The SAM-dependent methyltransferase EgtD, an enzyme involved in ergothioneine biosynthesis, has been crystallized and native data have been collected as well as anomalous diffraction data at the K edge of a crystal of seleno-l-methionine-labelled protein.
Keywords: biosynthesis, ergothioneine, mycobacteria, selenium SAD
Abstract
Ergothioneine is an amino-acid betaine derivative of histidine that was discovered more than one century ago. Despite significant research pointing to a function in oxidative stress defence, the exact mechanisms of action of ergothioneine remain elusive. Although both humans and bacterial pathogens such as Mycobacterium tuberculosis seem to depend on ergothioneine, humans are devoid of the corresponding biosynthetic enzymes. Therefore, its biosynthesis may emerge as potential drug target in the development of novel therapeutics against tuberculosis. The recent identification of ergothioneine-biosynthetic genes in M. smegmatis enables a more systematic study of its biology. The pathway is initiated by EgtD, a SAM-dependent methyltransferase that catalyzes a trimethylation reaction of histidine to give N(α),N(α),N(α)-trimethylhistidine. Here, the recombinant production, purification and crystallization of EgtD are reported. Crystals of native EgtD diffracted to 2.35 Å resolution at a synchrotron beamline, whereas crystals of seleno-l-methionine-labelled protein diffracted to 1.75 Å resolution and produced a significant anomalous signal to 2.77 Å resolution at the K edge. All of the crystals belonged to space group P212121, with two EgtD monomers in the asymmetric unit.
1. Introduction
N(α),N(α),N(α)-Trimethyl-2-thiohistidine is a ubiquitous small metabolite found in all kingdoms of life (1; Fig. 1 ▶), with the potential exception of archaea. This compound was discovered in ergot fungi and was therefore named ergothioneine (Tanret, 1909 ▶). Since then, ergothioneine has been isolated from human tissue (Melville et al., 1957 ▶), plants, including important foodstuffs, fungi and mycobacteria (Genghof, 1970 ▶) as well as cyanobacteria (Pfeiffer et al., 2011 ▶). The identification of ergothioneine-biosynthetic genes revealed that ergothioneine is in fact biosynthesized by a broad range of bacteria, most sequenced basidomycetes, some ascomycetes and even by several plants (Seebeck, 2010 ▶). Humans absorb ergothioneine from their diet and maintain up to millimolar concentrations in specific tissues such as kidney, liver and the central nervous system (Hartman, 1990 ▶). Key to this inhomogeneous distribution is probably the ergothioneine-specific transporter protein OCTN1 (Gründemann et al., 2005 ▶). The elimination of this protein from cultured HeLa cells through RNA interference induces oxidative stress (Paul & Snyder, 2010 ▶). On the other hand, gain-of-function mutations of this transporter are associated with Crohn’s disease (Peltekova et al., 2004 ▶). These observations suggest that ergothioneine is an important player in human physiology, but the relationship between cellular ergothioneine levels and health is complex. Ergothioneine seems to protect the fungus Neurospora crassa from peroxide-induced stress, even though it is remarkably inert towards peroxides in vitro (Bello et al., 2012 ▶). The fact that human pathogens such as Mycobacterium tuberculosis biosynthesize and apparently require ergothioneine opens the possibility that this pathway provides a potential target for novel chemotherapeutics.
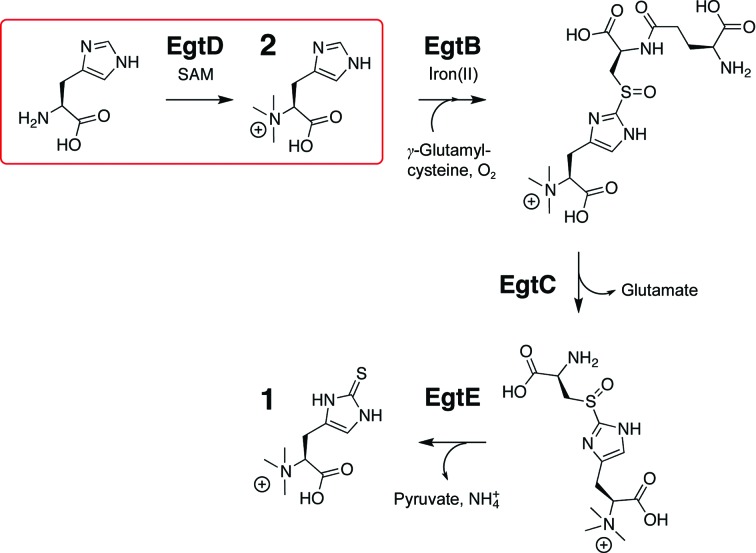
Figure 1.
Biosynthesis of ergothioneine (1) from histidine requires S-adenosylmethionine (SAM), γ-glutamylcysteine and atmospheric oxygen as co-substrates and is catalyzed by EgtABCDE. In the first step, the SAM-dependent methyltransferase EgtD catalyzes histidine-betaine synthesis (2).
In order to study the biology of ergothioneine, we recently characterized the biosynthetic gene cluster egtABCDE from M. smegmatis (Seebeck, 2010 ▶). In vitro reconstitution of the methyltransferase EgtD, the sulfoxide synthase EgtB and the amidohydrolase EgtC demonstrated their central role in thiohistidine biosynthesis (Fig. 1 ▶). EgtD catalyzes the first, committing step by transferring three methyl groups from three equivalents of S-adenosylmethionine (SAM) to the α-amino group of histidine. The resulting histidine betaine (2; Fig. 1 ▶) is the preferred substrate of EgtB, which does not act on histidine directly. Deletion of egtD in M. smegmatis abolishes ergothioneine biosynthesis (Sao Emani et al., 2013 ▶), suggesting that EgtD is a valid target to sabotage ergothioneine biosynthesis in vivo.
Although SAM-dependent methyltransferases constitute a well characterized class of enzymes (Gana et al., 2013 ▶; Martin & McMillan, 2002 ▶), the EgtD sequence, previously annotated as domain of unknown function 2260 (DUF2260), does not fit well to any known methyltransferase. The closest EgtD homologue with known function is EasF (24% sequence identity to EgtD), which catalyzes the N(α)-methylation of dimethylallyl tryptophan in ergot alkaloid biosynthesis in Claviceps species (3 and 4; Supplementary Fig. S11; Lorenz et al., 2007 ▶). However, this enzyme has not yet been crystallized. With a sequence identity of 16%, the closest structurally characterized homologue is the human methyltransferase Ad-003 (PDB entry 2ex4; Structural Genomics Consortium, unpublished work), which in turn belongs to a family of enzymes that catalyze the dimethylation of ribosomal proteins at N-terminal proline residues (Webb et al., 2010 ▶).
As a first step on the path to understanding bacterial ergothioneine production, we address the structure–function relationships of its producing enzymes. To unveil the role of EgtD in ergothioneine biosynthesis and at the same time characterize the structure of DUF2260-type proteins for the first time, we produced EgtD from M. smegmatis in Escherichia coli and crystallized the purified protein using the following methodology.
2. Materials and methods
2.1. Production of recombinant EgtD
The open reading frame of egtD (UniProtKB entry EGTD_MYCS2) was amplified from M. smegmatis genomic DNA (strain ATCC 607; DSMZ 43465). The forward primer (5′-GCGCATATGGCGCTCTCACTGGCCAACTACCTA-3′) includes an NdeI restriction site (bold) and the reverse primer (5′-GCGCTCGAG TCACCGCACCGCCAGCGACAACCs-3′) contains an XhoI restriction site (bold) and a stop codon (italics). After restriction, the gene was cloned into the modified pET-19b vector pET19m (Novagen) digested with the same enzymes. Cloning resulted in an EgtD fusion construct with an N-terminal His6 tag followed by a TEV (Tobacco etch virus) protease cleavage site and was checked by DNA sequencing (the full sequence of the pET19m_His6-TEV-EgtD construct is mghhhhhhaENLYFQ|GHMALSLANYLAADSAAEALRRDVRAGLTAAPKSLPPKWFYDAVGSDLFDQITRLPEYYPTRTEAQILRTRSAEIIAAAGADTLVELGSGTSEKTRMLLDAMRDAELLRRFIPFDVDAGVLRSAGAAIGAEYPGIEIDAVCGDFEEHLGKIPHVGRRLVVFLGSTIGNLTPAPRAEFLSTLADTLQPGDSLLLGTDLVKDTGRLVRAYDDAAGVTAAFNRNVLAVVNRELSADFDLDAFEHVAKWNSDEERIEMWLRARTAQHVRVAALDLEVDFAAGEEMLTEVSCKFRPENVVAELAEAGLRQTHWWTDPAGDFGLSLAVR, where lower-case letters indicate the affinity tag and italic letters and | indicates the TEV protease cleavage site).
Heterologous expression of EgtD employed E. coli BL21(DE3) cells in LB medium supplemented with 100 µg ml−1 ampicillin. The cells were grown to an OD600 of 0.8 and were then induced with 0.5 mM IPTG for 16 h at 293 K. The cells were harvested by centrifugation, resuspended in buffer A (300 mM NaCl, 50 mM Na2HPO4 pH 8.0) and lysed by sonication. After centrifugation for 1 h at 48 000g, the protein was purified using Ni2+–NTA Agarose beads (Qiagen) by washing the beads with buffer A containing 10 mM imidazole followed by elution with buffer A supplemented with 250 mM imidazole. Cleavage of His6-tagged EgtD with recombinant His6-tagged TEV protease proceeded in a dialysis bag (cutoff 10 kDa) against 20 mM Tris–HCl, 150 mM NaCl pH 7.5 overnight at 277 K. After the removal of precipitated proteins from the dialysate, uncleaved EgtD and TEV were separated in a second purification with Ni2+–NTA Agarose (Qiagen). The tag-removed EgtD was concentrated and loaded onto a size-exclusion chromatography column (Superdex 75 26/60, GE Healthcare) equilibrated in the same buffer. EgtD eluted with a retention volume of 170 ml, which suggested it to be a monomer in solution (calculated molecular weight of 35.2 kDa). This was confirmed by analytical size-exclusion experiments using an S200 10/300 GL column (GE Healthcare; Supplementary Fig. S2). Purified EgtD was concentrated to 25 mg ml−1 (calculated ∊280 = 36 440 M −1 cm−1), flash-frozen in liquid nitrogen and stored at 193 K if not used immediately.
Production of seleno-l-methionine-labelled EgtD was achieved by cultivation of E. coli BL21(DE3)-RIL cells in artificial medium suppressing l-methionine biosynthesis (LeMaster & Richards, 1985 ▶) and with buffers containing 5 mM β-mercaptoethanol. To yield purer protein in the first affinity-chromatography step, a Ni2+ HiTrap Chelating HP column connected to an ÄKTAprime system (GE Healthcare) was used to allow elution of EgtD in a continuous gradient of 10–250 mM imidazole. Further purification steps were performed as for the native protein. The identity of the proteins and the incorporation of seleno-l-methionine were confirmed by ESI mass-spectrometric analysis.
2.2. Crystallization
Initial crystallization conditions of tag-removed EgtD (in 20 mM Tris–HCl pH 7.5, 150 mM NaCl) were determined by the sitting-drop vapour-diffusion method with The JCSG Core Suites I–IV, The PACT Suite and The PEGs Suite (Qiagen). Screens were set up in a 96-well plate at 293 K using a dispensing robot (Mosquito, TTP Labtech). The drops consisted of 100 nl protein solution and 100 nl reservoir solution equilibrated against 40 µl reservoir solution. The protein was screened at three different concentrations (20, 15 and 10 mg ml−1). Crystals of different morphology appeared with various precipitants, which were optimized with respect to pH and component concentrations. A hanging drop set up in 24-well plates was used for optimization of the initial hits. The final condition for the native protein consisted of 0.5 µl protein solution at 20 mg ml−1 EgtD mixed with 0.5 µl reservoir [16–18%(w/v) PEG 8000, 20%(w/v) glycerol, 0.16 M magnesium acetate, 0.08 M sodium cacodylate pH 6.2–6.7] equilibrated against 500 µl reservoir solution. Crystals of the seleno-l-methionine-labelled protein were obtained by changing the buffer to MES but keeping the composition of the crystallization solution otherwise similar [18%(w/v) PEG 8000, 20%(w/v) glycerol, 0.08 M MES pH 6.3, 0.16 M magnesium acetate]. High-quality crystals appeared overnight and no differences in crystallization behaviour were observed between fresh and frozen protein.
2.3. Data collection and processing
The crystals were flash-cooled in liquid nitrogen for data collection without additional cryoprotecting agents and tested at an X-ray home source at 100 K using a copper rotating anode (λ = 1.54 Å) and a MAR345 image-plate detector (MAR Research). Well diffracting crystals were sent to beamline PXII of the SLS (Swiss Light Source at the Paul Scherrer Institute, Villigen, Switzerland) for data collection. A native data set was collected at a wavelength of 0.9830 Å as 1000 non-overlapping 0.25° frames using a Pilatus 6M detector (Dectris). Since molecular replacement using BALBES (Long et al., 2008 ▶) was not successful for this native EgtD data set, diffraction data were collected from seleno-l-methionine-labelled crystals of EgtD at 100 K using the same setup. The wavelength was set to 0.9786 Å to collect data at the Se K edge (12.659 keV). A single anomalous dispersion (SAD) data set was collected from the seleno-l-methionine-labelled protein crystal as 1440 non-overlapping 0.25° frames. A fluorescence scan was not performed. The data set was indexed and integrated with XDS (Kabsch, 2010 ▶) and scaled using AIMLESS (Evans, 2006 ▶, 2011 ▶) in the CCP4 suite (Winn et al., 2011 ▶). To determine the anomalous cutoff of the data, phenix.xtriage from the PHENIX package was used (Zwart et al., 2005 ▶; Adams et al., 2011 ▶). Using HKL2MAP, extraction of the anomalous signal was achieved with SHELXC and Se atoms were located with SHELXD (Schneider & Sheldrick, 2002 ▶; Pape & Schneider, 2004 ▶; Sheldrick, 2010 ▶). Details of the data-collection and processing statistics can be found in Table 1 ▶.
Table 1. Data-collection and processing statistics.
Values for the highest resolution shell are given in parentheses. All data were collected on beamline PXII at the Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland.
| Data set | Se-SAD† | Se | Native |
|---|---|---|---|
| Wavelength (Å) | 0.9786 | 0.9786 | 0.9830 |
| Resolution range (Å) | 48.72–2.77 (2.92–2.77) | 48.72–1.75 (1.78–1.75) | 49.1–2.35 (2.43–2.35) |
| Space group | P212121 | P212121 | P212121 |
| Unit-cell parameters | |||
| a (Å) | 71.8 | 71.8 | 71.8 |
| b (Å) | 75.5 | 75.5 | 76.9 |
| c (Å) | 138.7 | 138.7 | 139.0 |
| α = β = γ (°) | 90.0 | 90.0 | 90.0 |
| Mosaicity‡ (°) | 0.115 | 0.115 | 0.061 |
| Completeness (%) | 99.1 (98.6)† | 97.4 (95.8) | 100 (100) |
| Total No. of reflections | 250555 (39024) | 1013484 (57635) | 302720 (30322) |
| No. of unique reflections | 19504 (2761) | 74528 (3977) | 32867 (3182) |
| Multiplicity | 6.9 (7.4)† | 13.6 (14.5) | 9.2 (9.5) |
| Mean I/σ(I) | 43.6 (28.1)§ | 18.8 (2.5) | 10.2 (2.5) |
| R anom ¶ (%) | 2.7 | — | — |
| R merge †† (%) | 5.4 (9.1) | 10.6 (127.6) | 18.9 (112.8) |
| R p.i.m ‡‡ (%) | 1.9 (3.3) | 3.0 (34.5) | 6.5 (38.1) |
| Overall B factor from Wilson plot (Å2) | 18.8 | 19.3 | 20.1 |
Friedel mates were treated as separate reflections.
Mosaicity values as reported by XDS (Kabsch, 2010 ▶).
The data were cut at mean I/σ(I) = 28.1 in the highest resolution shell because the anomalous signal is significant to 2.77 Å resolution according to phenix.triage from PHENIX (Zwart et al., 2005 ▶; Adams et al., 2011 ▶).
R
anom = 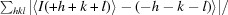
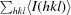 .
.
R
merge = 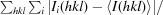
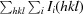 , where Ii(hkl) is the intensity of the ith observation of the reflection with index hkl.
, where Ii(hkl) is the intensity of the ith observation of the reflection with index hkl.
R
p.i.m. = 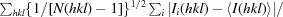
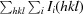 , where N(hkl) is the number of observations of the reflection with index hkl (Weiss, 2001 ▶).
, where N(hkl) is the number of observations of the reflection with index hkl (Weiss, 2001 ▶).
3. Results and discussion
Methyl-group transfer is an important post-translational modification of various proteins, but also occurs in DNA, RNA and small molecules. In most cases, one or two methyl groups are transferred, whereas EgtD is one of the few crystallized enzymes that catalyze the transfer of three methyl groups to the same substrate. In proteins, trimethylation is catalyzed, for example, by diphthine synthases, which trimethylate a pre-modified histidine in the translation elongation factor 2 (EF-2) precursor (Kishishita et al., 2008 ▶; Moehring et al., 1984 ▶). Although the diphthine synthases and EgtD catalyse somewhat similar reactions, the two enzymes share no discernable sequence homology. In contrast to diphthine synthases, which are homodimers, EgtD elutes as a monomer from size-exclusion chromatography columns. The retention time and apparent molecular weight of EgtD perfectly agree with the calculated molecular weight (MWcalc) of 35.2 kDa (Supplementary Fig. S2).
The purification yield was 25 mg native EgtD and 4.5 mg seleno-l-methionine-labelled EgtD from 1 l of culture, respectively. Verification of selenium incorporation using ESI mass spectrometry resulted in a 234 Da shift with respect to native EgtD. This demonstrates that all five methionine residues have been labelled with seleno-l-methionine.
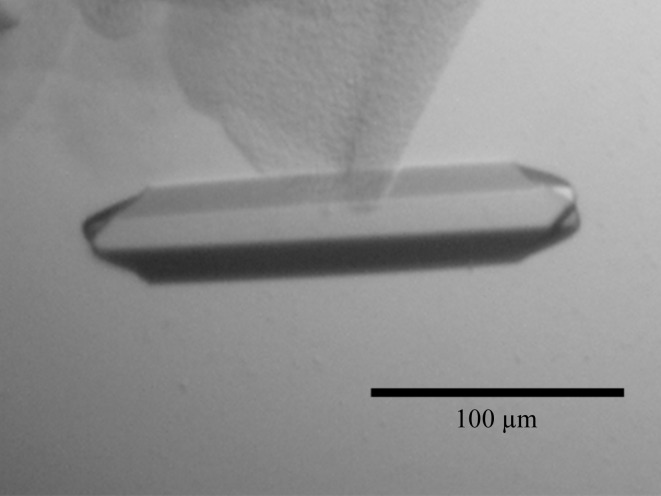
In the initial crystallization condition of native EgtD (as indicated in §2.2), rod-shaped crystals appeared overnight and were optimized with respect to pH and precipitant concentration. Furthermore, the initial condition was successfully used with the seleno-l-methionine-labelled EgtD. In additional optimization rounds, the cacodylate buffer was replaced with a variety of organic buffer substances to avoid interference with the anomalous signal of selenium. All conditions yielded crystals of 200 × 30 × 30 µm in size (Fig. 2 ▶). Additional cryoprotection of EgtD crystals was not required since the reservoir solution contained sufficient concentrations of precipitants.
Figure 2.
Crystal of native EgtD.
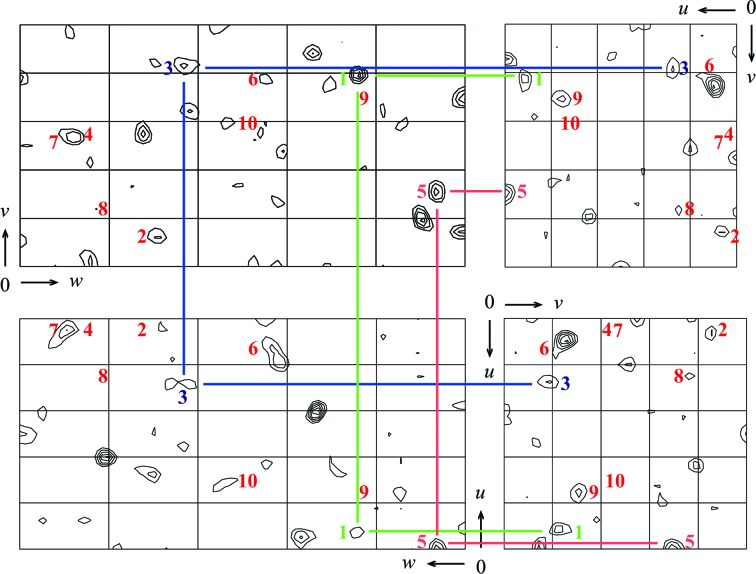
Initially, we collected a native data set for EgtD. Indexing and integration with XDS (Kabsch, 2010 ▶) revealed that the investigated EgtD crystals belonged to space group P212121, suggesting the presence of two EgtD molecules in the asymmetric unit (solvent content 52.5%). None of the methyltransferase structures available in the PDB allowed structure solution of EgtD by molecular replacement, even using the molecular-replacement engine BALBES (Long et al., 2008 ▶). We therefore initiated experimental phasing by producing seleno-l-methionine-labelled EgtD. These crystals diffracted better than those obtained with the unlabelled protein, allowing data collection to 1.75 Å resolution. Analysis with phenix.xtriage from the PHENIX package (Zwart et al., 2005 ▶; Adams et al., 2011 ▶) detected a significant anomalous signal to 2.77 Å resolution. A Patterson search with SHELXD (Schneider & Sheldrick, 2002 ▶) located the positions of ten anomalous scatterers, as expected for two EgtD monomers in the asymmetric unit (Supplementary Fig. S3), yet not all of the self-vectors of these atoms can be unambigously identified in the Harker sections of the anomalous Patterson difference map, most likely owing to the presence of additional cross-vectors (Fig. 3 ▶). We are currently using these data for initial phase determination and will report the structure elsewhere.
Figure 3.
Harker sections of an anomalous difference Patterson map calculated from the Se-SAD data reported in Table 1 ▶. The map cutoff was set to 2σ. Numbers indicate the expected peaks (self-vectors) deduced from the positions of the ten anomalous scatterers located with SHELXD (Supplementary Fig. S3). Coloured lines indicate the corresponding Harker vectors of three anomalous scatterers that can immediately be deduced from this plot.
Supplementary Material
Supporting Information.. DOI: 10.1107/S2053230X1400805X/no5051sup1.pdf
Acknowledgments
The authors are indebted to R. S. Goody, who enabled the initiation of this project at the MPI for Molecular Physiology in Dortmund. The authors would like to thank the X-ray communities of the Max Planck Institutes for Medical Research and Molecular Physiology (Heidelberg and Dortmund, Germany) for help with data collection and S. Gentz for assistance with ESI mass spectrometry. The Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) is acknowledged for providing generous access to their beamlines. This project was supported by the Swiss National Science Foundation. AV is supported by the HZI Graduate School for Infection Research and FPS is supported by the ‘Professur für Molekulare Bionik’.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: NO5051).
References
- Adams, P. D. et al. (2011). Methods, 55, 94–106.
- Bello, M. H., Barrera-Perez, V., Morin, D. & Epstein, L. (2012). Fungal Genet. Biol. 49, 160–172. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, P. R. (2011). Acta Cryst. D67, 282–292. [DOI] [PMC free article] [PubMed]
- Gana, R., Rao, S., Huang, H., Wu, C. & Vasudevan, S. (2013). BMC Struct. Biol. 13, 6. [DOI] [PMC free article] [PubMed]
- Genghof, D. S. (1970). J. Bacteriol. 103, 475–478. [DOI] [PMC free article] [PubMed]
- Gründemann, D., Harlfinger, S., Golz, S., Geerts, A., Lazar, A., Berkels, R., Jung, N., Rubbert, A. & Schömig, E. (2005). Proc. Natl Acad. Sci. USA, 102, 5256–5261. [DOI] [PMC free article] [PubMed]
- Hartman, P. E. (1990). Methods Enzymol. 186, 310–318. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kishishita, S., Shimizu, K., Murayama, K., Terada, T., Shirouzu, M., Yokoyama, S. & Kunishima, N. (2008). Acta Cryst. D64, 397–406. [DOI] [PubMed]
- LeMaster, D. M. & Richards, F. M. (1985). Biochemistry, 24, 7263–7268. [DOI] [PubMed]
- Long, F., Vagin, A. A., Young, P. & Murshudov, G. N. (2008). Acta Cryst. D64, 125–132. [DOI] [PMC free article] [PubMed]
- Lorenz, N., Wilson, E. V., Machado, C., Schardl, C. L. & Tudzynski, P. (2007). Appl. Environ. Microbiol. 73, 7185–7191. [DOI] [PMC free article] [PubMed]
- Martin, J. L. & McMillan, F. M. (2002). Curr. Opin. Struct. Biol. 12, 783–793. [DOI] [PubMed]
- Melville, D. B., Eich, S. & Ludwig, M. L. (1957). J. Biol. Chem. 224, 871–877. [PubMed]
- Moehring, T. J., Danley, D. E. & Moehring, J. M. (1984). Mol. Cell. Biol. 4, 642–650. [DOI] [PMC free article] [PubMed]
- Pape, T. & Schneider, T. R. (2004). J. Appl. Cryst. 37, 843–844.
- Paul, B. D. & Snyder, S. H. (2010). Cell Death Differ. 17, 1134–1140. [DOI] [PMC free article] [PubMed]
- Peltekova, V. D., Wintle, R. F., Rubin, L. A., Amos, C. I., Huang, Q., Gu, X., Newman, B., Van Oene, M., Cescon, D., Greenberg, G., Griffiths, A. M., St George-Hyslop, P. H. & Siminovitch, K. A. (2004). Nature Genet. 36, 471–475. [DOI] [PubMed]
- Pfeiffer, C., Bauer, T., Surek, B., Schömig, E. & Gründemann, D. (2011). Food Chem. 129, 1766–1769.
- Sao Emani, C., Williams, M. J., Wiid, I. J., Hiten, N. F., Viljoen, A. J., Pietersen, R.-D. D., van Helden, P. D. & Baker, B. (2013). Antimicrob. Agents Chemother. 57, 3202–3207. [DOI] [PMC free article] [PubMed]
- Schneider, T. R. & Sheldrick, G. M. (2002). Acta Cryst. D58, 1772–1779. [DOI] [PubMed]
- Seebeck, F. P. (2010). J. Am. Chem. Soc. 132, 6632–6633. [DOI] [PubMed]
- Sheldrick, G. M. (2010). Acta Cryst. D66, 479–485. [DOI] [PMC free article] [PubMed]
- Tanret, C. (1909). C. R. Acad. Sci. 49, 222–224.
- Webb, K. J., Lipson, R. S., Al-Hadid, Q., Whitelegge, J. P. & Clarke, S. G. (2010). Biochemistry, 49, 5225–5235. [DOI] [PMC free article] [PubMed]
- Weiss, M. S. (2001). J. Appl. Cryst. 34, 130–135.
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zwart, P. H., Grosse-Kunstleve, R. W. & Adams, P. D. (2005). CCP4 Newsl. Protein Crystallogr. 43, contribution 7.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.. DOI: 10.1107/S2053230X1400805X/no5051sup1.pdf