Abstract
LINE-1 (L1) elements are the only active and autonomous transposable elements in humans. The core retrotransposition machinery is a ribonucleoprotein particle (RNP) containing the L1 mRNA, with endonuclease and reverse transcriptase activities. It initiates reverse transcription directly at genomic target sites upon endonuclease cleavage. Recently, using a direct L1 extension assay (DLEA), we systematically tested the ability of native L1 RNPs to extend DNA substrates of various sequences and structures. We deduced from these experiments the general rules guiding the initiation of L1 reverse transcription, referred to as the snap-velcro model. In this model, L1 target choice is not only mediated by the sequence specificity of the endonuclease, but also through base-pairing between the L1 mRNA and the target site, which permits the subsequent L1 reverse transcription step. In addition, L1 reverse transcriptase efficiently primes L1 DNA synthesis only when the 3′ end of the DNA substrate is single-stranded, suggesting so-far unrecognized DNA processing steps at the integration site.
Keywords: reverse transcription, reverse transcriptase, target-primed reverse transcription, TPRT, non-LTR retrotransposon, LINE-1, L1, poly(A) tail, endonuclease
L1 Elements are Endogenous Mutagens in the Human Genome
Transposable elements account for half to two-thirds of the human genome.1 Among them, LINE-1 (L1) non-LTR retrotransposons form the only autonomous and active family and are the most abundant, representing 17% of our DNA. Each individual genome contains hundreds of potentially active L1 copies, and hundreds of thousands of defective copies, which are truncated, fragmented, and/or mutated.2 The active copies can proliferate via an RNA-mediated copy-and-paste mechanism, called retrotransposition. L1 insertions are intrinsically mutagenic, however their actual impact on gene expression depends on their specific site of integration. Intergenic or deep intronic insertions can often have no detectable effects on genes. In contrast, insertions in exons or regulatory sequences have the potential to profoundly alter gene expression/function, by disrupting coding- or cis-regulating sequences, or by carrying cis-regulating sequences (transcription factor binding sites, cryptic splicing and polyadenylation sites, etc.).2,3 Hence, germline L1 insertions sporadically cause de novo genetic diseases,2,4 and somatic L1 retrotransposition in cancer has been shown to contribute to tumor genome dynamics, including driver mutations.5-10 Therefore, exploring the mechanisms that influence L1 target choice is crucial to our understanding of L1-driven genome plasticity.
L1 Retrotransposition can be Initiated through Two Different Pathways
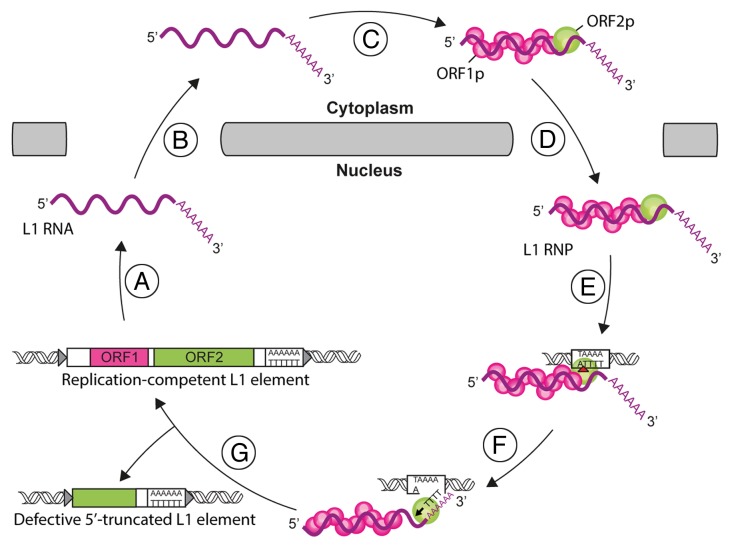
The L1 replication cycle starts with the synthesis of a bicistronic mRNA coding for the two L1 proteins, ORF1p and ORF2p (Fig. 1). ORF1p is a 40 kDa RNA-binding protein able to form trimers.11-13 It exhibits nucleic acid chaperone activity,14 the function of which has not been elucidated. ORF2p is a large, 149 kDa, protein with endonuclease (EN) and reverse transcriptase (RT) activities.15,16 Both ORF1p and ORF2p bind the L1 RNA to form a stable ribonucleoprotein particle (RNP), the core of the L1 retrotransposition machinery.17,18 The L1 RNP can mediate two different integration processes. In the canonical pathway, called target-primed reverse transcription (TPRT),19 the L1 EN activity produces a nick at the recognized target site in the chromosomal DNA.16 The RT moiety then extends this liberated 3′-OH group, using the L1 RNA as a template.20 Reverse transcription is primed within the poly(A) tail of the L1 RNA.21 L1 EN preferentially cuts DNA at a consensus sequence 5′-TTTTA-3′, with nicking occurring at the TpA bond.16,22-24 In an alternative pathway, named endonuclease-independent (ENi) retrotransposition or non-classical L1 insertion (NCLI), reverse transcription is initiated at pre-existing DNA lesions, without the need for endonuclease cleavage.25,26 A particular case of this pathway is retrotransposition at telomeres, the natural extremity of chromosomes.27 Regardless of whether a particular retrotransposition event is initiated by EN or not, the subsequent steps of the reaction, such as second strand DNA synthesis or ligation of the 3′ ends of the neo-synthesized DNA to the target DNA, have not been explored yet.
Figure 1. The L1 life-cycle. L1 replication starts by the transcription of a bicistronic mRNA (A). The L1 RNA is exported to the cytoplasm (B). ORF1p and ORF2p proteins are translated and bind to the L1 RNA to form L1 ribonucleoprotein particles (RNP) (C). The L1 RNP is imported into the nucleus (D). Integration and reverse transcription occur at the genomic target site. First, the L1 endonuclease (EN) activity nicks the target DNA (red arrowhead, E). Then, the L1 reverse transcriptase (RT) initiates the reverse transcription of L1 RNA (black arrowhead, F). The mechanisms involved in the final steps of this process and the resolution of the integration are unresolved yet (G). Partial reverse transcription can lead to 5′-truncated L1 copies.
The Snap-Velcro Model of L1 Reverse Transcription Initiation
Recently, we explored the mechanism of L1 reverse transcription initiation using a direct L1 extension assay (DLEA).28 In this approach, native L1 RNP expressed in—and enriched from—human cells are incubated with oligonucleotide primers of various sequences or structures, and with radioactive dTTP only, for a very short time (less than 5 min). The products are then resolved on sequencing gels to directly visualize the extension of the primer. Due to the short incubation time and the use of dTTP only, it focuses on the initiation of reverse transcription. Advantages of this method, compared with previously PCR-based techniques, include its versatility with regards to the primers that can be used and its quantitative nature. A limitation of DLEA is the absence of sequence information and its lower sensitivity.
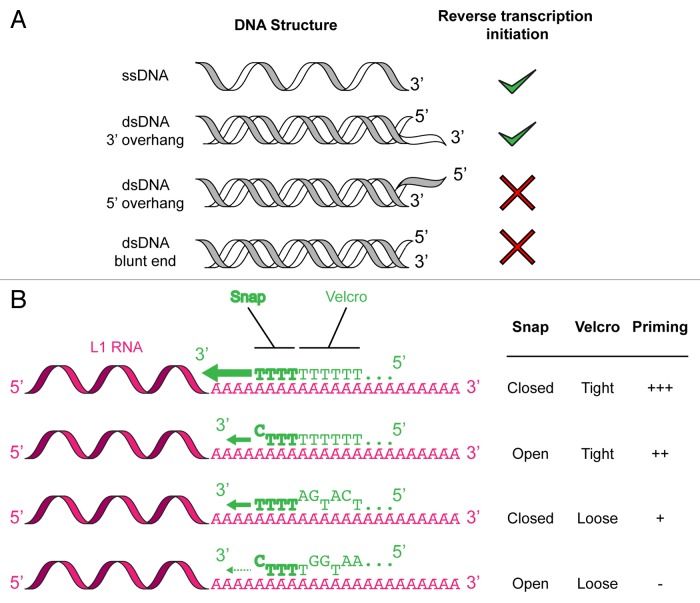
One of the unresolved questions related to L1 reverse transcription priming was whether—or to which degree—the 3′ end of the nicked genomic DNA needs to be accessible and to base-pair with the poly(A) tail of the L1 RNA. Indeed, R2, a related non-LTR retrotransposon which has been used to establish the basis of the TPRT model, does not require such a complementarity.29,30 Although the consensus sequence released upon L1 EN cleavage (5′-TTTT-3′) could in principle anneal to the poly(A) tail of the L1 RNA, it is extremely short for maintaining a stable interaction and the actual sequences cleaved by the L1 EN can significantly differ from the consensus sequence.22-24 To directly address this question, we quantified the efficiency of extension of more than 65 primers by DLEA. Based on the results of these experiments, and on additional analyses of the distribution of polymorphic L1 insertions in the human genome, we proposed the snap-velcro model for L1 reverse transcription initiation.28 This model is detailed in Figure 2. The efficiency of reverse transcription initiation is influenced by the 10 last nucleotides of the target DNA. The 4 last nucleotides (the snap) contribute the most to this process. Reverse transcription priming is the most efficient when the snap corresponds to 4 Ts (snap closed). However suboptimal sequences with terminal or internal mismatches can be tolerated (snap open). For terminal mismatches, the efficiency of extension depends on the nature of the base ending the primer (T > C > A > G). These suboptimal sequences can be more efficiently extended if mismatches are compensated by an increased number of matching Ts in the upstream 6 nt (velcro strap fastened). Finally an important aspect of our results is that priming only occurs if the DNA substrate is single-stranded. Indeed, double-stranded DNA substrates are extended only if they end with a 3′ overhang. If the 3′ extremity of the target DNA is embedded in duplex DNA, either as a blunt- or as a 3′-recessed end, no extension could be detected under the DLEA conditions employed.
Figure 2. Features of the snap-velcro model of L1 reverse transcription priming. (A) Reverse transcription priming only occurs if the DNA substrate is single-stranded. (B) Reverse transcription priming requires base-pairing between the L1 RNA (pink) poly(A) tail and the target-site DNA (green). The snap (bold green) corresponds to the last 4 nucleotides at the 3′ of the DNA primer. The velcro (light green) contains the 6 bases upstream of the snap. The snap is considered as closed if 4 nucleotides are T. The velcro is tightly fastened if the position-weighted T-density is superior to 0.5 (see ref. 28 for the detailed numerical model). The snap-velcro status predicts the efficiency of L1 reverse transcription priming (green arrow).
Consequences of the Snap-Velcro Model Related to Primer-Template Sequence Match
The snap-velcro model indicates that complementarity between the L1 poly(A) tail and the last 10 nucleotides of the target DNA is important for priming of L1 reverse transcription, yet allows sufficient flexibility to accommodate a wide range of potential target sites. It explains a long-standing observation that L1 elements are often flanked by imperfect T-rich sequences significantly longer than expected for the recognition site of the L1 endonuclease.23,24,31,32 This model also implies that L1 target-site selection relies not only on the sequence specificity of the EN nicking reaction, but also on the subsequent ability of the RT to efficiently extend the cleaved product. This observation has practical and technological implications. Non-LTR retrotransposons fall into two main classes: i) the stringent elements, which always insert into discrete and defined genomic locations with high sequence specificity (such as R1, R2, Tx1L, SART1, Tras1), and ii) more promiscuous elements, which insert into multiple locations within a very short and degenerate sequence (such as L1). EN domain swapping between two stringent retrotransposons is sufficient to exchange their respective target site selectivity both in vitro and in vivo.33 Similarly, structure-driven domain swapping between a stringent EN and the L1 EN moiety has succeeded in modifying its target selectivity in vitro.34 However, such EN variants, when reintroduced into a complete L1 element and tested in vivo, are unable to redirect L1 insertions to altered target sites. Instead, they continue to insert in T-rich stretches,34 indicating that other determinants downstream of the initial EN cleavage contribute to L1 target site selection. Our results suggest that L1 RT priming specificity could be one of these determinants. Therefore engineering L1 to achieve site-specific integration in vivo might require to take into account both EN and RT specificities.
Consequences of the Snap-Velcro Model Related to the Accessibility of the Target DNA
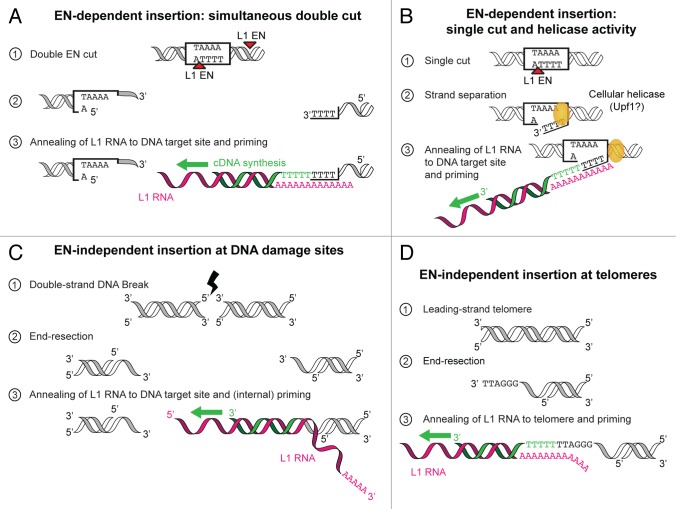
A second important feature of the snap-velcro model is the requirement for a single-stranded 3′ overhang to initiate reverse transcription at the target site. The original TPRT model stipulates that retrotransposition is initiated by a nick in the target DNA.19,20,35 How does the 3′-OH extremity of the substrate, which is embedded in the duplex DNA, become available for L1 RT? We can envision several possibilities. First, the L1 EN produces double-stranded staggered cuts rather than nicks. In vitro, plasmid DNA can indeed be linearized upon prolonged incubation with an isolated recombinant EN domain.16 Whether L1 ORF2p acts as a monomer or a multimer in the context of the L1 RNP is unknown. However, many other reverse transcriptases form dimers, including the R2 RT or human telomerase.36,37 Dimerization of ORF2p could lead to concomitant cleavage of bottom and top strands, while maintaining the two target site DNA extremities together (Fig. 3A). Of note, the average length of the target-site duplication, which reflects the distance between the top and bottom strand cuts, is 15 nucleotides,31 a length compatible with the minimal size of the single-stranded region (6 nt). A second hypothetical mechanism involves a strand-transfer or a DNA helicase activity (Fig. 3B). Although ORF1p has been proposed to perform this task through its nucleic acid chaperone activity,14 the presence of this protein in native L1 RNPs was not sufficient under our experimental conditions to prime reverse transcription with duplex DNA substrates. Recent efforts have identified a number of cellular proteins associated with the L1 retrotransposition machinery.38,39 Among them, Upf1 an RNA- and DNA-dependent 5′-3′ helicase implicated in nonsense-mediated mRNA decay, and also in telomeric DNA replication,40 is a serious candidate. It is noteworthy that our attempts to extend duplex DNA by L1 RNP were performed in the absence of ATP, an essential cofactor of helicase activities. Thus, to explore the mechanism allowing annealing of the target site DNA to the L1 poly(A) tail, future follow-up DLEA experiments could be performed with the additional inclusion of ATP. Finally, unlike a TPRT reaction, DLEA uncouples EN cleavage and RT priming. Thus we cannot exclude that, in vivo, the product of the nicking reaction would somehow be channeled to the RT active site, being unwounded in the process.
Figure 3. Hypothetical mechanisms allowing L1 RNA base-pairing with target-site DNA. (A) ORF2p dimerization leads to simultaneous staggered cuts through its EN activity. The resulting extremities have 3′ overhangs, which can anneal to the L1 RNA and prime L1 cDNA synthesis. (B) L1 EN initially starts with a single cut, but a DNA-dependent helicase unwinds the target site DNA strands, enabling L1 cDNA synthesis. (C) Upon double-strand DNA break, DNA repair factors resect these ends and generate 3′ overhangs. These new extremities not necessarily end with Ts as for EN sites. Consequently, base-pairing generally occurs at internal sites within the L1 RNA which show spurious matches with the damaged site. (D) Telomeres naturally end with 3′ overhangs. Red arrowhead, EN cut; green, cDNA; pink, L1 RNA.
In this perspective, DLEA reactions are more similar to reverse transcription taking place in the ENi pathway.25,26 The nature of the preexisting lesions used by L1 RT to prime reverse transcription is still unclear, in particular whether they are single- or double-stranded. Telomeres are frequent integration sites in this pathway,27,41 and notably they end with a 3′ overhang, which is also the preferred substrate of telomerase42 (Fig. 3D). Another interesting observation is that ENi-retrotransposition only occurs efficiently in cells defective for both p53 and non-homologous end-joining (NHEJ).25,43 Impairing NHEJ results in end-resection and extremities with 3′ overhangs, which ultimately leads to homologous recombination-mediated repair.44 We speculate that NHEJ inhibition could be necessary to permit the generation of these 3′ overhangs and thereby reverse transcription priming in the ENi pathway (Fig. 3C). Possible end-processing factors include the MRN complex (Mre11, Rad50, Nbs1), CtIP, Exo1, BLM or Dna2. Poly(ADP-ribose) polymerase 1 (PARP-1) was shown to interact with both ORF2p38 and with Mre11.45 Therefore it could potentially recruit both end-resection factors and the L1 machinery at sites of DNA damage.
Because DLEA uses native L1 RNP enriched from cells, it can be combined with shRNA-mediated knock-down of specific cellular factors. This approach will help in the future to elucidate the role of cellular factors in retrotransposition.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Dominic van Essen for critical reading. This work was funded by INSERM and INCa (Avenir program, agreement #2009-340), the European Research Council (Retrogenomics, #243312) and the French Government (National Research Agency, ANR) through the “Investments for the Future” (LABEX SIGNALIFE, #ANR-11-LABX-0028-01). CM was the recipient of a PhD fellowship from the Fondation ARC.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/28907
References
- 1.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hancks DC, Kazazian HH., Jr. Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaer K, Speek M. Retroelements in human disease. Gene. 2013;518:231–41. doi: 10.1016/j.gene.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Kazazian HH, Jr., Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–6. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 5.Shukla R, Upton KR, Muñoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–11. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, Burns MB, Harris RS, Sigmon DF, Casella A, Erlanger B, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22:2328–38. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Cancer Genome Atlas Research Network Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–71. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–61. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–5. [PubMed] [Google Scholar]
- 10.Rodić N, Burns KH. Long interspersed element-1 (LINE-1): passenger or driver in human neoplasms? PLoS Genet. 2013;9:e1003402. doi: 10.1371/journal.pgen.1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–9. [PMC free article] [PubMed] [Google Scholar]
- 12.Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci U S A. 2003;100:13815–20. doi: 10.1073/pnas.2336221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khazina E, Truffault V, Büttner R, Schmidt S, Coles M, Weichenrieder O. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat Struct Mol Biol. 2011;18:1006–14. doi: 10.1038/nsmb.2097. [DOI] [PubMed] [Google Scholar]
- 14.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–75. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathias SL, Scott AF, Kazazian HH, Jr., Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–10. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 16.Feng Q, Moran JV, Kazazian HH, Jr., Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–16. doi: 10.1016/S0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 17.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–60. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 18.Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6:6. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 20.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–27. doi: 10.1016/S0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 22.Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–93. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–25. doi: 10.1016/S0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 24.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–38. doi: 10.1016/S0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 25.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–65. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 26.Sen SK, Huang CT, Han K, Batzer MA. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res. 2007;35:3741–51. doi: 10.1093/nar/gkm317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–12. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 28.Monot C, Kuciak M, Viollet S, Mir AA, Gabus C, Darlix JL, Cristofari G. The specificity and flexibility of L1 reverse transcription priming at imperfect T-tracts. PLoS Genet. 2013;9:e1003499. doi: 10.1371/journal.pgen.1003499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol. 1995;15:3882–91. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luan DD, Eickbush TH. Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol Cell Biol. 1996;16:4726–34. doi: 10.1128/mcb.16.9.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3:h0052. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasior SL, Preston G, Hedges DJ, Gilbert N, Moran JV, Deininger PL. Characterization of pre-insertion loci of de novo L1 insertions. Gene. 2007;390:190–8. doi: 10.1016/j.gene.2006.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi H, Fujiwara H. Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. EMBO J. 2002;21:408–17. doi: 10.1093/emboj/21.3.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Repanas K, Zingler N, Layer LE, Schumann GG, Perrakis A, Weichenrieder O. Determinants for DNA target structure selectivity of the human LINE-1 retrotransposon endonuclease. Nucleic Acids Res. 2007;35:4914–26. doi: 10.1093/nar/gkm516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen SM, Ye J, Eickbush TH. RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc Natl Acad Sci U S A. 2006;103:17602–7. doi: 10.1073/pnas.0605476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen SM, Eickbush TH. R2 target-primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol Cell Biol. 2005;25:6617–28. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauerwald A, Sandin S, Cristofari G, Scheres SH, Lingner J, Rhodes D. Structure of active dimeric human telomerase. Nat Struct Mol Biol. 2013;20:454–60. doi: 10.1038/nsmb.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor MS, Lacava J, Mita P, Molloy KR, Huang CR, Li D, Adney EM, Jiang H, Burns KH, Chait BT, et al. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell. 2013;155:1034–48. doi: 10.1016/j.cell.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodier JL, Cheung LE, Kazazian HH., Jr. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 2013;41:7401–19. doi: 10.1093/nar/gkt512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azzalin CM. UPF1: a leader at the end of chromosomes. Nucleus. 2012;3:16–21. doi: 10.4161/nucl.18929. [DOI] [PubMed] [Google Scholar]
- 41.Kopera HC, Moldovan JB, Morrish TA, Garcia-Perez JL, Moran JV. Similarities between long interspersed element-1 (LINE-1) reverse transcriptase and telomerase. Proc Natl Acad Sci U S A. 2011;108:20345–50. doi: 10.1073/pnas.1100275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc Natl Acad Sci U S A. 1996;93:10712–7. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MC, Muotri AR, Mu Y, Carson CT, Macia A, Moran JV, Gage FH. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc Natl Acad Sci U S A. 2011;108:20382–7. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–8. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haince JF, McDonald D, Rodrigue A, Déry U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]