Abstract
Purpose
Epstein-Barr virus (EBV)-positive diffuse large B-cell lymphoma (DLBCL) of the elderly is a variant of DLBCL with worse outcome that occurs most often in East Asian countries and is uncommon in the Western hemisphere. We studied the largest cohort of EBV+ DLBCL, independent of age, treated with R-CHOP in developed Western countries.
Experimental design
A large cohort (n=732) of patients with DLBCL treated with R-CHOP chemotherapy are included from the multicenter Consortium. This study group has been studied for expression of different biomarkers by immunohistochemistry, genetic abnormalities by fluorescent in situ hybridization and mutation analysis, genomic information by gene expression profiling (GEP) and gene set enrichment analysis (GSEA).
Results
Twenty-eight patients (4.0%) were positive for EBV with a median age of 60.5 years. No clinical characteristics distinguished patients with EBV+ DLBCL from patients with EBV-negative DLBCL. Genetic aberrations were rarely seen. NF-κB p50, phosphorylated STAT-3 and CD30 were more commonly expressed in EBV+ DLBCLs (P<.05). Significant differences in survival were not observed in patients with EBV-positive DLBCL versus EBV-negative DLBCL. CD30 co-expression appeared to confer inferior outcome although statistical significance was not achieved. GEP showed a unique expression signature in EBV-positive DLBCL. GSEA revealed enhanced activity of the NF-κB and JAK/STAT pathways.
Conclusions
The clinical characteristics of patients with EBV+ versus EBV-negative DLBCL are similar and EBV infection does not predict a worse outcome. EBV+ DLBCL, however, has a unique genetic signature. CD30 expression is more common in EBV+ DLBCL and, when present, is associated with an adverse outcome.
Keywords: Epstein-Barr virus, diffuse large B-cell lymphoma, CD30, p50, p65, c-Rel, phosphorylated STAT3
Introduction
Epstein-Barr virus (EBV) positive (+) diffuse large B-cell lymphoma (DLBCL) of the elderly is defined in the 2008 World Health Organization (WHO) classification as an EBV+ monoclonal large B-cell lymphoproliferative disorder in immunocompetent patients > 50 years (1). The median age of patients with these tumors is 71 years (range, 50–91), but younger patients can be affected (2, 3). The reported prevalence of EBV is more frequent in East Asia and Mexico compared with Western countries, but this concept has been challenged (4–8). Of note, no consensus has been reached regarding the definition or criteria, including a methodology to detect EBV infection, separation of EBV-positive tumor cells from EBV-positive background cells, cutoff for EBV-positive cells for EBV positivity. The lack of consensus may impact differences in the reported prevalence. Lymph nodes are commonly involved (~70%), and extranodal sites of involvement are not uncommon (3).
Although prognostic significance has not been demonstrated, two morphologic variants – monomorphic and polymorphic - have been recognized. Recently, Montes-Moreno et al subdivided the polymorphic subtype based on the relative proportion of large neoplastic cells and the presence of Hodgkin Reed-Sternberg-like cells into 3 groups: 1) canonical large B-cell neoplasm; 2) DLBCL with Hodgkin lymphoma-like features; and 3) DLBCL with polymorphic lymphoproliferative disorder-like features (9). Immunophenotypically, most cases have activated B-cell phenotype. EBV latent membrane protein 1 (LMP1) is expressed more than 90% of cases, which denotes EBV latency type II. EBV nuclear antigen 2 (EBNA2), seen in EBV latency type III, is expressed approximately 15–30% of cases. Specific cytogenetic and genomic aberrations are not known.
In reports from the East Asia, South America and Europe, a poor outcome has been observed in patients with EBV+ DLBCL of the elderly (5, 8, 10, 11). In these studies, however, many patients were treated with cyclophosphamide, hydroxydoxorubicin, vincristine and prednisone (CHOP) and not Rituximab (R)-CHOP. Therefore, the effect of R-CHOP immunochemotherapy in patients with EBV+ DLBCL of the elderly is largely unknown. In this study, we assessed the effect of EBV infection in a large cohort of DLBCL patients treated with R-CHOP in Western developed countries.
Materials and Methods
Patients
A total of 732 cases (training set, n=500 and validation set, n=232) of de novo DLBCL treated with R-CHOP were evaluated. Formalin-fixed and paraffin-embedded lymphoma samples were brought into tissue microarrays (TMA) as part of the International DLBCL Rituximab-CHOP Consortium Program Study. All cases were reviewed by a group of hematopathologists (A.T., M.B.M., M.A.P., and K.H.Y.) and were diagnosed according to the WHO criteria. DLBCLs transformed from a low-grade B-cell lymphoma or associated with acquired immunodeficiency (e.g. human immunodeficiency virus infection), primary cutaneous DLBCLs, primary central nervous system DLBCLs, and primary mediastinal large B-cell lymphomas were excluded. We did not exclude patients younger than 50 years. Morphologic variants of EBV+ DLBCL were classified as described by Montes-Moreno et al (9). This study was conducted in accordance with Declaration of Helsinki and was approved by the IRBs of all participating collaborative institutions (12, 13). The overall study was approved by the Institutional Review Board at The University of Texas MD Anderson Cancer Center in Houston, Texas, USA.
Immunohistochemistry and In Situ Hybridization Methods
Tissue microarrays were constructed as described previously (12, 13). Immunohistochemical analysis (IHC) for various markers and in situ hybridization (ISH) for Epstein-Barr virus-encoded RNA (EBER) were performed. Evaluated IHC markers were B-cell lymphoma 2 (BCL2), B-cell lymphoma 6 (BCL6), CD10, CD30, Forkhead box protein P1 (FOXP1), Germinal Center B cell-expressed Transcript-1 (GCET1), MDM2, MDM4, Multiple Myeloma Oncogene 1 (MUM1), Epstein-Barr Virus Latent Membrane Protein 1 (LMP1), Epstein-Barr Virus nuclear antigen 2 (EBNA2), Myc, Nuclear factor-κB (NF-κB) components (p50, p65, RelB and c-Rel), p53, and phosphorylated signal transducer and activator of transcription 3 (pSTAT3). Receiver-operating characteristic (ROC) curve analysis described previously (14) was utilized to assess a cutoff with maximum sensitivity and specificity for each marker. When an optimal cutoff could not be determined by ROC curve analysis, a conventional cutoff value for individual markers was decided based on a literature review. The cutoff scores for these markers were as follows: 10% for LMP1, MDM2, MDM4 and EBER; 20% for CD30 and p53; 30% for CD10, BCL6 and pSTAT3; 40% for Myc; 60% for GCET1, MUM1, and FOXP1; 70% for BCL2. Any nuclear expression of each NF-κB component was considered positive.
Gene Expression Profiling and Gene Set Enrichment Analysis
Total RNA was extracted from 474 formalin-fixed, paraffin-embedded tissue samples in the training set using the HighPure RNA Extraction Kit (Roche Applied Science, Indianapolis, IN) and subjected to gene expression profiling (GEP) as described previously (14). We used the DQN algorithm, which is the non-central trimmed mean of differences between perfect match and mismatch intensities with quantile normalization, for data analysis and classification (15). DQN was normalized with beta distribution and a Bayesian model was used to determine the classification probability. The methodology developed in this study has been validated with the Lymphoma Leukemia Molecular Profiling Program dataset in the Gene Expression Omnibus Genomics Spatial Event database #10846, which has 181 CHOP-treated and 233 R-CHOP-treated DLBCL patients. We obtained an 80% concordance rate of classification for all 3 classes, germinal center B-cell (GCB), activated B-cell (ABC), and unclassified, and a 97% rate for GCB and ABC (excluding unclassified). Cell-of-origin (COO) classification was established by GEP (considered the “gold standard”) and/or in combination with IHC data. When COO is not classifiable with GEP, it was determined by IHC according to Visco-Young (the first choice) and Choi (the second choice) algorithms (14, 16). The correlation between GEP and IHC for COO classification was 86.3% overall (17).
Gene set enrichment analysis was performed with GSEA application (Broad Institute at MIT, Cambridge, MA) using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway gene sets and the curated gene sets from the Molecular Signature Database. Gene sets with a false discovery rate q value <0.05 after performing 1,000 permutations were considered to be significantly enriched.
Fluorescence In Situ Hybridization for BCL2, BCL6, MYC, MDM2 and MDM4 and Sequencing of TP53
Fluorescence in situ hybridization (FISH) analysis was performed on formalin-fixed, paraffin-embedded tissue sections using BCL2 and BCL6 dual-color break-apart probes (Vysis, Downers Grove, IL), MYC locus-specific IGH/MYC/CEP8 tricolor dual-fusion probes and a locus-specific MYC dual-color break-apart probe (Vysis, Downers Grove, IL), and MDM2 and MDM4 probes as described previously (12, 14). TP53 sequencing was performed using extracted genomic DNA from formalin-fixed, paraffin-embedded tissue in the training set. The coding sequence (exons 2–11) and splicing sites were sequenced using p53 AmpliChip (Roche Molecular Systems, Pleasanton, CA) as described (13). Sequences data was compared with the TP53 reference sequence (NC_000017.10) in the Genbank database for data analysis.
Statistical Analysis
Clinical and laboratory features were compared with the Fisher’s exact test for categorical variables and Mann-Whitney U test for continuous variables. Overall survival (OS) and progression-free survival (PFS) were defined from the date of diagnosis to the date of last follow-up or death and from the date of diagnosis to the date of progression or death, respectively. Survival distributions were estimated with the Kaplan-Meier method, with difference compared by the log-rank test. Multivariate analysis was performed using the Cox proportional hazards regression model. All differences with p<0.05 were considered to be statistically significant. SPSS Statistics V21.0 (Chicago, IL, USA) was used for statistical analyses.
Results
Clinicopathologic and Genetic features of EBV+ DLBCL
EBER was positive in 28 (4.0%) and negative in 675 (96.0%) cases of DLBCL. When a cutoff of ≥30% or ≥50% was used for EBER expression, EBER was positive for 24 (3.3%) and 16 (2.2%). Seven patients were <50 years. For 29 cases EBER expression could not be assessed due to exhaustion of tissue blocks. LMP1 was positive in 16 (66.7%) and EBNA2 was positive in 6 (26.1%) cases of EBER+ cases. Based on expression of EBER, LMP1, and other EBV viral antigens, eight (34.8%), 9 (39.1%) and 6 cases (26.1%) were for EBV latency I, II and III, respectively. Expression of LMP1 or EBNA2 in EBER+ DLBCL did not show worse outcome in OS (P=.583 and P=.750 for LMP1 and EBNA2, respectively). Latency type did not demonstrate a significant overall survival stratification (P=.793).
Various clinical characteristics were compared between EBER+ and EBER-negative DLBCLs and there were no significant differences (Table 1). Morphologically, four cases were monomorphic subtype, 8 cases were polymorphic subtype, canonical large B-cell neoplasm variant, 8 cases were polymorphic subtype, Hodgkin-like variant and 8 cases were polymorphic subtype, lymphoproliferative disorder-like variant (Figure S1) according to Montes-Moreno’s subclassification scheme (9). There was no significant difference in OS and PFS among different morphologic variants (P=.915 and P=.931, respectively). Eleven cases were GCB and 17 cases were ABC subtype.
Table 1.
Clinical characteristics and outcome of 732 cases of de novo DLBCL with R-CHOP regimen
| All patients (n=732) | EBV+ (n=28) | EBV− (n=675) | P value | |
|---|---|---|---|---|
| Median age | 63.0 (16.0–95.0) | 60.5 (35.0–86.0) | 63.5 (16.0–95.0) | 0.426 |
| Gender | ||||
| Male | 421 (57.5%) | 18 (64.3%) | 387 (57.3%) | |
| Female | 311 (42.5%) | 10 (35.7%) | 288 (42.7%) | 0.560 |
| Age | ||||
| ≤60 | 313 (42.8%) | 14 (50.0%) | 283 (41.9%) | |
| >60 | 419 (57.2%) | 14 (50.0%) | 392 (58.1%) | 0.438 |
| B symptoms | ||||
| Absence | 423 (64.4%) | 14 (58.3%) | 397 (64.8%) | |
| Presence | 234 (35.6%) | 10 (41.7%) | 216 (35.2%) | 0.521 |
| ECOG performance status | ||||
| <2 | 529 (84.5%) | 19 (73.1%) | 488 (85.2%) | |
| ≥2 | 97 (15.5%) | 7 (26.9%) | 85 (14.8%) | 0.099 |
| Stage | ||||
| I–II | 329 (46.4%) | 11 (39.3%) | 305 (46.8%) | |
| III–IV | 380 (53.6%) | 17 (60.7%) | 347 (53.2%) | 0.562 |
| Extranodal sites | ||||
| <2 | 529 (79.0%) | 22 (84.6%) | 487 (79.1%) | |
| ≥2 | 141 (21.0%) | 4 (15.4%) | 129 (20.9%) | 0.626 |
| LDH | ||||
| Normal | 246 (37.7%) | 8 (29.6%) | 228 (38.1%) | |
| Elevated | 407 (62.3%) | 19 (70.4%) | 371 (61.9%) | 0.423 |
| IPI score | ||||
| 0–2 | 401 (59.3%) | 16 (57.1%) | 368 (59.4%) | |
| 3 to 5 | 275 (40.7%) | 12 (42.9%) | 252 (40.6%) | 0.846 |
| Tumor size (cm) | ||||
| <6 | 354 (64.1%) | 10 (58.8%) | 335 (64.7%) | |
| ≤6 | 198 (35.9%) | 7 (41.2%) | 183 (35.3%) | 0.615 |
| Treatment response | ||||
| CR/PR | 612 (89.2%) | 24 (88.9%) | 562 (89.2%) | |
| No response | 74 (10.8%) | 3 (11.1%) | 68 (10.8%) | 1.000 |
ECOG; Eastern Cooperative Oncology Group, IPI; International Prognostic Index, CR; Complete Remission, PR; Partial Remission
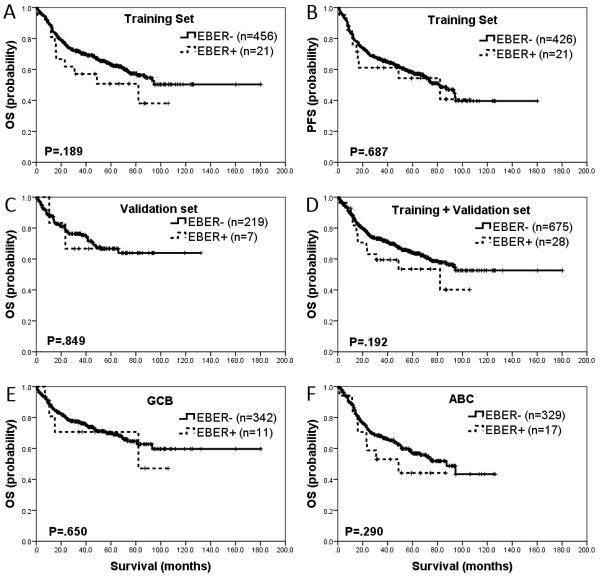
With a median follow-up of 42.1 months, patients with EBV+ DLBCL did not show any significant differences from EBV-negative DLBCL patients with respect to OS and PFS in the training set (P=.189 and P=.687, respectively) (Figure 1-A and 1-B). This finding was reproduced in the validation set (P=.849) (Figure 1-C) and in the combined set (P=.192) (Figure 1-D). Information on PFS was not available in the validation set. Combining the training and validation sets, the 5-year OS was 54% for patients with EBV+ DLBCL compared with 64% for patients with EBV-negative DLBCL (P=.321). The 5-year PFS was 57% and 58% in these groups, respectively (P=.867). When broken down into GCB and ABC subtypes, OS was not significantly different between patients with EBV+ versus EBV-negative DLBCL (P=.650 and P=.290, respectively) (Figures 1-E and 1-F). Excluding the 7 patients who were <50 years, there was no significant difference in OS and PFS between EBV+ versus EBV-negative DLBCL (P=.173)
Figure 1. Impact of Epstein-Barr virus (EBV) infection on overall and progression-free survival of patients with diffuse large B-cell lymphoma (DLBCL).
A and B. In the training set (n=500), overall survival (OS) and progression free survival (PFS) were not significantly different between EBV+ DLBCL and EBV− DLBCL (P=.189 and P=.687, respectively). C. Lack of difference in OS between EBV+ DLBCL and EBV− DLBCL was reproduced (P=.849) in the validation set (n=232). D. Combining the training set and validation set, OS was not significantly different between EBV+ DLBCL and EBV− DLBCL (P=.192). E and F. EBV positivity in DLBCL also failed to show any significant difference in cases of germinal center B-cell (GCB) versus activated B-cell (ABC) phenotype, OS (P=.650 and P=.290, respectively).
At the protein level, BCL6 was more commonly expressed in EBV-negative DLBCLs compared to EBV+ cases (76.1% vs. 50%, P =.003) (Table 2). All the other markers did not show any significant difference between the two groups. In EBV+ cases, LMP1 expression did not demonstrate significant correlation (P>.05) with either Myc protein expression or Myc/BCL2 double protein expression. All tested genetic aberrations involving BCL2, BCL6, MYC, MDM2, MDM4 and TP53 were infrequently observed in EBV+ DLBCLs. Of note, no cases of EBV+ DLBCL showed rearrangements of both MYC and BCL2. In multivariate analysis, EBV positivity did not have an increased hazard ratio (Table 3).
Table 2.
Protein expression and genetic aberrations of 732 cases of de novo DLBCL with R-CHOP regimen
| All patients (n=732) | EBV+ (n=28) | EBV− (n=675) | p value | |
|---|---|---|---|---|
| COO class | ||||
| GCB | 370 (51.0%) | 11 (39.3%) | 342 (51.0%) | - |
| ABC | 355 (49.0%) | 17 (60.7%) | 329 (49.0%) | 0.251 |
| CD30 | 118 (16.4%) | 12 (42.9%) | 103 (15.5%) | 0.001 |
| NF-κB | ||||
| p50 | 237 (36.9%) | 15 (65.2%) | 220 (35.9%) | 0.007 |
| p65 | 191 (28.6%) | 7 (25.9%) | 182 (28.8%) | 0.831 |
| c-Rel | 148 (23.2%) | 5 (21.7%) | 139 (22.8%) | 1.000 |
| pSTAT3 | 221 (35.5%) | 13 (56.5%) | 206 (34.9%) | 0.044 |
| BCL2 protein | 359 (49.9%) | 11 (39.3%) | 339 (50.6%) | 0.254 |
| BCL6 protein | 537 (75.1%) | 14 (50.0%) | 507 (76.1%) | 0.003 |
| p53 protein | 260 (36.9%) | 8 (34.8%) | 240 (36.5%) | 1.000 |
| MDM2 protein | 202 (42.3%) | 12 (63.2%) | 179 (40.7%) | 0.059 |
| MDM4 protein | 468 (97.7%) | 18 (94.7%) | 432 (97.7%) | 0.374 |
| Myc protein | 440 (60.9%) | 17 (60.7%) | 413 (61.7%) | 1.000 |
| Myc and BCL2 | 237 (33.0%) | 8 (28.6%) | 225 (33.7%) | 0.685 |
| BCL2 rearrangement | 79 (13.3%) | 1 (5.0%) | 76 (13.4%) | 0.497 |
| BCL6 rearrangement | 120 (25.2%) | 1 (6.3%) | 118 (26.2%) | 0.083 |
| TP53 mutation | 109 (22.0%) | 2 (10.0%) | 102 (22.6%) | 0.271 |
| MDM2 aberration | 3 (0.8%) | 1 (7.1%) | 2 (0.6%) | 0.108 |
| MDM4 aberration | 2 (0.5%) | 1 (7.1%) | 1 (0.3%) | 0.073 |
| MYC rearrangement | 40 (8.4%) | 2 (11.1%) | 38 (8.4%) | 0.660 |
| MYC/BCL2 double* | 11 (1.8%) | 0 (0%) | 11 (1.9%) | 1.000 |
Cutoff for each protein biomarker was found in the “Material and Methods”.
Both MYC and BCL2 are rearranged.
Table 3.
Multivariate analysis of clinicopathologic parameters in 732 cases of de novo DLBCL with R-CHOP regimen
| OS | PFS | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age >60 | 1.697 | 1.090–2.642 | 0.019 | 1.525 | 0.934–2.491 | 0.092 |
| Gender | 0.848 | 0.596–1.206 | 0.358 | 0.975 | 0.670–1.418 | 0.894 |
| B symptoms | 1.116 | 0.771–1.615 | 0.561 | 1.193 | 0.802–1.774 | 0.385 |
| ECOG 2 | 1.936 | 1.244–3.012 | 0.003 | 1.738 | 1.073–2.816 | 0.025 |
| Stage III or IV | 2.757 | 1.615–4.708 | <0.001 | 2.552 | 1.445–4.509 | 0.001 |
| Extranodal sites ≥2 | 1.603 | 1.038–2.476 | 0.033 | 1.794 | 1.112–2.894 | 0.017 |
| Serum LDH, Elevated | 1.477 | 0.974–2.240 | 0.066 | 1.724 | 1.087–2.732 | 0.021 |
| IPI score ≥3 | 0.992 | 0.526–1.871 | 0.981 | 0.817 | 0.398–1.676 | 0.582 |
| Size ≥6 cm | 1.166 | 0.818–1.663 | 0.396 | 1.230 | 0.839–1.804 | 0.288 |
| EBER + | 1.590 | 0.691–3.658 | 0.276 | 1.480 | 0.642–3.411 | 0.358 |
CI; confidence interval, ECOG; Eastern Cooperative Oncology Group, IPI; International Prognostic Index
Expression of NF-κB components and phosphorylated STAT3 in EBV+ DLBCL
Components of NF-κB (p50, p65 and c-Rel) and pSTAT3 were assessed by IHC. Expression of p50 was more commonly observed in EBV+ DLBCLs compared with EBV-negative DLBCLs (65.2% vs. 35.9%, P=.007). In contrast, expression of p65 and C-rel showed no difference between the two groups. These findings could suggest that the canonical NF-κB pathway is activated utilizing both p65 and c-Rel as dimerization partners for p50. Of note, nuclear expression of canonical NF-κB molecules was also expressed in GCB subtype of EBV+ DLBCLs (55.6%, 54.8% and 33.3% for p50, p65 and c-Rel, respectively), suggesting EBV-associated canonical NF-κB induction. However, single LMP1 or EBNA2 did not show significant correlation with any of the NF-κB molecules (all P>.05). pSTAT3 was more commonly expressed in EBV+ DLBCLs compared to EBV-negative DLBCLs (56.5% vs. 34.9%, P=.044). However, pSTAT3 did not show significant correlation with LMP1 (P>.05).
CD30 Expression Predicts Adverse Outcome in Patients with EBV+ DLBCL
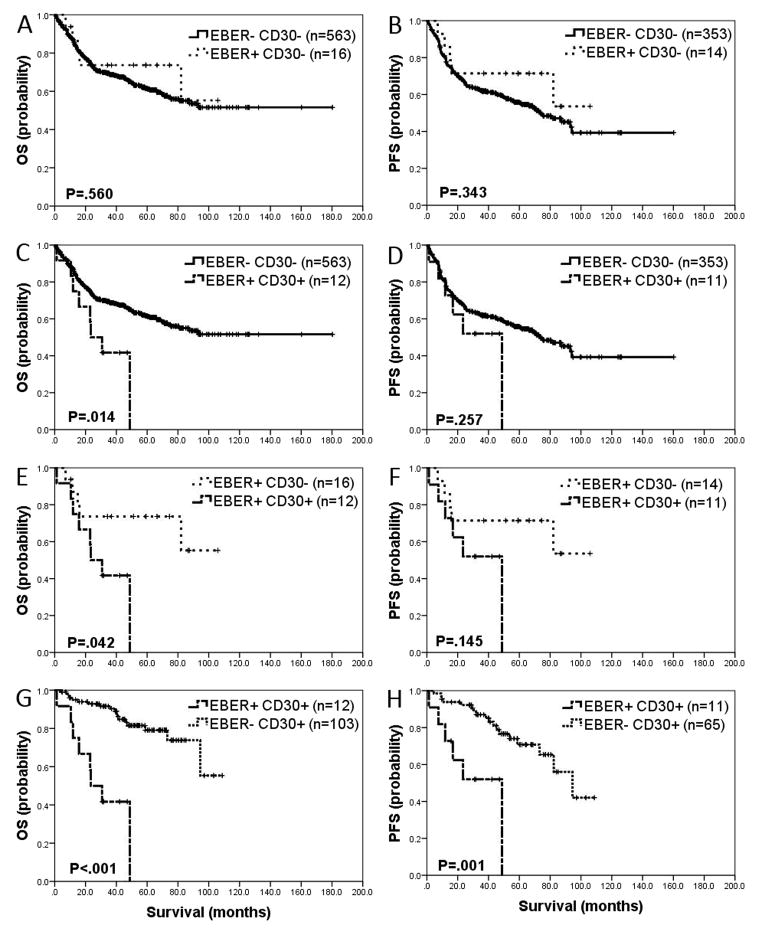
Our group previously found co-expression of CD30 and EBER in de novo DLBCLs harbor poor prognosis (12). To evaluate the effect of CD30 in EBV+ DLBCL, we analyzed four groups based on expression of EBER and CD30. CD30 expression was significantly higher in EBV+ DLBCLs compared with EBV-negative DLBCLs (42.9% vs. 15.5%, respectively, P=.001). When EBER+/CD30-negative, OS and PFS were not significantly different from EBER-negative/CD30-negative DLBCL (P=.560 and P=.343 for OS and PFS, respectively) (Figure 2-A and 2-B). However, when CD30 was co-expressed with EBER in DLBCL, a worse outcome in OS, but not PFS, was observed compared with EBER−/CD30− (P=.014 and P=.257 for OS and PFS, respectively) (Figure 2-C and 2-D). Comparing EBER+/CD30+ DLBCL with EBER+/CD30-negative DLBCL, inferior outcome in OS, but not in PFS, was seen in the former (P=.042 and P=.145 for OS and PFS, respectively) (Figure 2-E and 2-F). EBER+/CD30+ DLBCLs had significantly poor outcome compared with EBER-negative/CD30+ counterparts (P<.001 and P=.001 for OS and PFS, respectively) (Figure 2-G and 2-H).
Figure 2. Impact of CD30 expression and Epstein-Barr virus (EBV) infection on overall survival and progression-free survival of diffuse large B-cell lymphoma (DLBCL) patients.
A and B. Compared to patients with EBER− CD30− DLBCL, patients with EBER+ CD30− patients did not have significantly different OS (P=.560) and PFS (P=.343). C and D. Co-expression of EBER and CD30 in DLBCL showed worse OS (P=.014) compared to patients with EBER− CD30− DLBCL. However, PFS was not significantly different (P=.257) between the two groups. E and F. Worse outcome was observed in patients with EBER+ CD30+ DLBCL compared with EBER+ CD30− DLBCL in OS (P=0.042), but not in PFS (P=.145). G and H. Patients with EBER+ CD30+ DLBCL have significantly worse OS and PFS compared to patients with EBER− CD30+ DLBCL (P<.001 and P=.001, respectively).
Gene Expression Signature and Gene Set Enrichment Analysis of EBV+ DLBCL
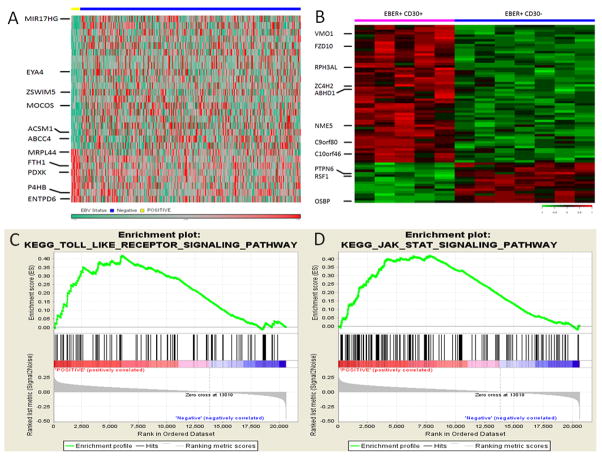
We evaluated GEP signatures of EBV+ and EBV-negative DLBCL to better understand molecular mechanisms involved in EBV+ DLBCL (Figure 3-A). A total of 24 genes were differentially expressed between the two groups. In EBV+ DLBCL, 7 genes were upregulated and 17 genes were downregulated (Table S1). Among the upregulated genes, FTH1, PDXK, MRPL44, P4HB and ENTPD6 were noteworthy. FTH1 is a known NF-κB target gene that denotes an activated NF-κB pathway. PDXK is positively related to cell proliferation. MRPL44 involves cell cycle regulation and P4HB and ENTPD6 are involved in cell metabolism. Among the downregulated genes, MIR17HG, MOCOS and EYA4 were prominent. Downregulation of MIR17HG has been shown in CD30+ DLBCL by Hu et al (12), and our finding underscores an association between CD30 expression and EBV infection. MOCOS sulfurates the molybdenum cofactor, essential for the enzyme activity of aldehyde oxidase. Since aldehyde oxidase is a major enzyme involving drug metabolism and clearance, downregulation of MOCOS could be a possible mechanism of chemotherapeutic resistance (18). EYA4 is a gene involved in DNA repair and EYA4 downregulation has been associated with poor clinical outcome in patients with non-Hodgkin lymphoma (19).
Figure 3. Gene expression profiling and gene set enrichment assay (GSEA) in EBV+ diffuse large B-cell lymphoma (DLBCL).
A. A unique gene expression signature was found in EBV+ DLBCL compared with EBV-negative DLBCL. B. DLBCLs with co-expression of EBER and CD30 showed a gene expression signature distinct from cases with EBER+ CD30−. C and D. GSEA validated enhanced activity of NF-κB pathway and JAK/STAT pathway in EBV+ DLBCL.
We further conducted GEP in EBER+/CD30+ DLBCL and EBER+/CD30-negative DLBCL. A total of 68 genes were differentially expressed between the two groups (Figure 3-B). Among 44 upregulated genes in EBER+/CD30+ DLBCL, NME5 has an anti-apoptotic effect, C10orf46 promotes cell proliferation, and ZC4H2 induces cell cycle progression. Among 14 genes downregulated in EBER+/CD30+ DLBCL, RSF1, PTPN6 and OSBP were conspicuous. Downregulation of RSF1, which normally represses the NF-κB pathway, might contribute to NF-κB activation. PTPN6 and OSBP normally dephosphorylate substrates and downregulation of these genes could enhance a variety of signal transduction pathways.
We used KEGG gene sets for GSEA. The Toll-like receptor (TLR) and RIG-1-like receptor signaling pathways were significantly enriched in EBV+ DLBCL (Figure 3-C). These pathways can activate the NF-κB pathway, providing indirect evidence of enhanced NF-κB activity in EBV+ DLBCL. The GSEA also demonstrated enrichment of the JAK/STAT signaling pathway, compatible with enhanced expression of pSTAT3 in EBV+ DLBCL (Figure 3-D). This finding suggests that STAT3 might be activated by EBV, similar to STAT3 activation by LMP1 in nasopharyngeal carcinoma (20).
Discussion
In a cohort of 732 patients with de novo DLBCL treated with R-CHOP, the prevalence of EBV was 4.0%. A prior report from Korea showed that EBER expression (cutoff ≥20%) was associated with >60 years, advanced Ann Arbor stage, ≥2 extranodal site involvement, high intermediate/high IPI, and B symptoms (5). In the present study, however, unique clinical characteristics were not found in EBV+ DLBCLs. Considering EBV+ de novo DLBCL patients from this study is composed of Caucasian patients, the difference between prior data, largely studied on Asian EBV+ de novo DLBCL patients, from ours might be related to different characteristics of host factors and ethnic background. For example, polymorphisms in chemokine or cytokine such as CCR5-Δ32, CCR2-64I, SDF1-3′A, IL4-589T and IL10-5′592A are observed with different frequency between European Caucasians and East Asians (21–23). Additionally, different allele frequency in human leukocyte antigen (HLA) might explain the different clinical manifestations in EBV+ DLBCL compared to Asian patients. Interestingly, one study showed that CCR5-Δ32 polymorphism, which is found in 5–15% of Caucasians but not seen in Asians, reduced the risk of DLBCL and follicular lymphoma in Caucasian men, partly explaining different prevalence of EBV+ DLBCL in developed Western countries (24).
Similar to the host factors, geographical variation of EBV strains might also contribute to the different prevalence and clinical behavior of EBV+ DLBCL. Frequency of Type 1 and type 2 EBV, based on genetic polymorphisms in EBNA2 and EBNA3 genes, is different according to geographical region (25). Genome sequencing of three different EBV strains (B95-8, GD1 and AG876) showed that LMP1 sequence in GD1 (a Chinese patient of nasopharyngeal carcinoma) was markedly different from that in B95-8 (a North American patient of IM) and AG876 (a Western African patient of Burkitt lymphoma), suggesting tumorigenic potential of LMP1 might be different based on geographic location (26). Additionally, polymorphism in the BamHI F region in EBV genome, “f” variant, is more frequently observed in patients with nasopharyngeal carcinoma in Southern China compared to patients in North America (27). One study showed that nasopharyngeal carcinoma with “f” variant demonstrated more frequent expression of phosphorylated STAT3, p53, BCL2 and higher Ki-67 proliferation rate compared to F prototype, suggesting genetic variation in EBV genome can contribute aggressiveness of tumor (28). Taken together, different genetic variation in EBV genome is seen in different geographical regions and it might attribute to different behavior of the disease. Although distinct clinical features were not observed, interesting pathologic and molecular features were observed in EBV+ DLBCL. Our data showed slightly higher frequency (39.3%) of the GCB type in EBV+ DLBCL compared to prior data (22% – 32%) (5, 8, 9, 11). Unlike others who used Hans or Choi algorithm for determining cell-of-origin, we used GEP as gold standard with immunohistochemical method as complimentary method. Of note, the Hans and Choi algorithm have reported 71% and 88% concordance and 96% and 89% concordance with GCB and ABC by GEP classification of DLBCL, respectively (16, 29).
Compared to previous data, LMP1 expression in EBV+ DLBCL was low (66.7%, n=8) in our study. Six of them showed EBER expression in ≥30% tumor cells, suggesting EBER-positive cells would be less likely to be reactive cells. Of the remaining two patients with EBER expression <30%, one case expressed CD30. This case was a splenic mass (5 cm), classified as a lymphoproliferative disorder-like large cell variant of polymorphic subtype. Coagulative necrosis and frequent mitotic figures were also observed. The other case was a retroperitoneal lymph node lesion with involvement in the bowel. The lymph node architecture was effaced by lymphoma, which was a monomorphic large cell subtype. Combining clinical manifestation and morphology, reactive process was excluded in these two patients.
Genetic aberrations involving BCL2, BCL6, MYC, MDM2, MDM4 and TP53 were infrequent (5~11%), reinforcing the concept that the oncogenic effects of EBV might supplant the need for chromosomal or genetic abnormalities in lymphomagenesis (9). Protein expression of the respective genes was not significantly different, except for less frequent expression of BCL6 in EBV+ DLBCLs. Although the underlying mechanism of decreased expression of BCL6 is unclear, an inverse correlation between BCL-6 and LMP1 was recognized previously (30). A recent study showed EBV microRNAs such as EBV-miR-BART9 and EBV-miR-BART17-5p could down-regulate BCL6 expression in DLBCL (31).
Functional NF-κB protein exists as dimers composed of class I and class II molecules. Class I molecules, p50 and p52, contain nuclear localization regions without transcriptional action. Class II molecules, p65 (RelA), RelB and c-Rel, have a transcription activation domain. Nuclear accumulation of p50/p65 or p50/c-Rel dimer is observed in the canonical pathway. In comparison, p52/relB dimers translocate to the nucleus in the alternative pathway. A significant increase of p50 expression in EBV+ DLBCL suggests that NF-κB activation occurs via the canonical pathway. Considering NF-κB activation is more common in ABC DLBCL (32), nuclear expression of canonical NF-κB components in EBV+ GCB DLBCL suggests EBV-induced canonical NF-κB activation in the tumor cells. However, single expression of LMP1 or EBNA2 did not show significant correlations with any of the studied NF-κB molecules. Our findings suggest yet unknown mechanism(s) induced by EBV might provide an additive effect on NF-κB activation.
STAT3 is one of 7 different members of STAT family (STAT1, STAT2, STAT3, STAT4, STA5a, STAT5b and STAT6) of transcription factors and resides in an inactive form in the cytoplasm of non-stimulated cells. Activation of STAT3 is mediated by phosphorylation of a particular tyrosine residue (Tyr 705), which promotes dimerization through phosphotyrosine-SH2 domain interaction (33). The dimerized STAT3 migrates to the nucleus and acts as a transcription factor. Constitutively active STAT3 can be seen in cancers in breast, lung, pancreas and prostate (34). Data showed STAT3 could be activated by LMP1 (20, 35). Moreover, EBNA2 was shown to be a transcriptional co-activator of STAT3 (36). No correlation between pSTAT3 and LMP1 or EBNA2 was observed. Our findings might support that the synergistic effect of LMP1 and EBNA2 are required on STAT3 activation. pSTAT3 can physically interact with NF-κB dimer p50/p65 and the compound can recruit p300 histone acetyltransferase. In turn, p300 can acetylate activated (phosphorylated) p65 and increase its nuclear retention so that prolong transcriptional activity of NF-κB (37). Therefore, expression of pSTAT3 might potentiate activity of canonical NF-κB in EBV+ DLBCL, although its expression did not show statistically significant correlation with NF-κB p50.
Gene expression profiling showed EBV+ DLBCL is distinct from EBV-negative DLBCL at the molecular level. Distinctly upregulated genes in EBV+ DLBCL are involved in enhanced activity of NF-κB, cell proliferation, cell cycle progression, and cell metabolism. Meanwhile, downregulated genes suggest possible chemotherapeutic resistance, poor prognosis and EBV-associated CD30 expression. By GSEA, immunohistochemical evidence of activated NF-κB pathway was reproduced. Gene set enrichment of the JAT/STAT pathway, in tandem with significant overexpression of pSTAT3 in EBV+ DLBCL might provide a rationale for new therapeutic options.
Contrary to most previous reports, we showed EBER positivity does not predict poorer survival in patients with DLBCL in developed countries irrespective of COO stratification and entire cohort. Since all the cases in our cohort expressed CD20, we cannot entirely exclude a possibility that anti-CD20 agent overcome inferior outcome observed in the CHOP era because we did not have a well-selected cohort treated with CHOP to compare with. No large reliable cohort study has been performed in the Western countries for EBV+ de novo DLBCL patients treated with CHOP regimen that could be used for comparison. Theoretically, because all EBER+ and EBER− DLBCL cells express CD20 with similar intensity, we would believe that rituximab should excel its anti-lymphoma functions more or less similarly in both the EBER+ and EBER− DLBCL patients. Our results are in accord with a few reports that EBV+ DLBCL did not demonstrate poor outcome (4, 38, 39). Despite such hypothesis, rituximab has shown its variable anti-lymphoma functions in individual patients due to the heterogeneity of tumor cells or pathologic biology related to the genomic and epigenetic status within the tumor cells including variable CD20 antigen expression intensity, antibody-dependent cell-mediated cytotoxicity efficacy, microenvironment regulatory role, chemokine and cytokine expression, B-cell receptor signaling inhibition, CD30 activation and death signaling pathway regulation, and EBV-specific microRNA expression. In fact, co-expression of CD30 and EBER in DLBCL showed a worse outcome. This result suggests that 1) previously reported worse outcome in EBV+ DLBCLs could be associated with co-expression of CD30 and EBER, not just single expression of EBER, and 2) there might be geographical differences in EBV strains so that prognostic effects might be variable. We, therefore, will recommend include CD30 assay in our daily diagnostic practice for any EBV+ lymphoma patients.
We further showed by using GEP that EBER+ CD30+ DLBCL is a unique subgroup of EBV+ DLBCL. Compared with EBER+ CD30-negative DLBCL, DLBCL with co-expression of EBER and CD30 was characterized with increased activity of NF-κB pathway, cell proliferation, and cell cycle progression. Unique features of GEP in EBER+ CD30+ DLBCL suggest the need for a larger study focusing on a direct comparison of CD30 expression among patients with EBV+ DLBCLs.
Our findings also suggest a rationale for targeted therapy of EBV+ DLBCL. Brentuximab vedotin is a CD30-specific antibody-drug conjugate. Although a recent report of clinical trial with Brentuximab in patients with CD30+ DLBCL was not promising, the result of this trial might not be applicable to EBV+ CD30+ DLBCL, since only two patients were EBER+ (40). Our study suggests more specifically selected patients are needed for a clinical trial with Brentuximab. We also showed enhanced NF-κB activity in EBV+ DLBCL, even in the GCB type. Therefore, inhibitors of the NF-κB pathway would be good therapeutic options for these patients irrespective of COO. A phase I/II clinical trial with bortezomib plus R-CHOP in patients with DLBCL negated the adverse outcome of non-GCB subtype and showed similar survival rates in patients with GCB versus non-GCB subtype (41). Another NF-κB pathway inhibitor, MLN4924, was shown to induce apoptosis and tumor regression in ABC DLBCL cell lines and xenograft models (42). Currently, a clinical trial is underway with MLN4924 in DLBCL patients (NCT01415765). NEMO-binding domain peptide has also been shown to inhibit NF-κB target gene expression and reduce tumor burden in an in vivo mouse model (43). Additionally, in parallel with NF-κB inhibition, targeting or co-targeting JAK/STAT pathway might be an attractive strategy. As shown by Lam et al, a JAK inhibitor in combination with an IKK inhibitor killed ABC DLBCL cell lines more potently (44).
Two morphologic variants of EBV+ DLBCL, monomorphic and polymorphic, have been recognized. Montes-Moreno and colleagues have subdivided the polymorphic subtype into three groups based on the relative proportion of large neoplastic cells and Hodgkin Reed-Sternberg-like cells: 1) canonical large B-cell neoplasm; 2) DLBCL with Hodgkin lymphoma-like features; and 3) DLBCL with polymorphic lymphoproliferative disorder-like features (9). Although elucidation of these morphologic variants is of value to pathologists to facilitate recognition of EBV+ DLBCLs, in this study these morphologic variants did not have prognostic importance.
Our study has shown that EBV+ DLBCL is a genetically unique subset with increased canonical NF-κB and JAK/STAT pathway, but EBV infection itself does not predict inferior outcome in Western developed countries if CD30 is not expressed. This is the first comprehensive study in this uniquely classified group of lymphoma patients from Western countries, providing a strong rationale of targeted therapy on CD30 activation pathway for this subset of DLBCL patients.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
Epstein-Barr virus (EBV)-positive diffuse large B-cell lymphoma (DLBCL) is a genetically unique subgroup of DLBCL with activated canonical NF-κB and JAK/STAT pathways. CD30 is more commonly expressed in this subset compared to DLBCL without EBV infection. The disease has been known for inferior outcome compared to DLBCL without EBV infection, but significant proportion of data was gleaned from patients treated with cyclophosphamide, hydroxydoxorubicin, vincristine and prednisone (CHOP). In our group of patients in Western developed countries treated with rituximab combined with CHOP (R-CHOP), EBV infection determined by positive expression of Epstein-Barr virus encoded small RNA (EBER) in tumor cells did not predict inferior outcome. However, CD30 co-expression with EBER portended poor prognosis. This study provides a strong foundation for using anti-CD30 therapy, Brentuximab vedotin (Adcetris), in patients whose tumor co-express CD30 and EBER.
Acknowledgments
This work was supported by the Hematopathology Fellowship Award (C.Y.O.); Hematology/Oncology Scholarship Award (L.L.); Harold C. and Mary L. Daily Endowment Fellowships and Shannon Timmins Fellowship for Leukemia Research Award (Z.Y.X.-M.); the Zurich Stiftung zur Krebsbekaempfung (A.T.); The University of Texas MD Anderson Cancer Center Institutional Research Grant Award, an MD Anderson Lymphoma Specialized Programs of Research Excellence (SPORE) Research Development Program Award, an MD Anderson Myeloma SPORE Research Development Program Award, and MD Anderson Collaborative Research Funds with High-Throughput Molecular Diagnostics, Daiichi Sankyo and Roche Molecular Systems (K.H.Y.). This work was also partially supported by National Cancer Institute and National Institutes of Health grants (R01CA138688, 1RC1CA146299, P50CA136411 and P50CA142509), and by the MD Anderson Cancer Center Support Grant CA016672.
Footnotes
AUTHORSHIP CONTRIBUTION
C.Y.O. and K.H.Y. designed and conducted the research, collected clinical and follow-up data with the approval of the institutional review boards of the participating centers and the material transfer agreement performed the statistical analysis, and wrote the manuscript; Z.Y.X.-M., C.V., A.T., S.M.M., K.D., A.C., Y.Z., G.B., J.C., K.L.R., E.D.H., W.W.L.C., J.H.K., J.H., W.A., M.P., A.J.M.F., J.P.F., M.B.M., C.E.B.R., J.N.W., and M.A.P., L.J.M., and K.H.Y. contributed vital new reagents, resources, technology, and analytical tools; L.L., Z.Y.X.-M., C.V., A.T., S.M.M., K.D., A.C., Y.Z., G.B., K.L.R., E.D.H., W.W.L.C., J.H.K., J.H., W.A., M.P., A.J.M.F., J.P.F., M.B.M., J.N.W., and M.A.P., and K.H.Y. collected clinical and follow-up data, contributed resources, and contributed to the writing of the manuscript. C.Y.O., L.J.M., and K.H.Y. wrote the manuscript. All authors contributed vital strategies, participated in discussions, and provided scientific input.
AUTHORS’ DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
References
- 1.Nakamura S, Jaffe ES, Swerdlow SH, editors. EBV positive diffuse large B-cell lymphoma of the elderly. Lyon: International Agency for Research on Cancer; 2008. [Google Scholar]
- 2.Beltran BE, Morales D, Quinones P, Medeiros LJ, Miranda RN, Castillo JJ. EBV-positive diffuse large b-cell lymphoma in young immunocompetent individuals. Clin Lymphoma Myeloma Leuk. 2011;11:512–6. doi: 10.1016/j.clml.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Ok CY, Papathomas TG, Medeiros LJ, Young KH. EBV-positive diffuse large B-cell lymphoma of the elderly. Blood. 2013;122:328–40. doi: 10.1182/blood-2013-03-489708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wada N, Ikeda J, Hori Y, Fujita S, Ogawa H, Soma T, et al. Epstein-barr virus in diffuse large B-Cell lymphoma in immunocompetent patients in Japan is as low as in Western Countries. J Med Virol. 2011;83:317–21. doi: 10.1002/jmv.21967. [DOI] [PubMed] [Google Scholar]
- 5.Park S, Lee J, Ko YH, Han A, Jun HJ, Lee SC, et al. The impact of Epstein-Barr virus status on clinical outcome in diffuse large B-cell lymphoma. Blood. 2007;110:972–8. doi: 10.1182/blood-2007-01-067769. [DOI] [PubMed] [Google Scholar]
- 6.Hofscheier A, Ponciano A, Bonzheim I, Adam P, Lome-Maldonado C, Vela T, et al. Geographic variation in the prevalence of Epstein-Barr virus-positive diffuse large B-cell lymphoma of the elderly: a comparative analysis of a Mexican and a German population. Mod Pathol. 2011;24:1046–54. doi: 10.1038/modpathol.2011.62. [DOI] [PubMed] [Google Scholar]
- 7.Gibson SE, Hsi ED. Epstein-Barr virus-positive B-cell lymphoma of the elderly at a United States tertiary medical center: an uncommon aggressive lymphoma with a nongerminal center B-cell phenotype. Hum Pathol. 2009;40:653–61. doi: 10.1016/j.humpath.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Hoeller S, Tzankov A, Pileri SA, Went P, Dirnhofer S. Epstein-Barr virus-positive diffuse large B-cell lymphoma in elderly patients is rare in Western populations. Hum Pathol. 2010;41:352–7. doi: 10.1016/j.humpath.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Montes-Moreno S, Odqvist L, Diaz-Perez JA, Lopez AB, de Villambrosia SG, Mazorra F, et al. EBV-positive diffuse large B-cell lymphoma of the elderly is an aggressive post-germinal center B-cell neoplasm characterized by prominent nuclear factor-kB activation. Mod Pathol. 2012;25:968–82. doi: 10.1038/modpathol.2012.52. [DOI] [PubMed] [Google Scholar]
- 10.Uner A, Akyurek N, Saglam A, Abdullazade S, Uzum N, Onder S, et al. The presence of Epstein-Barr virus (EBV) in diffuse large B-cell lymphomas (DLBCLs) in Turkey: special emphasis on ‘EBV-positive DLBCL of the elderly’. APMIS. 2011;119:309–16. doi: 10.1111/j.1600-0463.2011.02736.x. [DOI] [PubMed] [Google Scholar]
- 11.Beltran BE, Castillo JJ, Morales D, de Mendoza FH, Quinones P, Miranda RN, et al. EBV-positive diffuse large B-cell lymphoma of the elderly: a case series from Peru. Am J Hematol. 2011;86:663–7. doi: 10.1002/ajh.22078. [DOI] [PubMed] [Google Scholar]
- 12.Hu S, Xu-Monette ZY, Balasubramanyam A, Manyam GC, Visco C, Tzankov A, et al. CD30 expression defines a novel subgroup of diffuse large B-cell lymphoma with favorable prognosis and distinct gene expression signature: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2013;121:2715–24. doi: 10.1182/blood-2012-10-461848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu-Monette ZY, Wu L, Visco C, Tai YC, Tzankov A, Liu WM, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2012;120:3986–96. doi: 10.1182/blood-2012-05-433334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Tzankov A, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26:2103–13. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu WM, Li R, Sun JZ, Wang J, Tsai J, Wen W, et al. PQN and DQN: algorithms for expression microarrays. J Theor Biol. 2006;243:273–8. doi: 10.1016/j.jtbi.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Choi WW, Weisenburger DD, Greiner TC, Piris MA, Banham AH, Delabie J, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494–502. doi: 10.1158/1078-0432.CCR-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–6. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garattini E, Terao M. The role of aldehyde oxidase in drug metabolism. Expert Opin Drug Metab Toxicol. 2012;8:487–503. doi: 10.1517/17425255.2012.663352. [DOI] [PubMed] [Google Scholar]
- 19.Alzein N, Mueller M, van Deursen J, Ahlquist D, Witzig T, Molina J. Epigenetic silencing of the EYA-4 gene in non-Hodgkin’s lymphoma (NHL). 2005 ASCO Annual Meeting; May 12–17 2005; Orlando, FL, USA. 2005. Abstract #9553. [Google Scholar]
- 20.Wang Z, Luo F, Li L, Yang L, Hu D, Ma X, et al. STAT3 activation induced by Epstein-Barr virus latent membrane protein1 causes vascular endothelial growth factor expression and cellular invasiveness via JAK3 And ERK signaling. Eur J Cancer. 2010;46:2996–3006. doi: 10.1016/j.ejca.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 21.O’Brien SJ, Moore JP. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol Rev. 2000;177:99–111. doi: 10.1034/j.1600-065x.2000.17710.x. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama EE, Hoshino Y, Xin X, Liu H, Goto M, Watanabe N, et al. Polymorphism in the interleukin-4 promoter affects acquisition of human immunodeficiency virus type 1 syncytium-inducing phenotype. J Virol. 2000;74:5452–9. doi: 10.1128/jvi.74.12.5452-5459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama EE, Meyer L, Iwamoto A, Persoz A, Nagai Y, Rouzioux C, et al. Protective effect of interleukin-4 -589T polymorphism on human immunodeficiency virus type 1 disease progression: relationship with virus load. J Infect Dis. 2002;185:1183–6. doi: 10.1086/339825. [DOI] [PubMed] [Google Scholar]
- 24.Bracci PM, Skibola CF, Conde L, Halperin E, Lightfoot T, Smith A, et al. Chemokine polymorphisms and lymphoma: a pooled analysis. Leuk Lymphoma. 2010;51:497–506. doi: 10.3109/10428190903518337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CM, Yu KJ, Mbulaiteye SM, Hildesheim A, Bhatia K. The extent of genetic diversity of Epstein-Barr virus and its geographic and disease patterns: a need for reappraisal. Virus Res. 2009;143:209–21. doi: 10.1016/j.virusres.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGeoch DJ, Gatherer D. Lineage structures in the genome sequences of three Epstein-Barr virus strains. Virology. 2007;359:1–5. doi: 10.1016/j.virol.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Lung ML, Chang GC, Miller TR, Wara WM, Phillips TL. Genotypic analysis of Epstein-Barr virus isolates associated with nasopharyngeal carcinoma in Chinese immigrants to the United States. Int J Cancer. 1994;59:743–6. doi: 10.1002/ijc.2910590605. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Han A, You S, Yang Q, Liang Y, Dong Y. The association of genomic variation of Epstein-Barr virus BamHI F fragment with the proliferation of nasopharyngeal carcinoma. APMIS. 2010;118:657–64. doi: 10.1111/j.1600-0463.2010.02642.x. [DOI] [PubMed] [Google Scholar]
- 29.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 30.Carbone A, Gaidano G, Gloghini A, Pastore C, Saglio G, Tirelli U, et al. BCL-6 protein expression in AIDS-related non-Hodgkin’s lymphomas: inverse relationship with Epstein-Barr virus-encoded latent membrane protein-1 expression. Am J Pathol. 1997;150:155–65. [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Perez D, Vargiu P, Montes-Moreno S, Leon EA, Rodriguez-Pinilla SM, Lisio LD, et al. Epstein-Barr virus microRNAs repress BCL6 expression in diffuse large B-cell lymphoma. Leukemia. 2012;26:180–3. doi: 10.1038/leu.2011.189. [DOI] [PubMed] [Google Scholar]
- 32.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–74. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21:11–9. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 35.Kung CP, Meckes DG, Jr, Raab-Traub N. Epstein-Barr virus LMP1 activates EGFR, STAT3, and ERK through effects on PKCdelta. J Virol. 2011;85:4399–408. doi: 10.1128/JVI.01703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muromoto R, Ikeda O, Okabe K, Togi S, Kamitani S, Fujimuro M, et al. Epstein-Barr virus-derived EBNA2 regulates STAT3 activation. Biochem Biophys Res Commun. 2009;378:439–43. doi: 10.1016/j.bbrc.2008.11.053. [DOI] [PubMed] [Google Scholar]
- 37.Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–93. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perepu U, Dogan A, Maurer M, Allmer C, Thompson C, Ansell S, et al. EBV(+) Diffuse Large B Cell Lymphoma Is Infrequent in Upper Central United States and Lacks Unique Clinical Characteristics or Adverse Prognosis Compared to EBV (−) Counterparts: Results From University of Iowa/Mayo Clinic SPORE. 54th ASH Annual Meeting and Exposition; December 8–11, 2012; Atlanta, GA, USA. 2012. Abstract #1604. [Google Scholar]
- 39.Ahn JS, Yang DH, Choi YD, Jung SH, Yhim HY, Kwak JY, et al. Clinical outcome of elderly patients with Epstein-Barr virus positive diffuse large B-cell lymphoma treated with a combination of rituximab and CHOP chemotherapy. Am J Hematol. 2013 doi: 10.1002/ajh.23507. [DOI] [PubMed] [Google Scholar]
- 40.Jacobsen ED, Advani RH, Oki Y, Sharman J, Horwitz SM, Forero-TOrres A, et al. A Phase 2 Study of Brentuximab Vedotin in Patients with Relapsed or Refractory CD30-Positive Non-Hodgkin Lymphomas: Interim Results. 54th ASH Annual Meeting and Exposition; December 8–11, 2012; Atlanta, GA, USA. 2012. Abstract #2746. [Google Scholar]
- 41.Ruan J, Martin P, Furman RR, Lee SM, Cheung K, Vose JM, et al. Bortezomib plus CHOP-rituximab for previously untreated diffuse large B-cell lymphoma and mantle cell lymphoma. J Clin Oncol. 2011;29:690–7. doi: 10.1200/JCO.2010.31.1142. [DOI] [PubMed] [Google Scholar]
- 42.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116:1515–23. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 43.Gaurnier-Hausser A, Patel R, Baldwin AS, May MJ, Mason NJ. NEMO-binding domain peptide inhibits constitutive NF-kappaB activity and reduces tumor burden in a canine model of relapsed, refractory diffuse large B-cell lymphoma. Clin Cancer Res. 2011;17:4661–71. doi: 10.1158/1078-0432.CCR-10-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lam LT, Wright G, Davis RE, Lenz G, Farinha P, Dang L, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111:3701–13. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.