematic biases in M related to a group of selected variables.
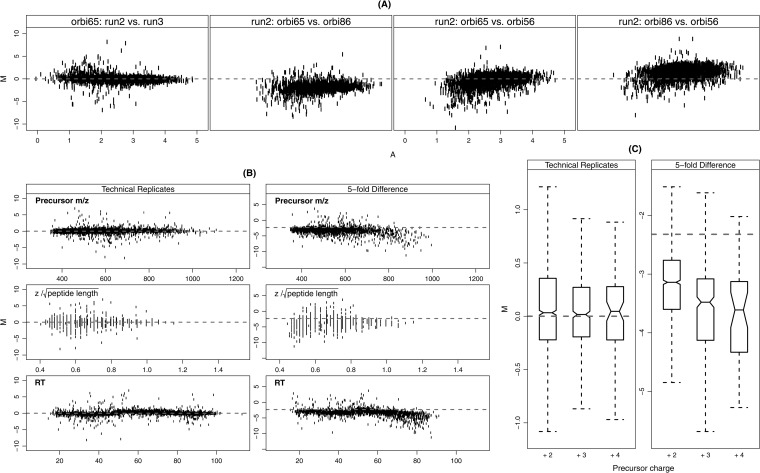
Fig. 2.
Systematic biases in ion current measurements measured by the relative intensities (M) and its relationship with selected variables. Relative intensity M is defined as the log2 ratio, M = log2 (IR1/IR2), where IR1 and IR2 are the intensities for the experimental run R1 and R2, respectively. The selected variables included the average abundance A = 0.5 [log10 (IR1) + log10 (IR2)], precursor m/z, , and retention time (RT). A, The relative intensity (M) versus abundance (A) within and across instruments. All runs are 300 ng/μl yeast samples (high). Panel 1: orbitrap 65 (2nd run) versus orbitrap 65 (3rd run); panel 2: orbitrap 65 (2nd run) versus orbitrap 86 (2nd run); panel 3: orbitrap 65 (2nd run) versus orbitrap 56 (2nd run); panel 4: orbitrap 86 (2nd run) versus orbitrap 56(2nd run). B, relative intensities (M) versus precursor m/z, , and retention time (RT). All runs are from Orbitrap 65 in Study 8. The technical replicate pair is the 2nd and the 3rd runs in the 300 ng/μl yeast samples (high). The fivefold difference pair is the 2nd run in the 300 ng/μl yeast sample (high) and the 2nd run in the 60 ng/μl yeast sample (low). The left column shows technical replicates pairs and the right column shows fivefold difference pairs. These plots illustrate that systematic bias is more significant between the high and low samples with fivefold difference. C, Boxplots of the relative intensities (M) under the three observed charge states (+2, +3, +4) on experimental runs from Orbitrap 65 in Study 8. The same experimental runs were used for the pairs of technical replicates and fivefold difference as in B. The boxplot bounds in the form of [IQR (median)] are as follows: technical replicates: +2 [0.58(0.03)], +3 [0.46(0.02)], +4 [0.49 (0.05)]; fivefold difference: +2 [0.84(−3.13)], +3 [1.05(−3.48)], +4 [1.20 (−3.61)]. The distribution similarity was tested by a two-sample Wilcoxon rank test. The distributions of M between the charge states in high versus low samples (fivefold difference) were statistically different (p value <0.001) with the exception of +3 compared with +4 (p value = 0.46). The distributions of M were not significantly different under different charge states for technical replicates (p value > 0.15).
Fig. 2A shows commonly used M-A plots for a comparison of runs. In these plots, the y axis represents relative intensities M and the x axis is used to order the peptides by average abundance defined as A = 0.5 [log10 (IR1) + log10 (IR2)]. Specifically, these plots can be used to identify systematic biases affecting absolute abundance (i.e. low signal and saturation effects). The first panel of Fig. 2A shows data from two technical replicates (the second and third runs in 300 ng/μl yeast sample, high) on the same instrument (Orbitrap 65 in Study 8). In the other three panels, M-A plots compare data from the second run in high sample to the same run on each of the three Orbitrap instruments used in Study 8.
Several observations can be made from this analysis. First, as expected, replicate runs from the same instrument fit the expected reference value (dashed line) much better than runs compared between different instruments. Second, dispersions were on average, greater at lower A values. Third, several inter-instrument (also interlaboratory) comparisons exhibited nonlinear trends and global and/or local departures from the expected reference value. Local deviations confound simple global corrections (i.e. scaling) during normalization because they are not representative of the entire run. It is also important to note that no two comparisons displayed exactly the same behavior, suggesting the importance of pair-wise analyses, particularly if data between instruments are compared. supplemental Fig. S1A in the Supplemental Data shows the similar results on an LTQ instrument.
From previous work, it was known that varying the injection amount leads to broad differences in a number of performance metrics: in particular, the number of singly charged relative to doubly charged peptide ion identifications increases with injection amount, as does the average precursor m/z (13). These observations indicate a reduced average peptide ion charge state at higher injection amounts, most likely because of limited proton availability during ESI.
With the intent of further investigating these observations on the individual peptide-level, we compared data from the Study 8 low and high samples to look for deviations in M from the expected reference value, given no or a fivefold difference in concentration between samples. In Fig. 2B, the technical replicates pair includes the second and the third runs in the 300 ng/μl yeast samples (high); the fivefold difference pair includes the second run in the 300/μ; ng yeast sample (high) and the second run in the 60 ng/μl yeast sample (low). Peptides are ordered according to peptide properties chosen here because they are known to be affected by sample concentration.
In this analysis, all three variables, precursor m/z, m/z, z/, and retention time, showed strong correlation with systematic biases, indicating that larger peptides and those eluting late in the gradient are more intense at higher concentrations. This suggests that sample loading selectively biases the intensities of the peptides with higher charge state potentials. The variable is the charge density standardized by the square root of the peptide length. The square root was used because z/peptide length exhibited strong correlation with the peptide length, which was also included in the group of variables discussed later.
To further investigate the possibility that loading systematically biases peptides with higher charge state potentials, M values of the above runs in Fig. 2B were separated by charge states. Fig. 2C summarizes the distributions of M values in boxplots. For technical replicates (second and third run from the high sample on Orbitrap 65), the median of boxplots on the relative intensities under three charge states were all close to the reference level 0 with IQR as 0.58, 0.46, and 0.49 for 2+, 3+, and 4+, respectively. For the pair with fivefold difference (second run from the high sample and second run from the low sample on Orbitrap 65), the boxplots showed variations in both the medians and the IQRs across charge states. Charge state 2+ had a median (-3.13) which was the closest to the reference line (−log2(5)) with the shortest IQR = 0.84. For 3+, the median was −3.48 with IQR = 1.05 and for 4+, the median was −3.61 with IQR = 1.20.
To determine if the distributions were significantly different, a two-sample Wilcoxon rank test was used because the bias on M values under different charge states is not normally distributed. As expected, the distributions between the charge states in high and low samples (5-fold difference) were statistically different (p value <0.001) with the exception of +3 compared with +4 (p value = 0.46). The distributions of peptide ion intensities were not significantly different under different charge states for technical replicates (p value > 0.15). These results indicated that precursor charge state is an important variable to be considered during data normalization, especially between different samples. Supplemental Fig. S1B in Supplemental data shows the similar analysis on an LTQ instrument.
Since retention time introduces a large systematic bias when data from samples at different concentrations were compared, median relative abundance deviations (Experimental Procedures) were calculated for each retention quartile from comparisons between the above runs on Orbitrap 65. The high median deviations in M illustrated the higher variability during the fourth quartile of retention time (Q4) observed for intersample comparisons in five-fold difference pair (dashed lines) relative to technical replicates (solid lines) (See supplemental Fig. S2 in the Supplemental Data). The investigation on the median deviance suggested that, although comparisons between low and high samples are somewhat artificial, significant biases may be unwittingly introduced by small loading differences, giving rise to retention time biases.
Development of a Normalization Model
The variables examined in Fig. 2B as well as abundance (A), peptide length, and the number of mobile protons (precursor charge minus number of H, K and R's + 1) were used to develop a normalization model (Experimental Procedures). The proposed method is an extension of normalization using a linear regression model. But instead of using only abundance, several bias variables are included as predictors. Supplemental Fig. S3 shows a schematic of the algorithm and supplemental Fig. S4 gives an example showing resultant regression curves and deviance values during stepwise normalization using two runs from Study 8.
Examining the Results of the Normalization Model
Fig. 3 shows the densities of M values before and after normalization using various approaches, including removing the mean of log ratios (around −2.7), regression against the abundance (A) and the proposed semiparametric regression models. The data used were a pair with fivefold difference from Study 8. The comparison in Fig. 3 indicated that models using only mean of M or abundance (A) as normalization factors, which have been widely used in proteomics and on microarray data, may have room for improvement when applied to peptide abundance data derived from ion current measurements.
Fig. 3.
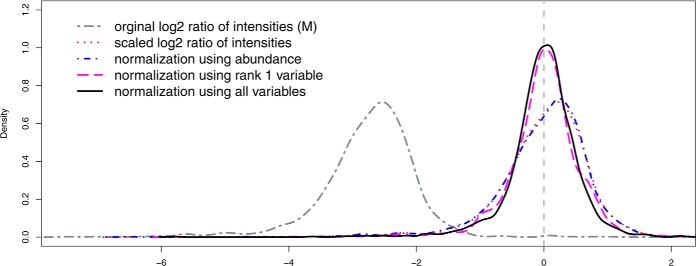
The densities of the relative intensities M (M = log2 (IR1/IR2)) under different normalization methods. The data used in the pair were the 1st run of 60 ng/μl yeast sample (low) and the 2nd run of 300 ng/μl yeast sample (high) on the ltq73 instrument in Study 8. The dark gray curve (“two-dash”) is for the original relative intensities M (before normalization). The red curve (“dotted”) is for the scaled relative intensities M by removing the sample mean of M (approximately −2.7 for the data used). The blue curve (“dot-dash”) is the normalized relative intensities M using peptide abundance (A) only. The purple curve (“long-dash”) is the normalized relative intensities M using Rank 1 variable only. The black curve (“solid”) is the normalized relative intensities M using all variables. The dashed gray line is the reference line at log2(M) = 0.
Examining the Influence of the Variables
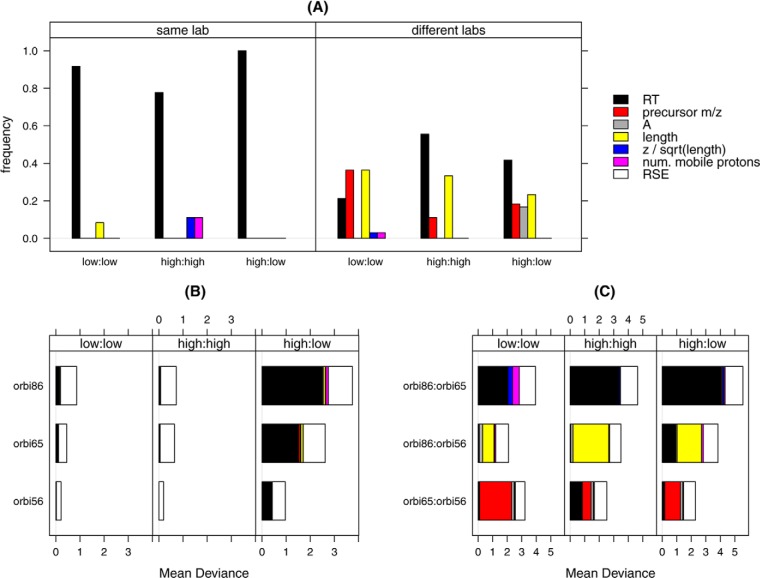
To cover many cases, the interlaboratory data from Study 8 were again used to approximate the relative influence of these variables with respect to peptide intensity deviations. The low and high concentration runs within and between labs were all compared. In this analysis, the frequency of each variable appearing in the rank 1 (most influential) position was calculated for all pairs of runs (Fig. 4A).
Fig. 4.
Ranking and mean deviances of the normalization variables. The data used were 19 experimental runs on the 3 Orbitrap instruments from Study 8, including 60 ng/μl (low) and 300 ng/μl (high) yeast samples. A, The frequency of the variables as Rank 1 for runs within the same lab or across different labs. B, The magnitude of the mean deviance adjusted by each variable, as well as the remaining mean deviance (represented by RSE) when experimental runs were from the same labs. C, The magnitude of the mean deviance adjusted by each variable as well as the remaining mean deviance (represented by RSE) when experimental runs were from different labs.
Within labs, RT was consistently ranked as the most influential variable and among the most influential variables when runs were compared between labs. The other two variables that became influential were precursor m/z and the peptide length. What is also notable from this analysis is the fact that within labs, average abundance (A) - the standardly used normalization variable - was never ranked as the most important variable. RT, length and precursor m/z were the most influential between labs. Again, although performing a differential expression analysis on runs between instruments (laboratories) may be somewhat artificial, it nevertheless highlighted the most variable aspects of the data. Similar results on LTQ instruments are included in supplemental Fig. S5A of the Supplemental Data.
The magnitude of the bias accounted for by each of the variables can be shown by the mean of the deviance (See Eq. (2) and (4) under “Experimental Procedures”). Fig. 4B shows the average of the mean of deviance (MD) within lab and Fig. 4C shows the average of the MD for each pair between labs. Several observations were made from this analysis. First, MD was larger for pairs across instruments than for those from the same instrument. Second, the largest reduction of the MD was introduced by the rank 1 variable. The remaining variables improved the MD only marginally. After removing systematic bias from the M values, the residual MD (RSE, white bars) was comparable at the two concentration levels, although pairs from the same labs had a smaller MD to those pairs from different labs. Similar results on LTQ instruments are presented in supplemental Fig. S5B and S5C of Supplemental Data.
The normalization algorithm developed for this work is directly applicable when the two runs are from an identical sample or are from samples with a known overall concentration difference, such as Study 8 data sets. In these samples, all peptide ions can be used in the normalization procedure because they can all be assumed to be randomly distributed around a known and identical reference level. When the two runs from biologically different samples are analyzed, some peptide ions are expected to remain quantitatively similar (common peptide ions), whereas significant changes are expected in others. Those peptide ions, whose intensities are suspected to be different under different biological conditions, should not be included in the normalization and ranking procedure, if possible. Their M values are adjusted by interpolation using the regression parameters obtained from the normalization-ranking procedure with the common peptide ions. The application of the proposed method to this type of data is shown below using Study 6 mock biomarker data.
Normalization Lowers the False Positive Rate in Mock Biomarker Experiments
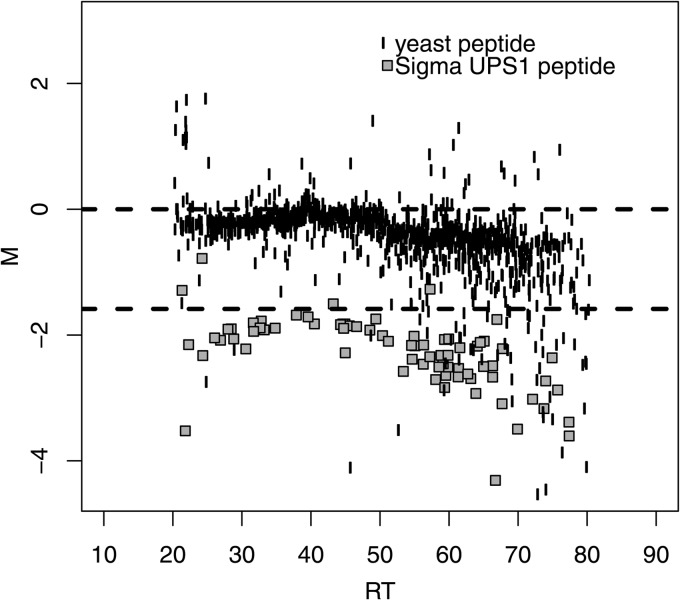
In order to assess the effectiveness of the newly developed normalization method, we used data from Study 6. In this study, several of the processed samples contained the SigmaUPS1 proteins spiked into the yeast reference material at known amounts, sequentially differing by factors of 3 (Experimental Procedures). To visualize the data prior to normalization, we plotted the M values of the yeast matrix peptide ions and Sigma UPS1 spike-ins separately versus RT (Fig. 5) using a pair of runs from Sample 6C and Sample 6D on Lab 1 in Study 6. The Sigma UPS1 peptides in this plot are clearly separated from the yeast matrix peptides and are present at close to the expected log2-ratio (approximately −1.58). However, because the spike-level is of relatively high concentration, a clear RT bias is visible at the end of the gradient.
Fig. 5.
The relative intensities (M) versus retention time (RT) of Sample 6C (yeast + UPS1 at 2. 2 fmol/μl) against Sample 6D (yeast + UPS1 at 6.7 fmol/μl) in Study 6. The yeast matrix peptide ions (unfilled circles) were expected to be centered on the reference line at 0. The peptide ions for Sigma UPS1 spike-ins (filled gray squares) were expected to be centered around the reference line at log2(M) = −log2(3) (approximately [−1.58]) because the Sigma UPS1 spike-ins differed by threefolds between the samples analyzed. Systematic bias existed in the observed peptide ions intensities for both the yeast matrix and the Sigma UPS1.
Study 6 data is a mock case for biologically different samples. That is the common peptide ions were known (yeast peptide ions). In a real data set, this information is usually not available. The selection of the common peptide ions for normalization is another important issue in comparative proteomics. Various methods have been developed in the literature (11, 15, 16–18). In the following results, we normalized and ranked the Study 6 data as if the identities of the yeast and Sigma UPS1 proteins were not known like in many real data sets. To select the set of common peptide ions, we used the global rank-invariant set selection methods in (18).
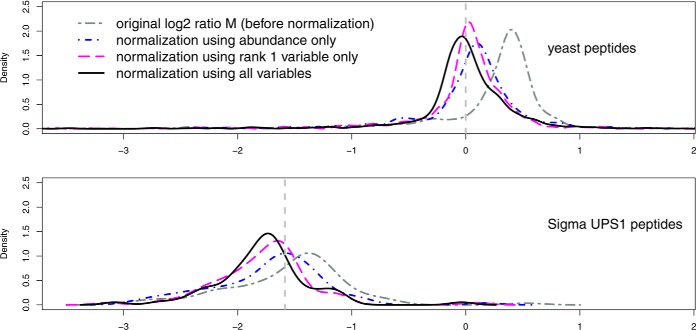
To examine the ability of the model to separate known spike-ins from matrix, each pair of runs from Sample 6C and 6D was normalized. In Fig. 6, the densities of the yeast (upper panel) and Sigma UPS1 (lower panel) M values in a pair (Run #8 of Sample 6C and Run #11 of Sample 6D on Orbitrap 65 in Study 6) are shown for the cases before normalization, normalization by abundance (A) only, by rank 1 variable only and by all variables used, respectively. Biases in M values were reduced more by the single rank 1 variable or by the combination of all six variables, than by that of the abundance (A).
Fig. 6.
Densities of relative intensities M (M = log2 (IR1/IR2)) under different normalization methods for run pairs Sample 6C (yeast + UPS1 at 2. 2 fmol/μl) against Sample 6D (yeast + UPS1 at 6.7 fmol/μl) (3-fold difference) in Study 6. In normalization and ranking, common peptides were selected based on the global invariant-ranking set (18). The top panel is for the yeast peptides ions, whose relative intensities were expected to be centered on the reference line at 0. The bottom panel is for the Sigma UPS1 spike-in peptides, whose relative intensities were expected to be centered on the threefold difference reference line at [–log2(3)] (approximately [-1.58]). The dark gray curve (two-dash) shows the original relative intensities M (before normalization). The blue curve (dot-dash) represents the normalized relative intensities M using peptide abundance (A). The purple curve (long-dash) is for the normalized relative intensities M using Rank1 variable only. The black curve (solid) is for the normalized relative intensities using all variables.
Gain in the stage of data preprocessing directly lead to better sensitivity and specificity for detection of the mock-biomarkers. Supplemental Fig. S6A shows ROC curves using fold-changes for spike-in concentration C versus D (Sample 6C with Run #7, 8, 9 and Sample 6D with Run #10, 11, 12 on Orbitrap 65 in Study 6). This figure shows the false positive rate (FPR = 1-specificity) and true positive rate (sensitivity), when using fold-change thresholds ranging from 1.5 to 6 at intervals of 0.5. Each of the runs from sample 6C (lower concentration) was used in the numerator to calculate M in a pair. The results are shown in panel (a) for pairs using run 7 as the base run; (b) for pairs using run 8 as the base run; (c) for pairs using run 9 as the base run. Normalization with all variables lead to a higher true positive rate, especially for fold-change thresholds below 4, and a consistently lower false positive rate compared with the results before normalization or normalization with the abundance (A) only. Also notably, the ROC curves after normalization with all variables did not vary much using different base runs, whereas base run choice significantly affected the ROC curves for data before normalization. At the true fold change (= 3), the FPRs were improved dramatically after normalization with all variables compared with those before normalization or normalization with the abundance only, independent of the large differences in base run (Table I).
Table I. The sensitivity and false positive rate (FPR = 1-specificity) with the threefold decision criterion for Sample 6C (yeast + UPS1 at 2.2 fmol/μl) against Sample 6D (yeast + UPS1 at 6.7 fmol/μl) from the first lab in Study 6. Each of the runs from Sample 6C (run #7, 8, 9) was used in the numerator (base run) to calculate the relative intensities (M) in a pair with the runs from Sample 6D (run #10, 11, 12). The results in Column “before” used data before normalization. The results in Column “With A only” used data normalized by the abundance (A) only. The results in Column “With all variables” used data normalized by all variables. The normalization was based on common peptides selected using the global rank-invariant set in (18).
| Sensitivity |
FPR |
|||||
|---|---|---|---|---|---|---|
| Before | With A only | With all variables | Before | With A only | With all variables | |
| Base run = 7 | 0.78 | 0.59 | 0.66 | 0.023 | 0.018 | 0.011 |
| Base run = 8 | 0.31 | 0.56 | 0.78 | 0.016 | 0.018 | 0.011 |
| Base run = 9 | 0.28 | 0.63 | 0.75 | 0.013 | 0.016 | 0.011 |
A rigorous discussion and comparison of different common peptide selection methods is beyond the scope of the current study. For a simple comparison in the current report, we also normalized and ranked variables based on known identities of peptide ions. That is, only the yeast peptide ions were used in normalization and ranking whereas the intensities of the spike-in Sigma UPS1 peptide ions were interpolated based on the models estimated using only yeast peptide ions. Results are included in Supplementary Data (supplemental Fig. S6B and supplemental Table S1). Though results still support the normalization by all variables, the selection of common peptide ions did have an impact on sensitivity and specificity in the biomarker discovery analysis. We suggest that the common set of peptide ions should be selected on the basis of biological consideration (16) or internal standards if possible. Data-driven methods may be used but caution is recommended.
DISCUSSION
The goal of this study was to identify and characterize systematic biases present in proteomics data, and then to attempt to develop a better normalization method. This was accomplished by examining the effects of loading on individual peptide ions as well as by monitoring other unidentified biases between technical replicates. Peptide property variables were used as reporters to identify biases which manifest as functions of retention time or one of several other variables. Callister et al. state that the most appropriate normalization techniques are ideally developed following identification and characterization of systematic biases (9). Here we described peptides with higher charge state potentials to be more sensitive to loading differences. Local deviations were also likely related to electrospray issues.
We also demonstrated that systematic biases are almost always present. Moreover, we observed that these effects are not predictable and local, nonlinear corrections are almost always necessary. A LOWESS regression model can be used to overcome the nonlinearity. However, it involves the choice of a fraction of neighboring samples (bandwidth) in the normalization step. Berger et al. demonstrate that bandwidth parameter choice significantly affects results (15). Semi-parametric regression avoids this problem by selecting the number of knots, resulting in statistical stability as long as the knots are dense enough (17, 19). Additionally, the semiparametric regression models are constructed within the framework of linear regression models. Linear model diagnostics and evaluation methods can be directly used; its computation can be easily implemented using the widely available mixed model packages.
We have developed an iterative normalization that does not require the user to be knowledgeable of the largest systematic biases present in their data prior to normalization. This was achieved by ranking variables along with stepwise normalization. Although this model proved effect in the analysis of the CPTAC Study 6 data, it will be necessary in follow-up studies to examine the effectiveness in the face of biological variability.
The products of this work are improvements to the NIST MSQC pipeline, which now performs the necessary calculations to be used for label-free analysis, and the R code to perform normalization. The pipeline has been tested with Thermo LTQ, Orbitrap and FT, and to a lesser extent, with Agilent QTOF and AB TripleTofTM 5600 data. The goal of this work was to unveil systematic biases in these proteomics data sets in order to help validate the appropriateness of these methods for quantitative workflows. Without first understanding the effects of systematic biases, interpretation of label-free data sets are subject to so-called “batch effects” and other incorrect assumptions about the samples underlying the data. These anomalies can lead to costly testing of irrelevant hypotheses, particularly in biomarker discovery efforts. Just as in microarray experiments, these biases can be effectively eliminated by applying appropriate statistical methods.
Supplementary Material
Footnotes
Author contributions: P.A.R. designed research; P.A.R. performed research; X.Y. and S.E.S. contributed new reagents or analytic tools; P.A.R., X.W., and S.E.S. analyzed data; P.A.R. and X.W. wrote the paper; N.S. and S.E.S. supervisor.
* This work was supported by IAA ACO13004 (“Proteomics Measurement Quality Assurance Program”), from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC) and National Institute of Standards and Technology. XW was partially supported by the Association for Women in Mathematics National Science Foundation Travel Grant and the Taft Travel Grant at the University of Cincinnati.
 This article contains supplemental Figs. S1 to S6 and Table S1.
This article contains supplemental Figs. S1 to S6 and Table S1.
Disclaimer: Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
1 The abbreviations used are:
- MS1
- full MS scan
- MS2
- tandem MS scan
- MS/MS
- tandem MS scan
- NCI
- National Cancer Institute
- NIST
- National Institute of Standards and Technology
- NIST MSQC
- NIST Mass Spectrometry Quality Control (Software)
- SOP
- standard operating procedure
- CPTAC
- Clinical Proteomics Technology Assessment for Cancer
- MD
- mean of deviance
- ROC
- receiver operating characteristic
- RT
- retention time
- PSM
- peptide-spectrum match
- SILAC
- stable isotope labeling with amino acids in cell culture
- GUI
- graphical user interface.
REFERENCES
- 1. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 2. Mueller L. N., Brusniak M. Y., Mani D. R., Aebersold R. (2008) An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res 7, 51–61 [DOI] [PubMed] [Google Scholar]
- 3. Florens L., Carozza M. J., Swanson S. K., Fournier M., Coleman M. K., Workman J. L., Washburn M. P. (2006) Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jaffe J. D., Keshishian H., Chang B., Addona T. A., Gillette M. A., Carr S. A. (2008) Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol. Cell. Proteomics 7, 1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Silva J. C., Gorenstein M. V., Li G.-Z., Vissers J. P. C., Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 6. Leek J. T., Scharpf R. B., Bravo H. C., Simcha D., Langmead B., Johnson W. E., Geman D., Baggerly K., Irizarry R. A. (2010) Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11, 733–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baggerly K. A., Coombes K. R., Morris J. S. (2005) Bias, randomization, and ovarian proteomic data: a reply to “producers and consumers.” Cancer Inform. 1, 9–14 [PMC free article] [PubMed] [Google Scholar]
- 8. Gregori J., Villarreal L., Méndez O., Sánchez A., Baselga J., Villanueva J. (2012) Batch effects correction improves the sensitivity of significance tests in spectral counting-based comparative discovery proteomics. J. Proteomics 75, 3938–3951 [DOI] [PubMed] [Google Scholar]
- 9. Callister S. J., Barry R. C., Adkins J. N., Johnson E. T., Qian W., Webb-Robertson B. J. M., Smith R. D., Lipton M. S. (2006) Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res. 5, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Webb-Robertson B.-J. M., Matzke M. M., Jacobs J. M., Pounds J. G., Waters K. M. (2011) A statistical selection strategy for normalization procedures in LC-MS proteomics experiments through dataset-dependent ranking of normalization scaling factors. Proteomics 11, 4736–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kultima K., Nilsson A., Scholz B., Rossbach U. L., Fälth M., Andrén P. E. (2009) Development and evaluation of normalization methods for label-free relative quantification of endogenous peptides. Mol. Cell. Proteomics 8, 2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paulovich A. G., Billheimer D., Ham A.-J. L., Vega-Montoto L., Rudnick P. A., Tabb D. L., Wang P., Blackman R. K., Bunk D. M., Cardasis H. L., Clauser K. R., Kinsinger C. R., Schilling B., Tegeler T. J., Variyath A. M., Wang M., Whiteaker J. R., Zimmerman L. J., Fenyo D., Carr S. A., Fisher S. J., Gibson B. W., Mesri M., Neubert T. A., Regnier F. E., Rodriguez H., Spiegelman C., Stein S. E., Tempst P., Liebler D. C. (2010) Interlaboratory study characterizing a yeast performance standard for benchmarking LC-MS platform performance. Mol. Cell. Proteomics 9, 242–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rudnick P. A., Clauser K. R., Kilpatrick L. E., Tchekhovskoi D. V., Neta P., Blonder N., Billheimer D. D., Blackman R. K., Bunk D. M., Cardasis H. L., Ham A.-J. L., Jaffe J. D., Kinsinger C. R., Mesri M., Neubert T. A., Schilling B., Tabb D. L., Tegeler T. J., Vega-Montoto L., Variyath A. M., Wang M., Wang P., Whiteaker J. R., Zimmerman L. J., Carr S. A., Fisher S. J., Gibson B. W., Paulovich A. G., Regnier F. E., Rodriguez H., Spiegelman C., Tempst P., Liebler D. C., Stein S. E. (2010) Performance metrics for liquid chromatography-tandem mass spectrometry systems in proteomics analyses. Mol. Cell. Proteomics 9, 225–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ruppert D. (2002) Selecting the number of knots for penalized splines. J. Comput. Graph. Stat. 11, 735–757 [Google Scholar]
- 15. Berger J. A., Hautaniemi S., Järvinen A.-K., Edgren H., Mitra S. K., Astola J. (2004) Optimized LOWESS normalization parameter selection for DNA microarray data. BMC Bioinformatics 5, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wit E., McClure J. (2004) Statistics for microarrays: Design, analysis and inference. John Wiley & Sons, Ltd., Chichester, UK [Google Scholar]
- 17. Wang W., Zhou H., Lin H., Roy S., Shaler T. A., Hill L. R., Norton S., Kumar P., Anderle M., Becker C. H. (2003) Quantification of Proteins and Metabolites by Mass Spectrometry without Isotopic Labeling or Spiked Standards. Anal. Chem. 75, 4818–4826 [DOI] [PubMed] [Google Scholar]
- 18. Pelz C. R., Kulesz-Martin M., Bagby G., Sears R. C. (2008) Global rank-invariant set normalization (GRSN) to reduce systematic distortions in microarray data. BMC Bioinformatics 9:520, doi:10.1186/1471-2105-9-520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruppert R. J. (2003) Semiparametric regression Cambridge University Press, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.