The tripartite motif-containing protein 2 (TRIM2) functions as an E3 ubiquitin ligase. The crystal structure of the NHL domain of TRIM2, which belonged to space group P21, with unit-cell parameters a = 43.6, b = 76.4, c = 107.4 Å, α = 90.0, β = 94.0 and γ = 90.0° has been determined.
Keywords: TRIM2, NHL domain
Abstract
The tripartite motif-containing protein 2 (TRIM2) functions as an E3 ubiquitin ligase. Loss of function of TRIM2 has been shown to result in early-onset axonal neuropathy. As a member of the TRIM–NHL family of proteins, TRIM2 has a conserved modular architecture that includes N-terminal RING finger and B-box domains, a middle coiled-coil domain and a C-terminal NHL domain. To characterize the functional role of its NHL domain from the perspective of structural biology, a truncation of human TRIM2 (residues 465–744) was expressed, purified and crystallized. Rod-shaped crystals were obtained that diffracted X-rays to 1.7 Å resolution. The crystals belonged to space group P21, with unit-cell parameters a = 43.6, b = 76.4, c = 107.4 Å, α = 90.0, β = 94.0, γ = 90.0°. A Matthews coefficient of 1.97 Å3 Da−1, corresponding to a solvent content of 37.6%, indicated the presence of three molecules per asymmetric unit, which was further confirmed by the phasing solution from molecular replacement.
1. Introduction
The tripartite motif-containing protein 2 (TRIM2), a member of the TRIM–NHL protein family (NHL is named after the proteins NCL1, HT2A and LIN-41; Reymond et al., 2001 ▶), is a 81 kDa multidomain protein composed of N-terminal RING finger and B-box domains, a middle coiled-coil domain and a C-terminal NHL domain. TRIM2 functions as an E3 ubiquitin ligase, with its ubiquitination activity confined to the RING finger domain (Balastik et al., 2008 ▶). It interacts with the unconventional motor protein myosin V (Ohkawa et al., 2001 ▶), the neuro-filament light subunit (NF-L; Balastik et al., 2008 ▶) and the Bcl-2-interacting mediator (Bim) of cell death (Thompson et al., 2011 ▶), regulating the ubiquitination of these proteins. TRIM2 has specifically been shown to play a role in polarization of neurons and axon outgrowth (Khazaei et al., 2011 ▶). Consequently, loss of function of TRIM2 results in axonal neuropathy (Ylikallio et al., 2013 ▶).
The TRIM–NHL family proteins contain conserved modular structures including RING finger, B-box and coiled-coil domains and an NHL domain (Reymond et al., 2001 ▶). The RING finger domain has a zinc-finger motif and is essential for ubiquitination of the substrate (Lorick et al., 1999 ▶; Zheng et al., 2000 ▶). The B-box domain has been reported to coordinate the binding of two Zn atoms and to enhance the activity of RING-type E3 ligases (Lorick et al., 1999 ▶; Massiah et al., 2006 ▶). Interactions between coiled-coil domains help TRIM family proteins to form a homo-multimer, which is necessary for the correct positioning of components as cargos of this protein family in intracellular trafficking (Grigoryan & Keating, 2008 ▶).
Despite the demonstration of the essential roles played by TRIM proteins, the exact functional role of the NHL domain in proteins belonging to the TRIM–NHL family still remains to be illustrated. It has been reported that TRIM2 interacts with myosin V in the central nervous system through its NHL domain (Ohkawa et al., 2001 ▶). However, the molecular basis of this interaction is unknown. Determination of the crystal structure of the TRIM2 NHL domain would allow elucidation of the details of its protein–protein interaction sites and provide insights into its functional mechanism. Here, we describe the expression, purification, crystallization and preliminary X-ray diffraction analysis of the C-terminal NHL domain of human TRIM2.
2. Materials and methods
2.1. Molecular cloning
The gene encoding the C-terminal NHL domain of human TRIM2 (residues 465–744) was amplified from a cDNA clone (MCG accession code BC011052) by PCR using the forward primer 5′-GGGCATATGGAGAATCCCATCGAAGACGA-3′ (NdeI restriction site in bold) and the reverse primer 5′-GGGAAGCTTTTACTGTAAGTATCGATAGACTT-3′ (HindIII restriction site in bold). PCR was performed on a thermal cycler (Bio-Rad) using the following protocol: step 1, 368 K for 30 s; step 2, 328 K for 30 s; step 3, 345 K for 90 s; repeat from step 2 for 35 cycles. The PCR product was subjected to a final amplification step of 10 min at 345 K. The resulting amplified product was gel-extracted, digested with NdeI and HindIII (TaKaRa) and subcloned into expression vector pET-28a (Novagen). The recombinant plasmid containing the coding sequence of the TRIM2 NHL domain was transformed into Escherichia coli strain BL21 (DE3) for expression.
2.2. Expression and purification of recombinant protein
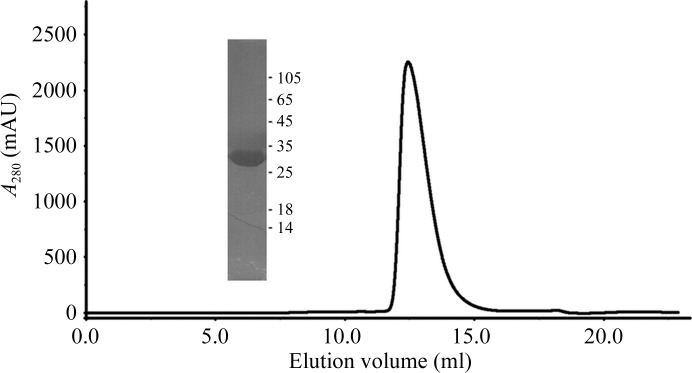
Cells were grown aerobically at 310 K for 4–5 h in Luria–Bertani medium supplemented with 50 µg ml−1 kanamycin. When the OD600 nm reached 0.8, expression of recombinant protein was induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (Sigma) and the culture was incubated for a further 20 h at 289 K. Cells were harvested by centrifugation at 4670g for 30 min at 277 K and resuspended in buffer A [50 mM Tris–HCl pH 8.0, 300 mM NaCl, 10%(v/v) glycerol]. Cells were disrupted by an EF-C3 Emulsiflex (Avestin) and the lysate was centrifuged at 30 700g for 30 min at 277 K to remove cell debris. The supernatant was loaded onto an Ni–NTA agarose column (Qiagen) pre-equilibrated with buffer A and then washed with five column volumes of buffer A containing 40 mM imidazole to remove nonspecifically bound proteins. Recombinant protein was eluted from the column using buffer A supplemented with 200 mM imidazole. The protein was concentrated and buffer-exchanged into 20 mM Tris–HCl pH 8.0, 50 mM NaCl, 10%(v/v) glycerol using centrifugal concentrators (10 kDa cutoff). The soluble protein was further purified using a HiTrap S anion-exchange column (GE Healthcare) at 289 K. After thorough washing, protein bound to the column was eluted with a linear gradient (0.1–1 M) of NaCl. Fractions containing protein were analyzed by 12% SDS–PAGE. Only those fractions that contained pure protein (as judged by SDS–PAGE) were pooled, concentrated and injected into a Superdex 75 gel-filtration column (GE Healthcare) pre-equilibrated with 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 10%(v/v) glycerol at 289 K (Fig. 1 ▶). The protein eluted as a single peak. Fractions containing pure 6×His-tagged TRIM2 NHL domain were pooled, concentrated to 10 mg ml−1 and stored at 277 K until further use.
Figure 1.
SDS–PAGE analysis (inset) and elution profile of the human TRIM2 NHL domain (residues 465–744) during gel-filtration chromatography using a Superdex G75 gel-filtration column. TRIM2 NHL was run on 12% SDS–PAGE and stained with Coomassie Blue to estimate its purity. The numbers on the right indicate the relative positions of protein molecular-mass markers (labelled in kDa).
2.3. Crystallization
Initial crystallization screening was carried out at 289 K by the hanging-drop vapour-diffusion method in 96-well plates (XtalQuest) using a Mosquito liquid-handling system (TTP Labtech). Each crystallization drop consisted of 200 nl protein solution (10 mg ml−1) mixed with 200 nl reservoir solution. The reservoir solution consisted of solutions from commercially available sparse-matrix crystallization screening kits (Crystal Screen, Crystal Screen 2, Index and PEG/Ion; Hampton Research). Drops were equilibrated over 100 µl reservoir solution. Rod-shaped crystals were obtained in one of the conditions consisting of 0.2 M potassium chloride, 20%(w/v) polyethylene glycol (PEG) 3350 pH 7.0 after 3 d of incubation at 289 K. Subsequent optimizations were carried out by varying the PEG 3350 and salt concentrations and the pH of the buffer. Final optimized crystals for data collection were obtained using a condition consisting of 0.2 M potassium chloride, 24%(w/v) PEG 3350 pH 7.5.
2.4. Data collection and processing
Crystals obtained after a series of optimization steps were flash-cooled in liquid nitrogen. Data collection was then performed at 100 K on beamline BL17U at Shanghai Synchrotron Radiation Facility (SSRF) using an ADSC Q315 detector. Crystals diffracted X-rays to 1.7 Å resolution and a data set was collected. The diffraction images were indexed, integrated, scaled and further processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). The structure of the extracellular domain of the serine/threonine protein kinase PknD from Mycobacterium tuberculosis (PDB entry 1rwl; Good et al., 2004 ▶), which shares 27% sequence identity with the TRIM2 NHL domain, was used as search model. Molecular replacement was performed using Phaser (McCoy et al., 2007 ▶) in the CCP4 suite (Winn et al., 2011 ▶).
3. Results and discussion
Truncated TRIM2 (residues 465–744), corresponding to the C-terminal NHL domain, was cloned and successfully overexpressed in E. coli. The protein was purified to homogeneity by employing a series of chromatographic techniques that included Ni affinity using an Ni–NTA agarose column (Qiagen), ion exchange using a HiTrap S anion-exchange column (GE Healthcare) and size-exclusion chromatography using a Superdex 75 gel-filtration column (GE Healthcare). The purity of the protein after gel filtration as estimated by SDS–PAGE was >95% (Fig. 1 ▶). Notably, during gel-filtration chromatography the protein eluted as a single peak (Fig. 1 ▶). An elution volume of 12.5 ml corresponded to an approximate molecular mass of 30 kDa based on calibration of the column using standard molecular-mass markers. Thus, the 30.2 kDa TRIM2 (residues 465–744) probably exists as a monomer in solution (Fig. 1 ▶).
Rod-shaped crystals of the TRIM2 NHL domain obtained after optimization of crystallization conditions (Fig. 2 ▶) diffracted X-rays to 1.7 Å resolution on beamline BL17U at SSRF (Fig. 2 ▶). Statistics of data collection are listed in Table 1 ▶. The crystals belonged to space group P21, with unit-cell parameters a = 43.6, b = 76.4, c = 107.4 Å, α = 90.0, β = 94.0, γ = 90.0°. A Matthews coefficient of 1.97 Å3 Da−1 (Matthews, 1968 ▶), corresponding to a solvent content of 37.6%, implies the presence of three molecules of the protein per asymmetric unit, although there is a possibility that only two molecules are present in the asymmetric unit, with a solvent content of 58.4%.
Figure 2.
(a) Typical crystals of the human TRIM2 NHL domain. (b) A typical diffraction image obtained from a crystal of the TRIM2 NHL domain. Circles are labelled with the resolution limits in Å.
Table 1. Data-collection and molecular-replacement phasing statistics for the TRIM2 NHL domain.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P21 |
| Unit-cell parameters (, ) | a = 43.56, b = 76.39, c = 107.35, = 90.0, = 94.0, = 90.0 |
| Wavelength () | 0.97889 |
| Resolution () | 35.841.70 (1.761.70) |
| Total reflections | 515125 (44807) |
| Unique reflections | 76074 (6997) |
| Data completeness (%) | 99.0 (91.5) |
| R merge † | 0.130 (0.917) |
| R p.i.m. ‡ | 0.055 (0.381) |
| Multiplicity | 6.8 (6.4) |
| I/(I) | 8.3 (2.8) |
| Molecular replacement§ | |
| No. of copies in asymmetric unit | 3 |
| Translation-function Z-score (TFZ) | 14.6 |
| No. of C clashes (PAK) | 0 |
| Log-likelihood gain (LLG) | 307 |
R
merge = 
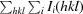 , where I(hkl) is the mean intensity of the i observations I
i(hkl) of reflection hkl.
, where I(hkl) is the mean intensity of the i observations I
i(hkl) of reflection hkl.
The precision-indicating merging R factor R
p.i.m. = 
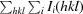 .
.
Scores for the best Phaser solution.
In subsequent studies, the data collected using these crystals were used for structure determination of the TRIM2 NHL domain by molecular replacement. A significant Phaser solution was obtained and the result confirmed the presence of three molecules in the asymmetric unit (Table 1 ▶). Model building and refinement of the TRIM2 NHL domain is currently in progress. The crystal structure of this TRIM2 fragment (residue 465–744) is likely to provide clues for the delineation of the exact function of the NHL domain and its mechanism of function at the molecular level.
Acknowledgments
This work was supported by grants from the State Key Development Program for Basic Research of the Ministry of Science and Technology of China (973 Project Grant Nos. 2014CB542802 and 2011CB910304), and the Strategic Priority Research Program of the Chinese Academy of Sciences, (Grant No. XDB080202008).
References
- Balastik, M., Ferraguti, F., Pires-da Silva, A., Lee, T. H., Alvarez-Bolado, G., Lu, K. P. & Gruss, P. (2008). Proc. Natl Acad. Sci. USA, 105, 12016–12021. [DOI] [PMC free article] [PubMed]
- Good, M. C., Greenstein, A. E., Young, T. A., Ng, H. L. & Alber, T. (2004). J. Mol. Biol. 339, 459–469. [DOI] [PubMed]
- Grigoryan, G. & Keating, A. E. (2008). Curr. Opin. Struct. Biol. 18, 477–483. [DOI] [PMC free article] [PubMed]
- Khazaei, M. R., Bunk, E. C., Hillje, A. L., Jahn, H. M., Riegler, E. M., Knoblich, J. A., Young, P. & Schwamborn, J. C. (2011). J. Neurochem. 117, 29–37. [DOI] [PubMed]
- Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S. & Weissman, A. M. (1999). Proc. Natl Acad. Sci. USA, 96, 11364–11369. [DOI] [PMC free article] [PubMed]
- Massiah, M. A., Simmons, B. N., Short, K. M. & Cox, T. C. (2006). J. Mol. Biol. 358, 532–545. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Ohkawa, N., Kokura, K., Matsu-Ura, T., Obinata, T., Konishi, Y. & Tamura, T. A. (2001). J. Neurochem. 78, 75–87. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S., Guffanti, A., Minucci, S., Pelicci, P. G. & Ballabio, A. (2001). EMBO J. 20, 2140–2151. [DOI] [PMC free article] [PubMed]
- Thompson, S., Pearson, A. N., Ashley, M. D., Jessick, V., Murphy, B. M., Gafken, P., Henshall, D. C., Morris, K. T., Simon, R. P. & Meller, R. (2011). J. Biol. Chem. 286, 19331–19339. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Ylikallio, E., Pöyhönen, R., Zimon, M., De Vriendt, E., Hilander, T., Paetau, A., Jordanova, A., Lönnqvist, T. & Tyynismaa, H. (2013). Hum. Mol. Genet. 22, 2975–2983. [DOI] [PubMed]
- Zheng, N., Wang, P., Jeffrey, P. D. & Pavletich, N. P. (2000). Cell, 102, 533–539. [DOI] [PubMed]