Abstract
Chronic exposure to drinking water disinfection byproducts has been linked to adverse health risks. The monohaloacetic acids (monoHAAs) are generated as byproducts during the disinfection of drinking water and are cytotoxic, genotoxic, mutagenic, and teratogenic. Iodoacetic acid toxicity was mitigated by antioxidants, suggesting the involvement of oxidative stress. Other monoHAAs may share a similar mode of action. Each monoHAA generated a significant concentration-response increase in the expression of a β-lactamase reporter under the control of the Antioxidant Response Element (ARE). The monoHAAs generated oxidative stress with a rank order of IAA > BAA >> CAA; this rank order was observed with other toxicological endpoints. Toxicogenomic analysis was conducted with a non-transformed human intestinal epithelial cell line (FHs 74 Int). Exposure to the monoHAAs altered the transcription levels of multiple oxidative stress responsive genes, indicating that each exposure generated oxidative stress. The transcriptome profiles showed an increase in TXNRD1 and SRXN1, suggesting peroxiredoxin proteins had been oxidized during monoHAA exposures. Three sources of reactive oxygen species were identified, the hypohalous acid generating peroxidase enzymes LPO and MPO, NADPH-dependent oxidase NOX5, and PTGS2 (COX-2) mediated arachidonic acid metabolism. Each monoHAA exposure caused an increase in COX-2 mRNA levels. These data provide a functional association between monoHAA exposure and adverse health outcomes such as oxidative stress, inflammation, and cancer.
INTRODUCTION
Disinfection of drinking water was a major public health achievement of the last century that substantially reduced outbreaks of waterborne disease [1]. During disinfection toxic byproducts (DBPs) are unintentionally generated when the disinfectant reacts with organic matter and inorganic precursors in the source water [2]. Over 600 individual DBPs have been identified in disinfected drinking water [3]; the mixture of DBPs generated varies depending on source water characteristics, disinfection method, DBP precursors and other factors [4, 5]. The United States Environmental Protection Agency (U.S. EPA) regulates 11 DBPs requiring distribution systems to monitor their levels [6]. DBP exposure increased the risk of adverse health outcomes including bladder cancer [7], colorectal cancer [8], and skin cancer [9]. Although DBP exposure during gestation was implicated in adverse pregnancy outcomes including fetal growth restriction [10-12] and congenital anomalies these associations are, at present, inconclusive [13].
The U.S. EPA estimated that the population risk to chlorinated water accounted for 2% to 17% of bladder cancer cases in the United States [14]. However, based on animal carcinogenicity data, the regulated DBPs, at levels measured in disinfected water, cannot account for the increased risk of cancer attributed to DBP exposure. This risk is possibly derived from additive or synergic effects of multiple DBPs [15, 16].
The mechanism(s) by which DBPs induce cancer are not understood. Oxidative stress is one mechanism that could explain these adverse health outcomes [17]. Multiple DBPs could contribute to an overall oxidative imbalance within cells or tissues. Biomarkers of oxidative stress have been reported after exposure to individual DBPs including 3-chloro-4-(dichloromethyl)-5-hydroxy-2-(5H)-furanone (MX) [18], bromate [19, 20], di- and trichloroacetate [21, 22], bromodichloroacetate, bromochloroacetate, dibromoacetate [22], chloroacetonitrile [23], dichloroacetonitrile [24], and also organic extracts from disinfected waters [25, 26].
The monohalogenated acetic acid DBPs (monoHAAs), including iodoacetic acid (IAA), bromoacetic acid (BAA), and chloroacetic acid (CAA), are genotoxic in human cells [27] and Chinese hamster ovary (CHO) cells [28-30], and mutagenic in Salmonella typhimurium [31], and CHO cells [32]. IAA was a potent inducer of malignant transformation of NIH3T3 cells which were tumorigenic in nude mice [33]. IAA induced reactive oxygen species (ROS), possibly by inhibiting glycolysis [34] causing a reduction of pyruvate, lowering ATP levels and inducing mitochondrial stress. ROS-induced genotoxicity may be the mechanism for the observed malignancy in NIH3T3 cells [33]. The genotoxicity and mutagenicity of IAA were mitigated by antioxidants [35]. Together these observations suggest that the genotoxicity and mutagenicity of IAA is derived from the generation of ROS. Recently we demonstrated that each monoHAA inhibited glycolysis by inhibiting the enzyme glyceraldehyde phosphate dehydrogenase (GAPDH); the inhibition kinetics strongly correlated with toxicological endpoints, suggesting a common mechanism for these compounds [36, 37].
ROS damage essential biomolecules and lead to cellular dysfunction and disease. Ageing, cancer, atherosclerosis, and neurodegenerative diseases are linked to oxidative stress [38]. Cellular response to oxidative and electrophilic stress is coordinated primarily through the redox sensitive transcription factor, nuclear factor E2-related factor 2 (Nrf2) [39] and antioxidant response elements (AREs), cis-acting factors present in the regulatory regions of ROS/electrophile detoxification genes [40]. This pathway allows cells to respond to oxidative or electrophilic stress by upregulating antioxidant genes, or genes involved in ROS detoxification. ARE-mediated reporter gene assays were used to screen for chemicals that induce oxidative stress [41] and were used as a biomarker of oxidative stress in a recent analysis of a water disinfection system [42].
The objective of this research was to gain additional information on the mechanism(s) of the toxicity of the monoHAAs. We employed an ARE β-lactamase reporter gene assay (ARE-blaassay) to determine if these compounds induce oxidative stress. Subsequently we used toxicogenomic analysis to determine if ROS were generated after monoHAA exposure and monitored specific genes associated with the generation of oxidative stress, or antioxidant response.
MATERIALS AND METHODS
Reagents
General laboratory chemicals were purchased from Fisher Scientific Co. (Itasca, IL) or Sigma Chemical Co. (St. Louis, MO). Growth medium and fetal bovine serum (FBS) were purchased from Hyclone Laboratories (Logan, UT). Human epidermal growth factor (EGF) was purchased from Sigma Chemical Co., BAA and CAA were purchased from Fluka Chemical Co. (St. Louis, MO) IAA was purchased from Aldrich Chemical Co (St. Louis, MO).
ARE-bla HepG2 Reporter Gene Assay
The CellSensor® ARE-bla HepG2 cell line (Life Technologies, Madison, WI) contains a β-lactamase reporter gene under control of the Antioxidant Response Element (ARE) stably integrated into HepG2 cells. Cells were maintained in DMEM with glutamax (Life Technologies) supplemented with 10% dialyzed FBS, 0.1 mM NEAA, 25 mM HEPES, 100U/mL Penn-strep and 5 μg/mL blasticidin at 37°C and 5% CO2. Cells were harvested using 0.25% trypsin, centrifuged, and suspended in assay medium (DMEM with glutamax supplemented with 1% dialyzed FBS, 0.1 mM NEAA, 25 mM HEPES and 100U/mL Penn-strep). Cells were plated at 2000/well/5 μL in 1536 well black-clear bottom plates (Greiner Bio-One North America, Monroe, NC) and incubated at 37°C, 5% CO2 for 5 h. Twenty-three nL of compounds or a positive control, β-napthoflavone, were transferred to each well using a pintool (Kalypsys, San Diego, CA). Cells were incubated at 37°C, 5% CO2 for 16 h. After the incubation period, 1 μL of CCF4 dye (Life Technologies) was added to each well and the plate was incubated at room temperature for 2 h. Fluorescence intensity at 460 and 530 nm emissions was measure at 405 nm excitation by an Envision plate reader (PerkinElmer, Boston, MA) followed by an addition of 4 μL/well of cell viability reagent (CellTiter-Glo, Promega, Madison, WI). The plates were incubated for 30 min at room temperature and luminescence was read using a ViewLux plate reader (Perkin-Elmer). For the ARE-bla assay, data were expressed as the ratio of the 460/530 emission values, normalized to the positive control response (23 μM β-napthoflavone, 100%). For cytotoxicity, data were normalized to 100% for the negative control, (DMSO) and to 0% for the basal cytotoxic control (92 μM tetraoctyl ammonium bromide).
Human Intestinal Epithelial Cells
Nontransformed human intestinal cells, line FHs 74 Int, were purchased from American Tissue Culture Collection (Manassass, VA.) Cells were received at passage 12, and used until passage 17 or 18. Cells were maintained in Hybri-care 46X medium supplemented with 10% FBS, 1% antibiotic (10 units/mL penicillin G sodium, 10 μg/mL streptomycin sulfate, 25 μg/mL amphotericin B, 0.85% saline), and 30 ng/mL human epidermal growth factor at 37°C in a humidified atmosphere of 5% CO2.
Cell Viability
To reduce artifactual transcriptome alterations derived from cell death, monoHAA concentrations were selected that produced equal genomic DNA damage in FHs 74 Int cells with minimal effect on cell viability [27]. Cell viability was measured both immediately and 24 h after HAA exposure as previously reported [27].
RNA Isolation, cDNA Synthesis, Real Time Quantitative PCR Analysis
Four days prior to treatment 4×105 FHs 74 Int cells were seeded into the wells of a 6 well plate. Prior to exposure the cells were washed twice in Hank’s balanced salt solution (HBSS). HAAs were added in culture medium without FBS. After either 30 min or 4 h exposure, the cells were washed and harvested with trypsin (0.05%) and centrifuged. Four h was chosen as the time period for late expressing genes since this time was standardized for the DBPs to determine the equivalent levels of DNA damage [27, 30, 43]. For early gene expression, we chose 30 min treatments because this was the earliest interval that we could technically repeat the procedure and demonstrate consistent gene expression. Aliquots of the cell suspension were retained for analyses of acute toxicity and genomic DNA damage with the single cell gel electrophoresis assay. After the supernatant was removed, RNA was isolated using a Qiagen RNeasy Mini Kit (Valencia, CA) following the manufacturers protocol. RNA integrity was determined using an Agilent 2100 Bioanalyzer (Santa Clara, CA). RNA integrity (RIN) numbers were determined for RNA from each exposure group and their concurrent negative controls. cDNA was prepared from RNA using SuperArray RT2 PCR Array First Strand Kit (SA Biosciences, Frederick, MD), cDNA samples were diluted in nuclease free water and stored at −20°C. Transcriptome profiles were analyzed using an Oxidative Stress and Antioxidant Defense pathway specific PCR array (SuperArray PAHs-065-24). A list of genes included in this PCR array is presented within their functional gene groupings (Table S1 in SI). Real-time PCR analysis was conducted using a two-step cycling program on a Stratagene Mx3000p thermocycler. Quality controls measuring genomic DNA contamination, reverse transcription, and PCR amplification efficiencies were included and analyzed. Because SYBR Green technology was utilized, melting curves were investigated for each gene analyzed. Wells with multiple amplification products were excluded.
Safety and Data Handling
Manipulations of toxic chemicals were conducted in certified biological/chemical stage-2 safety hoods. Average Ct values for each gene were calculated against the average of 5 housekeeping genes (B2M, HPRT1, RPL13A, GAPDH and ACTB) using the SA Biosciences PCR analysis software RT2 Profiler Data Analysis Template. This software also estimated fold changes using the ΔΔCt method comparing treated cells to concurrent negative controls. The RankProd algorithm from the R Bioconductor package was used to test significance [44], with P ≤ 0.05 considered significant. The raw and normalized data are available in the Gene Expression Omnibus (GEO) database [45] under the NCBI tracking system GSE49698 series accession number.
RESULTS AND DISCUSSION
ARE-dependent gene expression in a reporter gene assay demonstrated a strong response after cells were exposed to the monoHAAs. These responses implicated the generation of ROS as a function of monoHAA concentration. Previous studies suggested that the monoHAAs induced their cytotoxicity and genotoxicity by generating ROS based on the inhibition of GAPDH [36], the repression of ATP levels, pyruvate remediation of cell stress and genotoxicity [37], repression of genotoxicity by radical scavengers and antioxidants [35, 46], and the direct measurement of cellular ROS after monoHAA treatment [46]. We proposed that each of the monoHAAs share a similar ROS-mediated mechanism of toxicity and tested this hypothesis using an ARE reporter assay and toxicogenomic analyses.
Toxicogenomics measured as toxicant-induced modulation of gene expression is a powerful analytical tool for cell stress. When compared to concurrent control transcriptome profiles, metabolic pathways involved in the cellular responses to toxic agents can be identified and provide insights into the biological mechanisms of toxicity. Many in vitro toxicogenomic studies employ human or mammalian tumor cell lines because of their ease of growth and in some cases cells are exposed to cytotoxic concentrations to observe effects on gene expression. Tumor cell lines inherently exhibit aberrant gene expression. With cytotoxic concentrations, transcript profiles will reflect those of dead or dying cells. We avoided these complications by using nontransformed human cells, concurrent negative controls at each treatment time and noncytotoxic concentrations of each monoHAA. An additional concern is that much of the gene expression literature is based on whole genome arrays without qRT-PCR confirmation. Our experimental design to determine transcriptome profiles of the monoHAAs was based on the direct use of PCR gene arrays. Our previous data argued that the monoHAAs were genotoxic due to their ability to induce ROS. This was the hypothesis that we tested using a ROS-responsive gene array.
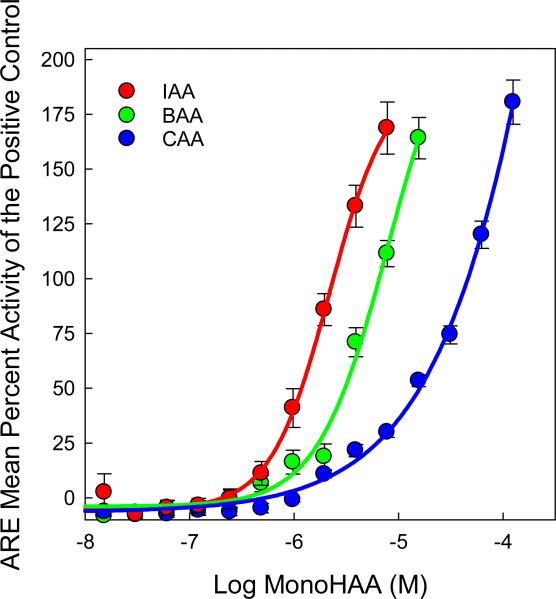
Besides the employment of a gene array for oxidative stress we used ARE transcription as an independent indicator. Briefly, the ARE-bla HepG2 cell line contains three stably integrated copies of the ARE derived from the human nicotinamide adenine dinucleotide phosphate (NADPH) quinone oxidoreductase 1 gene (NQO1) that drive the expression of a β-lactamase reporter gene [41]. Oxidative stress increases Nrf2 dependent and independent ARE-driven transcription of β-lactamase [41]. β-lactamase cleaves an exogenous substrate (coumarin) reducing the fluorescence resonance energy transfer (FRET) signal emitted by the intact substrate, while increasing the emission from the cleaved substrate. ARE-driven transcription of β-lactamase is quantified by measuring the emission ratio of 460:530 nm, the emission of the β-lactamase cleaved coumarin over the emission of the intact substrate. Increased production of ARE-linked β-lactamase shifts the fluorescence towards 460 nm and is indicative of oxidative stress. Each monoHAA generated concentration-dependent increases in ARE-dependent transcription of β-lactamase, as evidenced by the shift in fluorescence towards 460 nm; the data, normalized to the positive control (23 μM β-napthoflavone) are displayed in Figure 1. These data demonstrated that the monoHAAs induced the activation of the antioxidant response pathway suggesting the generation of oxidative stress by these DBPs. The EC50 values of IAA, BAA or CAA that induced ARE-dependent β-lactamase activity were 1.17, 3.16 and 16.2 μM, respectively. Using the ECIR1.5 metric (i.e. a 1.5 fold increase over the baseline) [47] the values for IAA, BAA or CAA were 0.99, 1.96 and 8.65 μM, respectively. A Pearson Product Moment Correlation analysis demonstrated that ARE expression by the monoHAAs in human HepG2 cells and genomic DNA damage induced by the monoHAAs in human FHs 74 Int cells [27] were highly correlated (r = 0.99; P ≤ 0.07). The response pattern of IAA > BAA >> CAA, observed in previous measurements of monoHAA-mediated toxicity, was conserved.
Figure 1.
Concentration-response curves for IAA, BAA and CAA for the induced expression of the ARE-controlled β-lactamase reporter gene in ARE-bla HepG2 cells. The data are presented as the mean percent activity (±SE) of the positive control (23 μM β-napthoflavone ).
ARE-dependent gene expression in a reporter gene assay demonstrated a strong response after cells were exposed to the monoHAAs implicating the generation of ROS as a function of monoHAA concentration. These responses also indicated that transcription rates of genes regulated by ARE could be altered by monoHAA exposure. Transcription factors activated by oxidative stress can bind to AREs and modify transcription of genes coding for proteins with antioxidant or ROS detoxification functions. While a review of ARE responsive genes is beyond the scope of this paper, proteins acting in the glutathione/glutathione reductase (GSH/GSR), thioredoxin/thioredoxin reductase (Txn/Txnrd) pathways, as well as superoxide dismutases (SOD), and catalase among others are all regulated by AREs [48]. The Human Oxidative Stress and Antioxidant Defense pathway specific PCR array contained members of each of these antioxidant pathways, along with additional genes involved with the production or metabolism of ROS and oxidative stress responsive genes. The oxidative stress responsive genes in the array were used collectively as an indicator of oxidative stress. Because it included several ARE-responsive genes, this array served as an additional measure of the effect of monoHAAs on ARE-driven transcription. This analysis also allowed us to evaluate the effect of the monoHAAs on 83 specific genes and several pro-, or antioxidant pathways, at the transcriptional level.
Toxicogenomic analysis quantifies mRNA and measures changes in transcription patterns in response to toxic insult. We exposed non-transformed human FHs 74 Int cells, to 22 μM IAA, 57 μM BAA, or 3.42 mM CAA, concentrations that generated equivalent genomic genotoxicity (approximately 50% tail DNA values measured by single cell gel electrophoresis) without causing significant acute cytotoxicity [27]. This experimental design reduced artifactual changes due to transcriptome profiles generated from dead or dying cells, or from altered gene expression/regulation that might be associated with cancer cell lines. Two exposure times, 30 min and 4 h, were chosen to evaluate early and late changes in transcription patterns. Each treatment was compared to a concurrent negative control. One gene, neutrophil cytosolic factor 1 (NCF1), was removed from analysis because multiple amplification products, as evidenced by multiple peaks in the melting point analysis, were detected.
Each of the monoHAAs altered transcription for multiple genes at both exposure times. Genes with statistically significant alterations in transcription (positive calls) are listed in Table 1 (30 min exposure) and Table 2 (4 h exposure). Positive calls were sorted into functional gene groupings including: glutathione peroxidases (GPx), peroxiredoxins (TPx), other peroxidases, other antioxidants, superoxide dismutases (SOD), other genes involved in superoxide metabolism, other genes involved in ROS metabolism, and oxidative stress responsive genes.
Table 1.
Gene Expression Level Changes vs. Concurrent Negative Controls after 30 min MonoHAA Exposure.
| IAA | BAA | CAA | |||||
|---|---|---|---|---|---|---|---|
| Gene Symbol | Functional Gene Group(s) | Fold Change | P-value | Fold Change | P-value | Fold Change | P-value |
| ALOX12 | Superoxide Metabolism | −2.286 | 0.015 | −1.972 | 0.013 | ||
| APOE | Antioxidant, Oxidative Stress Responsive | −1.725 | 0.040 | ||||
| CCL5 | Oxidative Stress Responsive | 3.588 | 0.001 | 2.424 | 0.004 | ||
| CYBA | Superoxide Metabolism | −1.519 | 0.045 | ||||
| CYGB | Peroxidase, Oxidative Stress Responsive | −1.941 | 0.038 | ||||
| DGKK | Oxidative Stress Responsive | 2.057 | 0.008 | ||||
| DHCR24 | Oxidative Stress Responsive | −1.832 | 0.050 | ||||
| DUOX1 | Peroxidase, Superoxide Metabolism, Oxidative Stress Responsive | −2.608 | 0.004 | −2.334 | 0.001 | ||
| EPHX2 | ROS Metabolism | 2.856 | 0.001 | 8.041 | < 0.001 | ||
| EPX | Peroxidase, Oxidative Stress Responsive | −1.857 | 0.017 | ||||
| GPX2 | Glutathione Peroxidase, Oxidative Stress Responsive | −4.396 | < 0.001 | ||||
| GSR | Antioxidant | −4.521 | <0.001 | −3.433 | 0.007 | ||
| KRT1 | Oxidative Stress Responsive | 3.549 | 0.008 | ||||
| LPO | Peroxidase, Oxidative Stress Responsive | 1.823 | 0.034 | 3.919 | 0.022 | ||
| MBL2 | Oxidative Stress Responsive | 2.702 | 0.038 | ||||
| MPO | Peroxidase, Oxidative Stress Responsive | 1.516 | 0.035 | 2.509 | 0.004 | ||
| NOS2A | Superoxide Metabolism | 7.000 | 0.002 | ||||
| NOX5 | Superoxide Metabolism | −1.782 | 0.042 | 3.901 | 0.004 | ||
| PIP3-E | Peroxidase, Oxidative Stress Responsive | 1.977 | 0.046 | 1.386 | 0.034 | ||
| PNKP | Oxidative Stress Responsive | −2.033 | 0.023 | −1.701 | 0.011 | ||
| PTGS2 (COX-2) | Peroxidase | 5.122 | < 0.001 | −2.410 | 0.004 | ||
| PXDNL | Peroxidase | −3.182 | 0.012 | ||||
| RNF7 | Oxidative Stress Responsive | 1.803 | 0.019 | ||||
| SCARA3 | Oxidative Stress Responsive | −1.840 | 0.013 | ||||
| SFTPD | ROS Metabolism | 1.973 | 0.017 | ||||
| SGK2 | Oxidative Stress Responsive | −2.422 | 0.007 | ||||
| SIRT2 | Oxidative Stress Responsive | −1.782 | 0.039 | −1.616 | 0.017 | ||
| STK25 | Oxidative Stress Responsive | −1.990 | 0.038 | ||||
| TPO | Peroxidase, Oxidative Stress Responsive | −2.339 | 0.012 | −2.377 | 0.002 | ||
| TXNDC2 | Antioxidant | 3.775 | 0.001 | −1.931 | 0.022 | ||
| TXNRD1 | Antioxidant | −1.543 | 0.049 | ||||
Table 2.
Gene Expression Level Changes vs. Concurrent Negative Controls after 4 h MonoHAA Exposure.
| IAA | BAA | CAA | |||||
|---|---|---|---|---|---|---|---|
| Gene Symbol | Functional Gene Group(s) | Fold Change | P-value | Fold Change | P-value | Fold Change | P-value |
| ALB | Antioxidant | 1.785 | 0.011 | ||||
| ALOX12 | Superoxide Metabolism | −3.125 | 0.020 | ||||
| ANGPT7L | Oxidative Stress Responsive | −5.900 | 0.001 | ||||
| AOX1 | ROS Metabolism | 1.785 | 0.017 | ||||
| CCL5 | Oxidative Stress Responsive | 1.981 | 0.011 | −2.074 | 0.018 | ||
| DUOX1 | Peroxidase, Superoxide Metabolism, Oxidative Stress Responsive | −11.610 | < 0.001 | −2.463 | 0.044 | ||
| DUSP1 | Oxidative Stress Responsive | 1.994 | 0.009 | 14.117 | < 0.001 | ||
| EPHX2 | ROS Metabolism | −2.737 | 0.004 | ||||
| EPX | Peroxidase, Oxidative Stress Responsive | −2.331 | 0.034 | −3.993 | 0.004 | −1.810 | 0.021 |
| GLRX2 | Oxidative Stress Responsive | −4.837 | 0.006 | ||||
| GPR156 | Peroxidase, Oxidative Stress Responsive | −2.504 | 0.032 | ||||
| GPX2 | Glutathione Peroxidase, Oxidative Stress Responsive | −2.957 | 0.019 | ||||
| GPX6 | Glutathione Peroxidase, Oxidative Stress Responsive | −2.377 | 0.016 | ||||
| GSR | Antioxidant | −2.683 | 0.012 | −2.671 | 0.037 | ||
| MTL5 | Oxidative Stress Responsive | 1.949 | 0.006 | ||||
| NOS2A | Superoxide Metabolism | −4.020 | 0.002 | −1.931 | 0.041 | ||
| NOX5 | Superoxide Metabolism | 1.976 | 0.008 | 2.323 | 0.016 | 3.830 | 0.001 |
| PIP3-E | Peroxidase, Oxidative Stress Responsive | −4.058 | 0.003 | 4.694 | 0.000 | ||
| PRDX3 | Peroxiredoxins | −3.040 | 0.014 | −1.823 | 0.040 | ||
| PRDX4 | Peroxiredoxins | −1.985 | 0.032 | ||||
| PTGS2 (COX-2) | Peroxidase | 5.108 | < 0.001 | 32.282 | < 0.001 | 8.153 | 0.000 |
| PXDNL | Peroxidase | −3.326 | 0.009 | 1.845 | 0.045 | ||
| SFTPD | ROS Metabolism | −3.476 | 0.009 | ||||
| SGK2 | Oxidative Stress Responsive | 1.433 | 0.033 | 3.587 | 0.007 | ||
| SRXN1 | Antioxidant, Oxidative Stress Responsive | 2.366 | 0.002 | 4.477 | 0.001 | 2.320 | 0.007 |
| TPO | Peroxidase, Oxidative Stress Responsive | −3.866 | 0.004 | ||||
| TTN | Peroxidase, Oxidative Stress Responsive | −3.665 | 0.003 | −5.467 | 0.001 | ||
| TXNRD1 | Antioxidant | 1.918 | 0.036 | 2.341 | 0.008 | ||
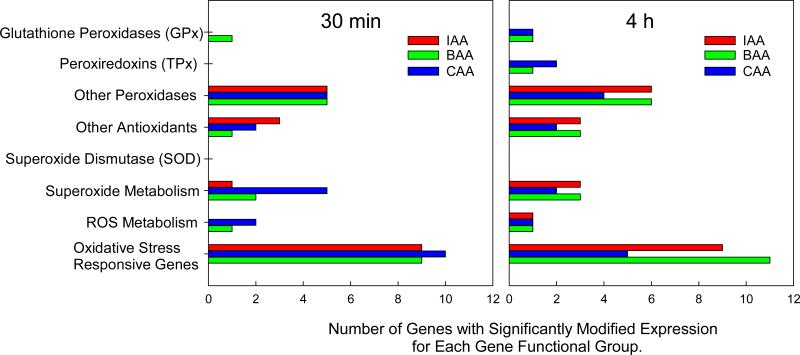
After a 30 min exposure to individual monoHAAs, 31 of the 83 genes assayed exhibited significantly altered expression levels, as compared to negative controls, with a total of 45 positive calls amongst all treated groups (Table 1, Figure 2). There was considerable overlap of altered gene expression, with 14 of the 31 (45.2%) positive calls generated by two monoHAAs. None of the altered gene expressions were shared by all 3 monoHAAs. Of the 45 positive calls, 26 (57.8%) were downregulated and 19 (42.2%) were upregulated (Table 1).
Figure 2.
The number of ROS-responding genes listed by functional groups that exhibited altered expression after 30 min or 4 h exposure to 22 μM IAA, 57 μM BAA, or 3.4 mM CAA. Some genes were counted more than once because of their involvement in multiple pathways.
For the 4 h exposure, a total of 47 gene expression levels were altered amongst all treatment groups, with transcription levels of 28 genes significantly altered (Table 2, Figure 2). Of the genes with significantly modified transcription levels, 11 (37.9%) were altered by 2 monoHAAs, and 4 (13.8%) were altered by all three monoHAAs. Of the 47 positive calls, 26 (55.3%) were downregulated and 21 (44.7%) were upregulated (Table 2).
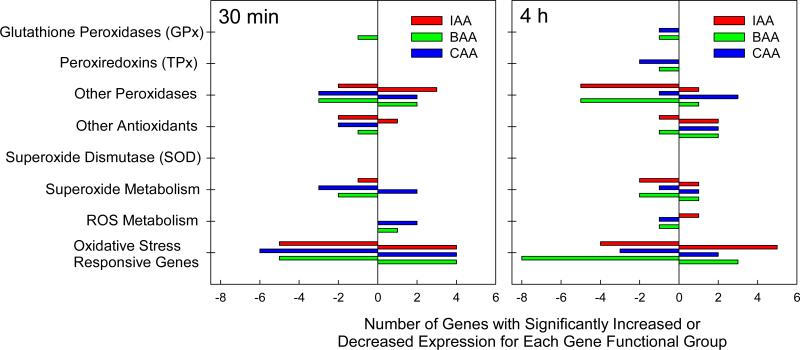
For each monoHAA exposure, at both time points, multiple oxidative stress responsive genes showed altered transcription patterns (Figures 2). Taking these genes collectively as an indicator, our hypothesis is supported that cells exposed to monoHAAs are responding to oxidative stress.
Several genes involved in antioxidant defense or detoxification are regulated by the Nrf2/ARE system at the transcription level [48]. Analysis of the transcriptome profiles showed that ARE-responsive genes exhibited modified transcription levels after monoHAA challenge, although no clear pattern of regulation emerged when all ARE-responsive genes were considered. Some of the ARE-responsive genes were upregulated: epoxide hydrolase (EPHX2) after 30 min exposure to BAA or CAA, sulfiredoxin (SRXN1) after 4 h exposures to IAA, BAA or CAA and thioredoxin reductase 1 (TXNRD1) after 4 h exposure to BAA or CAA. Other ARE-responsive genes were downregulated after 30 min: TXNRD1 with CAA, glutathione reductase (GSR) with IAA or BAA, as well as after 4 h: peroxiredoxin 3 (PRDX3) and GSR. ARE-regulated genes including SOD, and those peroxiredoxins and glutathione peroxidases not included in Tables 1 and 2 did not show altered transcription patterns after monoHAA exposures. These findings suggest that while monoHAAs activate ARE-driven transcription in the ARE-bla reporter system, the regulation of specific genes within the antioxidant pathways of the FHs cell line includes additional layers of complexity.
Within the GSH/GSR pathway, GPX2, GPX6 and GSR genes were downregulated (Tables 1 and 2). None of the SOD genes or catalase displayed altered transcription after monoHAA exposures at either time point. These data along with the altered transcription of SRXN1 and TXNRD1 suggested that peroxiredoxins (Prx) were the main target for ROS generated during monoHAA challenge.
Prx proteins reduce hydrogen peroxide, peroxynitrite, and lipid peroxides. Each of the 6 mammalian PRX genes was included in the gene array, however, only PRDX3 and PRDX4 were modified by monoHAA exposure. Peroxiredoxins contain 1 (Prx6) or 2 (Prx1-5) catalytic cysteine residue(s) that is/are oxidized during their peroxidase activity. TXNRD1 and SRXN1 encode proteins capable of reducing oxidized Prxs. Txnrd1 acts indirectly by regenerating reduced Txn after it has been oxidized while reducing Prx [49]. Sulfiredoxin directly reduces Prx1-4, the so-called typical 2 cysteine Prx [50]. Both TXNRD1 and SRXN1 were upregulated by BAA and CAA or all monoHAAs, respectively, after 4 h. However, none of the PRX genes were upregulated at any time point. PRDX3, was downregulated after 4 h exposure to BAA and IAA. The significance of the downregulation was unclear. However, post-transcriptional modifications that inactivate Prx provide localized increases in H2O2 concentration important for modifying signaling cascades. Prx3 specifically functions within the mitochondria to regulate apoptosis [51]. Prx proteins were modulated post-transcriptionally to allow local increases of H2O2 to modify growth factor signaling [52]. Metabolites of arachidonic acid (AA) metabolism oxidized and inhibited Prx1-3, suggesting a role in signaling pathways [53].
AA is liberated from membranes via calcium dependent cytosolic phospholipase A2 (cPla2) and is metabolized through cyclooxygenase (COX) or lipoxygenase (LOX) pathways yielding eicosanoid signaling molecules [54]. Both COX [55] and LOX [56] pathways generate superoxide and lipid peroxides as byproducts of AA metabolism. Free AA was measured in the medium of IAA treated neurons [57]. PLA2 inhibitors [58], antioxidants [34] and the calcium chelator BAPTA-AM [34] each reduced IAA-mediated toxicity. These results suggest a process involving increased intracellular [Ca2+], activation of cPla2, and ROS production that is involved in IAA-generated neurotoxicity. In this study we found each monoHAA exposure altered mRNA levels of COX to a greater degree than LOX enzymes. ALOX12 mRNA was decreased after BAA (30 min and 4 h) or CAA (30 min) exposures. Conversely, PTGS2 (COX-2) was downregulated by BAA after 30 min but upregulated by IAA at 30 min, and upregulated by all three monoHAAs after 4 h exposures (Tables 1 and 2). COX-2 exhibited the largest increase in each of the 4 h treatment groups, with a >32× increase in BAA treated cells.
In addition to the generation of ROS as a byproduct of AA metabolism, the upregulation of COX-2 suggests a possible inflammatory response which may impact human health. Chen et al showed that CAA activated the stress associated mitogen activated protein kinase (MAPK) p38 pathway via oxidative stress [59]. Activated p38 phosphorylates and inactivates tristetraproline preventing the degradation of both COX-2 and dual specific phosphatase 1 (DUSP1) mRNA [60]. Our data demonstrated that 4 h exposures to BAA or IAA increased COX-2 and DUSP1 mRNA levels, suggesting that activation of the p38 pathway may be a common response to monoHAA exposure. Cox-2 and p38 play critical roles in inflammation. Previous toxicogenomic analyses focused on DNA damage and repair pathways demonstrated that a 4 h exposure to IAA modified genes within the FcεRI receptor pathway [27]. FcεRI functions in phagocytosis and production of pro-inflammatory cytokines [61].
Inflammation has been linked to all phases of cancer [62]. The link between COX-2 and bladder and colon cancers, the cancers most commonly associated with DBP exposures [7, 8], was well documented. Prostaglandin E2, a byproduct of COX-2 mediated AA metabolism, increased colon cancer growth by increasing β-catenin signaling [63]. Furthermore, COX-2 inhibitors reduced growth of HT-29 and Caco-2 colon cancer cell lines [64], and inhibited haematogenous metastasis of colon cancer in mice [65]. COX-2 was detected in transitional cell carcinomas [66], and also squamous cell carcinomas [67] of the urinary bladder. Selective inhibitors of COX-2 reduced incidence of bladder cancer in male rats exposed to N-butyl-N-(4-hydroxybutyl) nitrosamine [68, 69]. AA metabolism also inhibited the tumor suppressor PTEN [54]. Although further studies are needed to verify an inflammatory effect in vivo, the data presented here could have important implications in linking DBP exposure with associated adverse health outcomes.
While previous studies showed ROS resulted from IAA exposures, the source(s) were not defined. Alterations in transcriptome profiles could provide insight into mechanisms by which ROS were generated during monoHAA exposure. Pro-oxidant encoding genes upregulated by two or more monoHAA exposures were considered potential sources of ROS. In addition to ROS generated through COX-2 catalyzed metabolism of AA, two additional sources of ROS were identified in the hypohalous acid generating peroxidases (myeloperoxidase (MPO) and lactoperoxidase (LPO)) and NADPH dependent oxidase 5 (NOX5) (Tables 1 and 2).
MPO was upregulated by a 30 min exposure to BAA or CAA; LPO was upregulated by CAA or IAA (Table 1). NOX5 was upregulated by CAA after 30 min, and by each of the monoHAAs after 4 h exposure (Tables 1 and 2). Noxs generate superoxide anion (O2−.) by transferring an electron from NADPH to molecular oxygen. NOXs work in tandem with Lpo, or Mpo proteins in the epithelial mucosa [70] or phagocytes [71], respectively, to generate hypohalous acids (HOX, where X represents I, Br, Cl, or thiocyanate (SCN)) for host defense. Nox enzymes were activated by cPla2 [72], and Nox5 is Ca2+-dependent [73]. Additional research is needed to evaluate the source(s) of ROS generated during monoHAA exposure and their contributions to the overall oxidative imbalance.
In summary, we employed the ARE-bla HepG2 reporter cell line as a biomarker for oxidative stress and observed an increase in ARE driven β-lactamase transcription in cells treated with each of the monoHAAs. Each exposure caused oxidative stress in the reporter cell line. The rank order IAA > BAA >> CAA found in the activation of the ARE response pathway is consistent with toxicological endpoints previously measured [27-29, 31, 32, 36, 37, 74]. Additionally we exposed non-transformed human embryonic intestinal epithelial cells (line FHs 74 Int) to equally genotoxic, but non-cytotoxic levels of the monoHAAs, IAA, BAA, and CAA for 30 min or 4 h and investigated the transcription of 83 genes involved in oxidative stress and antioxidant response. Each monoHAA at both exposure times caused significant changes in transcriptome profiles. The high number of modified oxidative stress responsive genes confirmed previous reports that IAA and CAA induced oxidative stress, and demonstrated that BAA shared this mechanism. The gene array data indicated a cellular response to oxidized Prx proteins, in that SRXN1 and TRXND1 were upregulated, whereas GSR was downregulated, and other antioxidant pathways were unchanged. The data also suggest a possible link between monoHAA exposure, inflammation, and cancer, with COX-2 connecting these three phenomena. The data indicated three possible sources of ROS, (1) HOX derived from peroxidase enzymes such as Mpo or Lpo, (2) O2−. derived from Nox5, and (3) ROS derived from the metabolism of AA via COX-2.
Supplementary Material
Figure 3.
The number of oxidative stress responsive genes listed by functional groups illustrating upregulation or downregulation after 30 min or 4 h exposure to 22 μM IAA, 57 μM BAA, or 3.4 mM CAA. Some genes were counted more than once because of their involvement in multiple pathways.
ACKNOWLEDGEMENTS
We appreciate support by the Center of Advanced Materials for the Purification of Water with Systems (Water-CAMPWS), a National Science Foundation Science and Technology Center, under Award CTS-0120978, J.P. was supported by a NIEHS Predoctoral Fellowship under Grant No. T32 ES007326. This work was partially supported through an interagency agreement (IAG Y3-ES-7020-01) from the National Institute of Environmental Health Sciences/National Toxicology Program to the National Center for Advancing Translational Sciences, National Institutes of Health.
Footnotes
SUPPORTING INFORMATION
The detailed procedure and the complete list of genes analyzed using the Oxidative Stress and Antioxidant Defense pathway specific PCR array (SuperArray PAHs-065-24) is presented in the Supporting Information. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cutler D, Miller G. The role of public health improvements in health advances: the twentieth-century United States. Demography. 2005;42(1):1–22. doi: 10.1353/dem.2005.0002. [DOI] [PubMed] [Google Scholar]
- 2.Krasner SW, Weinberg HS, Richardson SD, Pastor SJ, Chinn R, Sclimenti MJ, Onstad GD, Thruston AD., Jr. The occurrence of a new generation of disinfection by-products. Environ. Sci. Technol. 2006;40(23):7175–7185. doi: 10.1021/es060353j. [DOI] [PubMed] [Google Scholar]
- 3.Richardson SD, Plewa MJ, Wagner ED, Schoeny R, DeMarini DM. Occurrence, genotoxicity, and carcinogenicity of regulated and emerging disinfection by-products in drinking water: A review and roadmap for research. Mutat. Res. 2007;636:178–242. doi: 10.1016/j.mrrev.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Hua GH, Reckhow DA. Characterization of disinfection byproduct precursors based on hydrophobicity and molecular size. Environ. Sci. Techol. 2007;41(9):3309–3315. doi: 10.1021/es062178c. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Echigo S, Minear RA, Plewa MJ. Characterization and comparison of disinfection by-products of four major disinfectants. In: Barrett SE, Krasner SW, Amy GL, editors. Natural Organic Matter and Disinfection By-Products: Characterization and Control in Drinking Water. American Chemical Society; Washington, D.C.: 2000. pp. 299–314. [Google Scholar]
- 6.Environmental Protection Agency US. National primary drinking water regulations: Stage 2 disinfectants and disinfection byproducts rule. Federal Register. 2006;71(2):387–493. [Google Scholar]
- 7.Villanueva CM, Cantor KP, Cordier S, Jaakkola JJ, King WD, Lynch CF, Porru S, Kogevinas M. Disinfection byproducts and bladder cancer: a pooled analysis. Epidemiology. 2004;15(3):357–367. doi: 10.1097/01.ede.0000121380.02594.fc. [DOI] [PubMed] [Google Scholar]
- 8.Rahman MB, Driscoll T, Cowie C, Armstrong BK. Disinfection by-products in drinking water and colorectal cancer: a meta-analysis. Int. J. Epidemiol. 2010;39(3):733–745. doi: 10.1093/ije/dyp371. [DOI] [PubMed] [Google Scholar]
- 9.Karagas MR, Villanueva CM, Nieuwenhuijsen M, Weisel CP, Cantor KP, Kogevinas M. Disinfection byproducts in drinking water and skin cancer? A hypothesis. Cancer Causes Control. 2008;19(5):547–548. doi: 10.1007/s10552-008-9116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levallois P, Gingras S, Marcoux S, Legay C, Catto C, Rodriguez M, Tardif R. Maternal exposure to drinking-water chlorination by-products and small-for-gestational-age neonates. Epidemiology. 2012;23(2):267–276. doi: 10.1097/EDE.0b013e3182468569. [DOI] [PubMed] [Google Scholar]
- 11.Costet N, Garlantezec R, Monfort C, Rouget F, Gagniere B, Chevrier C, Cordier S. Environmental and urinary markers of prenatal exposure to drinking water disinfection byproducts, fetal growth, and duration of gestation in the PELAGIE birth cohort (Brittany, France, 2002-2006). Am. J. Epidemiol. 2012175(4):263–275. doi: 10.1093/aje/kwr419. [DOI] [PubMed] [Google Scholar]
- 12.Jeong CH, Wagner ED, Siebert VR, Anduri S, Richardson SD, Daiber EJ, McKague AB, Kogevinas M, Villanueva CM, Goslan EH, Luo W, Isabelle LM, Pankow JF, Grazuleviciene R, Cordier S, Edwards SC, Righi E, Nieuwenhuijsen MJ, Plewa MJ. The occurrence and toxicity of disinfection byproducts in European drinking waters in relation with the HIWATE epidemiology study. Environ. Sci. Technol. 2012;46(21):12120–12128. doi: 10.1021/es3024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nieuwenhuijsen MJ, Martinez D, Grellier J, Bennett J, Best N, Iszatt N, Vrijheid M, Toledano MB. Chlorination disinfection by-products in drinking water and congenital anomalies: review and meta-analyses. Environ. Health Perspect. 2009;117(10):1486–1493. doi: 10.1289/ehp.0900677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.U. S. Environmental Protection Agency Quantification of Cancer Risk from Exposure of Chlorinated Drinking Water; Office of Water, Office of Science and Technology. Health and Ecological Criteria Division; Washington, DC: 1998. [Google Scholar]
- 15.Simmons JE, Teuschler LK, Gennings C, Speth TF, Richardson SD, Miltner RJ, Narotsky MG, Schenck KD, Hunter ES, 3rd, Hertzberg RC, Rice G. Component-based and whole-mixture techniques for addressing the toxicity of drinking-water disinfection by-product mixtures. J.Toxicol.Environ.Health A. 2004;67(8-10):741–754. doi: 10.1080/15287390490428215. [DOI] [PubMed] [Google Scholar]
- 16.Yeatts SD, Gennings C, Wagner ED, Simmons JE, Plewa MJ. Detecting departure from additivity along a fixed-ratio mixture ray with a piecewise model for dose and interaction thresholds. J. Agric. Biol. Environ. Statistics. 2010;15(4):510–522. doi: 10.1007/s13253-010-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nieuwenhuijsen MJ, Grellier J, Smith R, Iszatt N, Bennett J, Best N, Toledano M. The epidemiology and possible mechanisms of disinfection by-products in drinking water. Philos. Transact. R. Soc. A. 2009;367(1904):4043–4076. doi: 10.1098/rsta.2009.0116. [DOI] [PubMed] [Google Scholar]
- 18.Yuan J, Liu H, Zhou LH, Zou YL, Lu WQ. Oxidative stress and DNA damage induced by a drinking-water chlorination disinfection byproduct 3-chloro-4-(dichloromethyl)-5-hydroxy-2 (5H)-furanone (MX) in mice. Mutat.Res. 2006;609:129–136. doi: 10.1016/j.mrgentox.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Delker D, Hatch G, Allen J, Crissman B, George M, Geter D, Kilburn S, Moore T, Nelson G, Roop B, Slade R, Swank A, Ward W, DeAngelo A. Molecular biomarkers of oxidative stress associated with bromate carcinogenicity. Toxicology. 2006;221(2-3):158–165. doi: 10.1016/j.tox.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, De Silva D, Sun B, Fisher J, Bull RJ, Cotruvo JA, Cummings BS. Cellular and molecular mechanisms of bromate-induced cytotoxicity in human and rat kidney cells. Toxicology. 2010;269(1):13–23. doi: 10.1016/j.tox.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Hassoun EA, Ray S. The induction of oxidative stress and cellular death by the drinking water disinfection by-products, dichloroacetate and trichloroacetate in J774.A1 cells. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2003;135(2):119–128. doi: 10.1016/s1532-0456(03)00082-6. [DOI] [PubMed] [Google Scholar]
- 22.Larson JL, Bull RJ. Metabolism and lipoperoxidative activity of trichloroacetate and dichloroacetate in rats and mice. Toxicol. Appl. Pharmacol. 1992;115(2):268–277. doi: 10.1016/0041-008x(92)90332-m. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed AE, Jacob S, Nouraldeen AM. Chloroacetonitrile (CAN) induces glutathione depletion and 8-hydroxylation of guanine bases in rat gastric mucosa. Journal of biochemical and molecular toxicology. 1999;13(3-4):119. doi: 10.1002/(sici)1099-0461(1999)13:3/4<119::aid-jbt1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed AE, Aronson J, Jacob S. Induction of oxidative stress and TNF-alpha secretion by dichloroacetonitrile, a water disinfectant by-product, as possible mediators of apoptosis or necrosis in a murine macrophage cell line (RAW). Toxicology in Vitro. 2000;14(3):199–210. doi: 10.1016/s0887-2333(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 25.Xie SH, Liu AL, Chen YY, Zhang L, Zhang HJ, Jin BX, Lu WH, Li XY, Lu WQ. DNA damage and oxidative stress in human liver cell L-02 caused by surface water extracts during drinking water treatment in a waterworks in China. Environ. Mol. Mutagen. 2010;51(3):229–235. doi: 10.1002/em.20537. [DOI] [PubMed] [Google Scholar]
- 26.Yuan J, Wu XJ, Lu WQ, Cheng XL, Chen D, Li XY, Liu AL, Wu JJ, Xie H, Stahl T, Mersch-Sundermann V. Chlorinated river and lake water extract caused oxidative damage, DNA migration and cytotoxicity in human cells. Int. J. Hyg. Environ. Health. 2005;208(6):481–488. doi: 10.1016/j.ijheh.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Attene-Ramos MS, Wagner ED, Plewa MJ. Comparative human cell toxicogenomic analysis of monohaloacetic acid drinking water disinfection byproducts. Environ. Sci. Technol. 2010;44(19):7206–7212. doi: 10.1021/es1000193. [DOI] [PubMed] [Google Scholar]
- 28.Plewa MJ, Simmons JE, Richardson SD, Wagner ED. Mammalian cell cytotoxicity and genotoxicity of the haloacetic acids, a major class of drinking water disinfection by-products. Environ. Mol. Mutagen. 2010;51:871–878. doi: 10.1002/em.20585. [DOI] [PubMed] [Google Scholar]
- 29.Plewa MJ, Wagner ED, Richardson SD, Thruston AD, Jr., Woo YT, McKague AB. Chemical and biological characterization of newly discovered iodoacid drinking water disinfection byproducts. Environ. Sci. Technol. 2004;38(18):4713–4722. doi: 10.1021/es049971v. [DOI] [PubMed] [Google Scholar]
- 30.Plewa MJ, Wagner ED. Mammalian Cell Cytotoxicity and Genotoxicity of Disinfection By-Products. Water Research Foundation; Denver, CO: 2009. p. 134. [Google Scholar]
- 31.Kargalioglu Y, McMillan BJ, Minear RA, Plewa MJ. Analysis of the cytotoxicity and mutagenicity of drinking water disinfection by-products in Salmonella typhimurium. Teratogen. Carcinogen. Mutagen. 2002;22(2):113–128. doi: 10.1002/tcm.10010. [DOI] [PubMed] [Google Scholar]
- 32.Zhang SH, Miao DY, Liu AL, Zhang L, Wei W, Xie H, Lu WQ. Assessment of the cytotoxicity and genotoxicity of haloacetic acids using microplate-based cytotoxicity test and CHO/HGPRT gene mutation assay. Mutat.Res. 2010;703(2):174–179. doi: 10.1016/j.mrgentox.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 33.Wei X, Wang S, Zheng W, Wang X, Liu X, Jiang S, Pi J, Zheng Y, He G, Qu W. Drinking water disinfection byproduct iodoacetic acid Induces tumorigenic transformation of NIH3T3 cells. Environ. Sci. Technol. 2013;47(11):5913–5920. doi: 10.1021/es304786b. [DOI] [PubMed] [Google Scholar]
- 34.Hernandez-Fonseca K, Cardenas-Rodriguez N, Pedraza-Chaverri J, Massieu L. Calcium-dependent production of reactive oxygen species is involved in neuronal damage induced during glycolysis inhibition in cultured hippocampal neurons. J. Neurosci. Res. 2008;86(8):1768–1780. doi: 10.1002/jnr.21634. [DOI] [PubMed] [Google Scholar]
- 35.Cemeli E, Wagner ED, Anderson D, Richardson SD, Plewa MJ. Modulation of the cytotoxicity and genotoxicity of the drinking water disinfection byproduct iodoacetic acid by suppressors of oxidative stress. Environ. Sci. Technol. 2006;40(6):1878–1883. doi: 10.1021/es051602r. [DOI] [PubMed] [Google Scholar]
- 36.Pals J, Ang J, Wagner ED, Plewa MJ. Biological mechanism for the toxicity of haloacetic acid drinking water disinfection byproducts. Environ. Sci. Technol. 2011;45:5791–5797. doi: 10.1021/es2008159. [DOI] [PubMed] [Google Scholar]
- 37.Dad A, Jeong CH, Pals J, Wagner ED, Plewa MJ. Pyruvate remediation of cell stress and genotoxicity induced by haloacetic acid drinking water disinfection byproducts. Environ. Mol. Mutagen. 2013 doi: 10.1002/em.21795. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007;39(1):44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. U S A. 1994;91(21):9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266(18):11632–11639. [PubMed] [Google Scholar]
- 41.Shukla SJ, Huang R, Simmons SO, Tice RR, Witt KL, Vanleer D, Ramabhadran R, Austin CP, Xia M. Profiling environmental chemicals for activity in the antioxidant response element signaling pathway using a high throughput screening approach. Environ. Health Perspect. 2012;120(8):1150–1156. doi: 10.1289/ehp.1104709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neale PA, Antony A, Bartkow ME, Farre MJ, Heitz A, Kristiana I, Tang JY, Escher BI. Bioanalytical assessment of the formation of disinfection byproducts in a drinking water treatment plant. Environ. Sci. Technol. 2012;46(18):10317–10325. doi: 10.1021/es302126t. [DOI] [PubMed] [Google Scholar]
- 43.Wagner ED, Plewa MJ. In: Microplate-based comet assay. In The Comet Assay in Toxicology. Dhawan A, Anderson D, editors. Royal Society of Chemistry; London: 2009. pp. 79–97. [Google Scholar]
- 44.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 45.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pals J, Ang J, Wagner ED, Plewa MJ. Environmental Mutagen Society 42nd Annual Meeting. Vol. 52. Wiley-Blackwell; Montreal, Canada: 2011. The role of glyceraldehyde-3-phosphate dehydrogenase in mono-halogenated acetic acid mediated toxicity. p. S79. [Google Scholar]
- 47.Escher BI, van Daele C, Dutt M, Tang JY, Altenburger R. Most oxidative stress response in water samples comes from unknown chemicals: the need for effect-based water quality trigger values. Environ. Sci. Technol. 2013;47(13):7002–7011. doi: 10.1021/es304793h. [DOI] [PubMed] [Google Scholar]
- 48.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 49.Chae HZ, Chung SJ, Rhee SG. Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 1994;269(44):27670–27678. [PubMed] [Google Scholar]
- 50.Woo HA, Jeong W, Chang TS, Park KJ, Park SJ, Yang JS, Rhee SG. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J. Biol. Chem. 2005;280(5):3125–3128. doi: 10.1074/jbc.C400496200. [DOI] [PubMed] [Google Scholar]
- 51.Chang TS, Cho CS, Park S, Yu S, Kang SW, Rhee SG. Peroxiredoxin III, a mitochondrion-specific peroxidase, regulates apoptotic signaling by mitochondria. J. Biol. Chem. 2004;279(40):41975–41984. doi: 10.1074/jbc.M407707200. [DOI] [PubMed] [Google Scholar]
- 52.Woo HA, Yim SH, Shin DH, Kang D, Yu DY, Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell. 2010;140(4):517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 53.Cordray P, Doyle K, Edes K, Moos PJ, Fitzpatrick FA. Oxidation of 2-Cys-peroxiredoxins by arachidonic acid peroxide metabolites of lipoxygenases and cyclooxygenase-2. J. Biol. Chem. 2007;282(45):32623–32629. doi: 10.1074/jbc.M704369200. [DOI] [PubMed] [Google Scholar]
- 54.Covey TM, Edes K, Fitzpatrick FA. Akt activation by arachidonic acid metabolism occurs via oxidation and inactivation of PTEN tumor suppressor. Oncogene. 2007;26(39):5784–5792. doi: 10.1038/sj.onc.1210391. [DOI] [PubMed] [Google Scholar]
- 55.Im JY, Kim D, Paik SG, Han PL. Cyclooxygenase-2-dependent neuronal death proceeds via superoxide anion generation. Free Radic. Biol. Med. 2006;41(6):960–972. doi: 10.1016/j.freeradbiomed.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Edderkaoui M, Hong P, Vaquero EC, Lee JK, Fischer L, Friess H, Buchler MW, Lerch MM, Pandol SJ, Gukovskaya AS. Extracellular matrix stimulates reactive oxygen species production and increases pancreatic cancer cell survival through 5-lipoxygenase and NADPH oxidase. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289(6):G1137–G1147. doi: 10.1152/ajpgi.00197.2005. [DOI] [PubMed] [Google Scholar]
- 57.Taylor BM, Fleming WE, Benjamin CW, Wu Y, Mathews WR, Sun FF. The mechanism of cytoprotective action of lazaroids I: Inhibition of reactive oxygen species formation and lethal cell injury during periods of energy depletion. J. Pharmacol. Exp. Ther. 1996;276(3):1224–1231. [PubMed] [Google Scholar]
- 58.Sun FF, Fleming WE, Taylor BM. Degradation of membrane phospholipids in the cultured human astroglial cell line UC-11MG during ATP depletion. Biochem. Pharmacol. 1993;45(5):1149–1155. doi: 10.1016/0006-2952(93)90261-t. [DOI] [PubMed] [Google Scholar]
- 59.Chen CH, Chen SJ, Su CC, Yen CC, Tseng TJ, Jinn TR, Tang FC, Chen KL, Su YC, Lee k I, Hung DZ, Huang CF. Chloroacetic acid induced neuronal cells death through oxidative stress-mediated p38-MAPK activation pathway regulated mitochondria-dependent apoptotic signals. Toxicology. 2013;303:72–82. doi: 10.1016/j.tox.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 60.Mancini AD, Di Battista JA. The cardinal role of the phospholipase A(2)/cyclooxygenase-2/prostaglandin E synthase/prostaglandin E(2) (PCPP) axis in inflammostasis. Inflamm. Res. 2011;60(12):1083–1092. doi: 10.1007/s00011-011-0385-7. [DOI] [PubMed] [Google Scholar]
- 61.Daeron M, Malbec O, Bonnerot C, Latour S, Segal DM, Fridman WH. Tyrosine-containing activation motif-dependent phagocytosis in mast cells. J. Immunol. Methods. 1994;152(2):783–192. [PubMed] [Google Scholar]
- 62.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castellone MD, Teramoto H, Williams BO, Druey KM, Gutkind JS. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science. 2005;310(5753):1504–1510. doi: 10.1126/science.1116221. [DOI] [PubMed] [Google Scholar]
- 64.Grosch S, Tegeder I, Niederberger E, Brautigam L, Geisslinger G. COX-2 independent induction of cell cycle arrest and apoptosis in colon cancer cells by the selective COX-2 inhibitor celecoxib. FASEB J. 2001;15(14):2742–2744. doi: 10.1096/fj.01-0299fje. [DOI] [PubMed] [Google Scholar]
- 65.Tomozawa S, Nagawa H, Tsuno N, Hatano K, Osada T, Kitayama J, Sunami E, Nita ME, Ishihara S, Yano H, Tsuruo T, Shibata Y, Muto T. Inhibition of haematogenous metastasis of colon cancer in mice by a selective COX-2 inhibitor, JTE-522. Br. J. Cancer. 1999;81(8):1274–1279. doi: 10.1038/sj.bjc.6694262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohammed SI, Knapp DW, Bostwick DG, Foster RS, Khan KN, Masferrer JL, Woerner BM, Snyder PW, Koki AT. Expression of cyclooxygenase-2 (COX-2) in human invasive transitional cell carcinoma (TCC) of the urinary bladder. Cancer Res. 1999;59(22):5647–5650. [PubMed] [Google Scholar]
- 67.Hassan HE, Mohamed AA, Bakhiet AO, Ahmed HG. Immunohistochemical expression of COX2 and iNOS in bladder cancer and its association with urinary schistosomiasis among Sudanese patients. Infect. Agent Cancer. 2013;8(1):9. doi: 10.1186/1750-9378-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okajima E, Denda A, Ozono S, Takahama M, Akai H, Sasaki Y, Kitayama W, Wakabayashi K, Konishi Y. Chemopreventive effects of nimesulide, a selective cyclooxygenase-2 inhibitor, on the development of rat urinary bladder carcinomas initiated by N-butyl-N-(4-hydroxybutyl)nitrosamine. Cancer Res. 1998;58(14):3028–3031. [PubMed] [Google Scholar]
- 69.Sereno J, Parada B, Reis F, Cunha FX, Teixeira-Lemos E, Garrido P, Pinto R, Rocha-Pereira P, Neto P, Ruivo J, Rodrigues-Santos P, Nunes S, Mota A, Figueiredo A, Teixeira F. Preventive but not curative efficacy of celecoxib on bladder carcinogenesis in a rat model. Mediators Inflamm. 2010;2010:380937. doi: 10.1155/2010/380937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim BW, Esworthy RS, Hahn MA, Pfeifer GP, Chu FF. Expression of lactoperoxidase in differentiated mouse colon epithelial cells. Free Radic. Biol. Med. 2012;52(9):1569–1576. doi: 10.1016/j.freeradbiomed.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hampton MB, Kettle AJ, Winterbourn CC. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood. 1998;92(9):3007–3017. [PubMed] [Google Scholar]
- 72.Brechard S, Plancon S, Tschirhart EJ. New insights into the regulation of neutrophil NADPH oxidase activity in the phagosome: a focus on the role of lipid and Ca(2+) signaling. Antioxidants & redox signaling. 2013;18(6):661–676. doi: 10.1089/ars.2012.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banfi B, Tirone F, Durussel I, Knisz J, Moskwa P, Molnar GZ, Krause KH, Cox JA. Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5). J. Biol. Chem. 2004;279(18):18583–18591. doi: 10.1074/jbc.M310268200. [DOI] [PubMed] [Google Scholar]
- 74.Hunter ES, Rogers EH, Schmid JE, Richard A. Comparative effects of haloacetic acids in whole embryo culture. Teratology. 1996;54(2):57–64. doi: 10.1002/(SICI)1096-9926(199606)54:2<57::AID-TERA1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.