Abstract
Thrips-borne tospoviruses cause severe damage to crops worldwide. In this investigation, tobacco lines transgenic for individual WLm constructs containing the conserved motifs of the L RNA-encoded RNA-dependent RNA polymerase (L) gene of Watermelon silver mottle virus (WSMoV) were generated by Agrobacterium-mediated transformation. The WLm constructs included: (i) translatable WLm in a sense orientation; (ii) untranslatable WLmt with two stop codons; (iii) untranslatable WLmts with stop codons and a frame-shift; (iv) untranslatable antisense WLmA; and (v) WLmhp with an untranslatable inverted repeat of WLm containing the tospoviral S RNA 3′-terminal consensus sequence (5′-ATTGCTCT-3′) and an NcoI site as a linker to generate a double-stranded hairpin transcript. A total of 46.7–70.0% transgenic tobacco lines derived from individual constructs showed resistance to the homologous WSMoV; 35.7–100% plants of these different WSMoV-resistant lines exhibited broad-spectrum resistance against four other serologically unrelated tospoviruses Tomato spotted wilt virus, Groundnut yellow spot virus, Impatiens necrotic spot virus and Groundnut chlorotic fan-spot virus. The selected transgenic tobacco lines also exhibited broad-spectrum resistance against five additional tospoviruses from WSMoV and Iris yellow spot virus clades, but not against RNA viruses from other genera. Northern analyses indicated that the broad-spectrum resistance is mediated by RNA silencing. To validate the L conserved region resistance in vegetable crops, the constructs were also used to generate transgenic tomato lines, which also showed effective resistance against WSMoV and other tospoviruses. Thus, our approach of using the conserved motifs of tospoviral L gene as a transgene generates broad-spectrum resistance against tospoviruses at the genus level.
Introduction
Members of the genus Tospovirus are the only plant-infecting viruses in the family Bunyaviridae. Tospoviruses are transmitted by thrips in a persistent manner and they infect more than 1090 species in 85 families of monocots and dicots, causing severe damage in many economically important crops around the world [1], [2], [3]. The virion of tospovirus is an enveloped quasi-spherical particle with a tripartite RNA genome with five open reading frames [4]. The genomic segments are named after their size, as S (Small), M (Medium) and L (Large). The S RNA is ambisense and it encodes a nonstructural RNA-silencing suppressor protein (NSs) and the nucleocapsid protein (N) [5], [6]. The M RNA is also ambisense and it encodes a cell-to-cell movement protein (NSm) and the envelope glycoproteins precursor (GPp) [7], [8]. The L RNA is negative sense and it encodes an RNA-dependent RNA polymerase (RdRp), also called L protein [9].
The classification of different tospovirus species is based on the evolutionary genetic relationship of their N gene sequences, host ranges and vector specificity [10], [11]. Earlier, different tospovirus species were categorized into serogroups based on their serological similarities [10] determined by immunodetectable epitopes of N proteins and detecting antibodies. The tendency of the serogroups of tospoviruses to align with the clades defined by the phylogenetic analyses of tospoviruses has been observed in several of our studies [12], [13], [14], [15], [16]. Serological detection of tospoviruses is useful for easy on-the-spot diagnosis of tospoviruses [17].
The most effective and environmentally sound control strategies rely on the availability of resistant cultivars. However, due to high divergence of tospoviruses and scarcity of natural resistant resources, efficient control measures are difficult to be developed by traditional breeding. The Sw-5 gene, first identified in tomato [18], [19], and Tsw gene in pepper [20] are widely used for resistance breeding against Tomato spotted wilt virus (TSWV). Unfortunately, several natural resistance-breaking strains of tospoviruses were reported all over the world, including in Australia, Brazil, Hawaii, Italy, South Africa, Spain, and USA [21], [22].
Since the concept of pathogen-derived resistance (PDR) was proposed [23] and confirmed by expressing the coat protein (CP) of Tobacco mosaic virus (TMV) in transgenic tobacco plants [24], transgenic resistance has become an important approach to protect various plant species against virus infection Transgenic tobacco plants that accumulated high levels of TSWV N protein exhibit broad-spectrum, but moderate level, resistance not only against the homologous isolate, but also against distantly related Impatiens necrotic spot virus (INSV) isolates [25]. However, the N protein-mediated protection can be overcome by increasing inoculum strength [26]. On the other hand, transgenic plants carrying untranslatable N [27], [28] or NSm gene [29] triggered RNA-mediated resistance to tospoviruses. Though RNA-mediated resistance provides higher degrees of resistance than protein-mediated resistance, it is specific against the homologous and closely related viruses [25], [27], [30], [31]. An artificial microRNA (amiRNA) approach targeting sequence elements within the conserved RdRp motifs of Watermelon silver mottle virus (WSMoV) L gene can successfully confer high degrees of transgenic resistance against the homologous virus [32]. To obtain resistance against multiple viruses, the N genes of TSWV, Tomato chlorotic spot virus (TCSV), and Groundnut ringspot virus (GRSV) were linked to generate transgenic tobacco plants with resistance to these three tospoviruses [33]. Similarly, a composite transgene containing small fragments from N genes of WSMoV, TSWV, GRSV and TCSV in a hairpin construct triggered RNA silencing for multiple resistances against the corresponding viruses [34]. Transgenic plants expressing an N protein-interacting peptide derived from the N open reading frame (ORF) of TSWV were also reported to confer high degrees of broad-spectrum resistance not only against TSWV, but also GRSV, and TCSV and Chrysanthemum stem necrosis virus (CSNV) [35]. However, N gene-mediated resistances, regardless of their direct origin from N transgenes [27], [28], [29], [33], [34] or from molecules (eg. peptides) targeting N gene sequences [35], may not be expected to be broad-spectrum and/or durable, because of the high degree of variation among the N gene sequences of tospoviral species and strains.
WSMoV, the type member of WSMoV clade [12], is one of the major limiting factors for cucurbit production in Taiwan [36] and other Asian countries [37], [38], [39]. The complete genome sequence of WSMoV has been determined [40], [41], [42], [43]. Recently, several new tospoviruses serologically related to WSMoV have been reported from India, China and East Asian countries [44], [45]. Comparison of the L protein sequence of WSMoV with those of other tospoviruses revealed a conserved region containing five RdRp motifs [43], [46], [47]. Based on the conserved region, genus-specific degenerate primers were designed for detecting most tospovirus species from greenhouse and field samples by reverse transcription-polymerase chain reaction (RT-PCR) [43], [48]. The objective of the present study was to develop broad-spectrum resistance against various tospoviral species using untranslatable transgenes designed from the highly conserved RdRp region of WSMoV L gene, through the post-transcriptional gene silencing (PTGS) mechanism.
Transformation of tobacco plants with the transgenes derived from the WSMoV L gene conserved region conferred broad-spectrum transgenic resistance not only against the WSMoV, but also against different tospovirus species from Asia type WSMoV, Iris yellow spot virus (IYSV) and Groundnut yellow spot virus (GYSV) clades and Euro-America type TSWV clade and INSV, which is considered as a distinct serotype [12]. When the same approach was extended to the real crop tomato, similar results were obtained. Thus, we conclude that the broad-spectrum resistance at the Tospovirus genus level generated by our approach is effective for the control of different tospovirus species infecting various crops.
Materials and Methods
Virus Sources
WSMoV [36] and Melon yellow spot virus (MYSV) [49] were collected from watermelon, and Peanut chlorotic fan-spot virus (renamed as Groundnut chlorotic fan-spot virus, GCFSV, by ICTV) [50] was collected from peanut in Taiwan. The high temperature-recovered isolate (HT-1) of Capsicum chlorosis virus (CaCV) collected from gloxinia in the United States was provided by H. T. Hsu [51]. TSWV NY, a New York isolate of TSWV, isolated from tomato was provided by R. Provvidenti, New York State Experiment Station, Geneva, Cornell University [42]. GRSV collected from tomato in Brazil [52] was provided by D. Gonsalves, Pacific Basin Agricultural Research Center, USDA, Hawaii. An isolate of INSV collected from impatiens in the United States [53] was provided by J. Moyer, North Carolina State University, USA. The iris isolate of IYSV from the Netherlands [54] and the tomato isolate of Tomato yellow ring virus (TYRV) from Iran [55] were provided by R. Kormelink, Wageningen University, The Netherlands (import permit: 96-V-54). Watermelon bud necrosis virus (WBNV), collected from watermelon in India [16], was provided by P. A. Rajagopalan, Mahyco Co., Jalna, India (import permit: 97-V-41). All these viruses were single lesion-isolated in the local lesion host Chenopodium quinoa Willd. and maintained in the systemic host Nicotiana benthamiana Domin. by mechanical inoculation under temperature-controlled (23–28°C) greenhouse conditions. Virus inocula were prepared by grinding infected leaves in 10 mM potassium phosphate buffer (pH 7.0) containing 10 mM sodium sulfite (1∶10 w/v).
Other viruses not belonging to the genus Tospovirus, used for comparison, include Zucchini yellow mosaic virus (ZYMV) TN-3 isolate [56], maintained in squash plants, Papaya ringspot virus (PRSV) YK isolate [57], maintained in papaya (Carica papaya L. cv. Tainung No. 2) plants, and Turnip mosaic virus (TuMV) CY-5 isolate [58] and Cucumber mosaic virus (CMV) [48] both maintained in N. benthamiana plants. One week after mechanical inoculation, inocula of these viruses were prepared by grinding infected leaf tissues in 10 mM potassium phosphate buffer (pH 7.2, 1∶20 w/v).
Sequence Alignment
Sequences of the reported full-length L RNAs for the comparison were obtained from GenBank database, through the following accession numbers: TSWV, (NC_002052); INSV, (NC_003625); Groundnut bud necrosis virus (GBNV), (NC_003614); WSMoV, (NC_003832); CaCV, (NC_008302); and MYSV, (NC_008306). Nucleotide sequences were compared by BestFit program of Seqweb v. 3.1 (Accelrys, Inc., San Diego, CA). Multiple sequence alignment of L genes was carried out using ClustalW+ program of Seqweb v. 3.1.
Construction of Translatable and Untranslatable Constructs Containing WSMoV L Gene Conserved Region
Total RNAs were isolated from leaves of the WSMoV-infected N. benthamiana plants by Ultraspec RNA isolation system (Biotex laboratories, Houston, TX). The primers WL3975(NcoI) (5′-GCCATGGAGCACACATACAAGCATATCGCC-3′, NcoI site underlined) and WL4928c(SacI) (5′-GAGCTCGAGTCGTTCTCTTCTCCTGGCAGC-3′, SacI site underlined) were used to amplify the nt 3975–4928 region of the vc strand of WSMoV L RNA, corresponding to the L gene conserved region containing five RdRp motifs, by RT-PCR. The amplified fragment, denoted WLm, was cloned in TOPO TA vector (Invitrogen, Carlsbad, CA), and after verifying the sequence, WLm was used as the template for subsequent modifications.
The primers WL3975NcoI (5′-GCCATGGAATAATAGGAGCACACATACAAGCATATCGCC-3′, NcoI site underlined and two termination codons italicized) and WLst3975NcoI (5′-GCCATGGATAATAGGAGCACACATACAAGCATATCGCC-3′, NcoI site underlined, frame-shifted termination codons italicized) individually coupled with WL4928cSacI were used to introduce two in-frame termination codons and two termination codons with a −1 frameshift at the 5′ end of the WLm fragment to generate the untranslatable fragments of WLmt and WLmts, respectively. Primers WL4928cNcoI (5′-CCATGGGTCGTTCTCTTCTCCTGGCAGC-3′, NcoI site underlined) and WL3975SacI (5′-GAGCTCGAGCACACATACAAGCATATCGCC-3′, SacI site underlined) were used to amplify WLm, containing opposite cloning sites for generating antisense fragment WLmA. The WLm, WLmt, WLmts and WLmA fragments were subsequently cloned into the plasmid pBGCP [59], a derivative of the binary vector pBI121 carrying the coat protein (CP) gene of PRSV with a β-glucuronidase (GUS) leader sequence, via NcoI and SacI sites, to generate pBGWLm, pBGWLmt, pBGWLmts and pBGWLmA, respectively (Figure 1).
Figure 1. Schematic representation of individual transgenes.
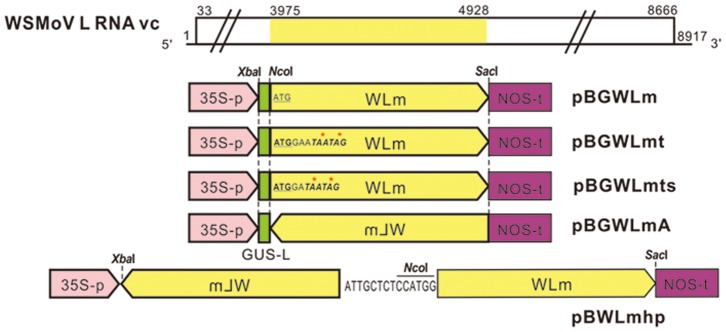
Nucleotide (nt) 3975 to 4928 region of the viral complementary (vc) strand of L RNA (yellow box) of Watermelon silver mottle virus (WSMoV) was amplified as the WLm fragment and constructed as translatable sense (pBGWLm), non-translatable sense (pBGWLmt and pBGWLmts), antisense (pBGWLmA) and inverted repeat (pBWLmhp) transgenes in the Ti binary vector pBI121. pBGWLmt and pBGWLmts contained identical sequence to pBGWLm, but two termination codons (italicized and asterisked) were added in-frame in pBGWLmt, and the same were added with a −1 frame shift in pBGWLmts. A linker sequence containing the tospoviral S RNA 3′-terminal consensus sequence (5′-ATTGCTCT-3′) and an NcoI site was added between antisense and sense WLm sequences for creating the inverted repeat in pBWLmhp. The restriction sites used for construction of transgenes are indicated. GUS leader sequence (GUS-L, green box), Cauliflower mosaic virus 35S promoter (35S-p, pink box) and nos terminator (NOS-t, purple box) are indicated.
Furthermore, a WLm-linker fragment amplified by the primers WL4928cXbaI (5′-ATGCTCTAGAGTCGTTCTCTTCTCCTGGCAGC-3′, XbaI site underlined) and WL3975Nco-linker [5′-CCGGCCATGG AGAGCAATGAGCACACATACAAGCA-3′ with an NcoI site (underlined) and a stretch of eight nucleotides (italicized) complementary to the tospoviral S RNA 3′-terminal consensus sequence] was cloned in TOPO TA vector. The S RNA 3′-terminal consensus sequence was included in the linker for further increasing the effectiveness of the transgene. The XbaI/NcoI-digested WLm-linker and NcoI/SacI-digested WLm fragments were mixed with XbaI/SacI-digested pBI121 for ligation to generate the construct pBWLmhp (Figure 1) that expressed a hairpin RNA from the inverted repeat transgene WLmhp.
Finally, all the transgene constructs were transferred into the Agrobacterium tumefaciens strain LBA 4404 by direct transformation with liquid nitrogen treatment [60].
Tobacco and Tomato Transformation
A. tumefaciens LBA 4404 containing individual constructs were used to transform tobacco plants of N. benthamiana, using small leaf pieces as described by Horsch et al., [61]. The plants of each line were originated and cloned from a single independent regenerated shoot.
For tomato transformation, the cotyledon explants were prepared from 14-day-old seedlings germinated from manually peeled and surface sterilized seeds of Solanum lycopersicum L. cv. Feminini Beauty (Evergrow Seed Co. Ltd., Taiwan) on half strength MS medium [62]. The explants were further pre-cultured on MS medium for two days. The pre-cultured cotyledon discs were infected with 1∶15 dilution of overnight grown Agrobacterium culture to introduce the individual transgene constructs by Agrobacterium-mediated transformation, as previously described [63]. Multiple shootlets micropropagated from a single shoot regenerated on the selection medium were cut and transferred to the rooting medium (MS medium with 0.5 mg/l IBA and 50 mg/l kanamycin) and defined as shootlets of a R0 line. After 1 wk, the rooted plantlets were transferred to vermiculite in plastic bags and maintained in growth chamber at 25–27°C with 16 hr photoperiod (53 µE m−2 s−1 photon irradiance). After acclimatization for 2 wk, the plantlets were transferred to a temperature-controlled (23–28°C) greenhouse.
Examination of Putative Transgenic Lines
To check the presence of individual transgenes in the generated transgenic N. benthamiana and tomato plants, total genomic DNA was extracted from leaves of non-transformed or transgenic plants by Genomic DNA Purification Kit (GMbiolab, Taichung, Taiwan), according to manufacturer’s instructions. The primer pair WL3975(NcoI)/WL4928c(SacI) was used for the transgenic N. benthamiana and tomato lines derived from pBGWLm, pBGWLmt, pBGWLmts, pBGWLmA and pBWLmhp. Primers PNPTII (5′-ATGATTGAACAAGATGGATTGCAC-3′) and MNPTII (5′-GAAGAACTCGTCAAGAAGGCGATA-3′) were designed to check the presence of the selection-marker neomycin phosphotransferase (nptII) gene linked with the transgenes in all transgenic plants. Fifty nanogram of extracted DNAs were used as templates, and PCR was conducted with 1 min for denaturation at 94°C, 2 min for annealing at 58°C (except 50°C for transgenic plants derived from pBWLmhp), and 3 min for synthesis at 72°C for 34 cycles, followed by a final extension at 72°C for 7 min. PCR products were analyzed by electrophoresis on 1% agarose gel.
Evaluation of Resistance to Homologous and Heterologous Tospoviruses
To evaluate the transgenic resistance to the homologous tospovirus, non-transgenic and putative transgenic N. benthamiana and tomato plants were mechanically inoculated with WSMoV under temperature controlled (23–28°C) greenhouse conditions. To evaluate the transgenic resistance against a heterologous tospovirus, the selected WSMoV-resistant transgenic tobacco R0 lines multiplied in vitro were mechanically inoculated with TSWV. To further evaluate the spectrum of the transgenic resistance, the in vitro multiplied WSMoV-resistant transgenic tobacco and tomato R0 lines were mechanically inoculated with individual tospoviruses which represent different clades of tospoviruses, including TSWV, GRSV, GCFSV and INSV, under the same greenhouse conditions. All inocula for challenging transgenic plants were prepared from TSWV, WSMoV, GRSV, INSV or GCFSV-infected N. benthamiana leaves, ground and diluted 50 to 100-fold (w/v) in 10 mM potassium phosphate buffer (pH 7.0) containing 10 mM sodium sulfite. Five plants of each transgenic line were inoculated with each tospovirus. Individual inocula were applied by rubbing the two fully-expended leaves with 600 mesh carborundum at the stage of 4–5 leaves. Because tospoviruses are not stable, for each inoculum, non-transgenic control plants were inoculated after inoculating the transgenic lines to ensure the infection. In some cases, the plants were inoculated twice with an interval of one week between the two inoculations. The plants were observed for 30 days for symptom development under the aforementioned greenhouse conditions.
For test the spectrum of resistance, the selected WSMoV-resistant transgenic tobacco lines were further mechanically challenged with other Asia-type tospoviruses, including WBNV, MYSV, TYRV, IYSV and CaCV, following the method described above. TuMV and CMV were used as out-group viruses for challenging the transgenic tobacco lines which exhibited broad-spectrum resistance against tospoviruses.
Indirect Enzyme-linked Immunosorbent Assay (ELISA)
Indirect ELISA was conducted following the method described by Yeh and Gonsalves [64] with required modifications. Leaf extracts prepared from four leaf disks (0.5 cm diameter) punched from different upper leaves of the tested plants 30 days post inoculation (dpi) were ground in coating buffer (50 mM sodium carbonate, pH 9.6, containing 0.01% sodium azide) at 1∶40 dilution and used for antigen coating. The antisera against the N protein of WSMoV, TSWV, GRSV, INSV or GCFSV [50] were used at a 1∶4000 dilution in conjugate buffer (PBS containing 0.05% Tween 20, 2% polyvinylpyrrolidone-40 and 0.2% ovalbumin). The alkaline phosphatase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) was used at 1∶5000 dilution as the secondary antibody. The incubation times for the reaction of crude antigens with the primary antibody and the reaction of rabbit IgG with the secondary antibody were both set at 37°C for 60 min. The absorbance at 405 nm was recorded using the Victor 1420 Multilabel Counter (PerkinElmer Life Sciences, Waltham, MA) 30 min after the addition of ρ-nitrophenyl phosphate substrate (Sigma-Aldrich, St. Louis, MO) that was dissolved in substrate buffer (9.7% diethanolamine and 0.02% sodium azide, pH 9.8). The threshold of a positive reaction was set at two-fold readings, as compared to that of the negative control.
Transcript and siRNA Detection by Northern Hybridization
For transgene transcript detection, total RNAs were isolated from leaves of non-transgenic or transgenic plants by Ultraspec RNA isolation system (Biotex laboratories) according to manufacturer’s instructions. Twenty microgram of total RNAs were separated in 1.0% agarose gels with formaldehyde, and then transblotted onto Hybond-N+ nylon membrane (Amersham Biosciences, Buckinghamshire, UK). After UV-crosslinking of the transferred RNAs to the nylon membrane, hybridization was done with the 32P-labeled DNA probe prepared from WLm by Primer-It II Random Primer Labeling Kit (Stratagene, La Jolla, CA). For siRNA detection, northern hybridization of total RNAs were performed as described previously [65], [66]. The hybridization signals were detected by autoradiography.
Southern Hybridization
Total genomic DNAs were extracted from leaves of non-transgenic or transgenic N. benthamiana plants by Genomic DNA Purification Kit (GMbiolab, Taichung, Taiwan), according to manufacturer’s instructions. Fifteen to twenty microgram of genomic DNAs were digested with SspI, separated at 120 V in a 0.8% agarose gel, transblotted onto Hybond-N+ nylon membrane (Amersham Pharmacia Biotech, UK), and hybridized with the 32P-labeled probe, as described above. The hybridization signals were detected by autoradiography.
Results
Relationship of WLm within L RNAs of Distantly Related Tospoviruses
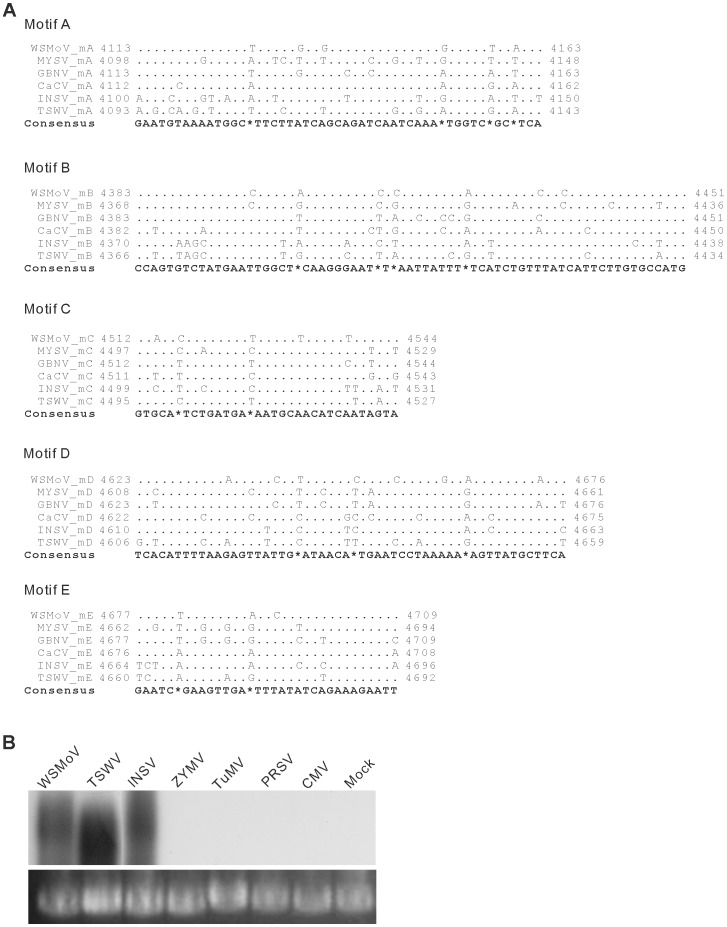
The sequence of the conserved region containing five RdRp motifs, motif A to E, of WSMoV was compared with those of TSWV, INSV, CaCV, GBNV and MYSV. This conserved region, amino acid (aa) 1315 to 1632 of WSMoV L protein, shares identities of 93.7%, 95.9%, 96.9%, 72.6% and 66.7% with those of MYSV, CaCV, GBNV, INSV and TSWV, respectively. The corresponding nucleotide sequence, nt 3975 to 4928 of the vc strand of WSMoV L RNA, shares identities of 82.9%, 82.6%, 79.4%, 69.0% and 66.4% with those of CaCV, GBNV, MYSV, TSWV and INSV, respectively (Table 1). High aa and nt identities of 76.5–100% and 75.5–93.8%, respectively, were found among the RdRp motifs of the tospoviruses compared (Table 1). Alignments of the nt sequences encoding individual RdRp motifs of WSMoV, GBNV, CaCV, MYSV, TSWV and INSV are shown in Figure 2A.
Table 1. Nucleotide (nt) and amino acid (aa) identities of the L conserved region, viral complementary position of the nt 3975 to 4928 of L RNA, containing five RNA-dependent RNA polymerase motifs of Watermelon silver mottle virus (WSMoV) compared with those of other tospoviruses.
| Virus | Conserved region | RdRp motif | ||||||||||
| A | B | C | D | E | ||||||||
| nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | |
| CaCV | 82.9 | 95.9 | 88.2 | 100 | 84.1 | 100 | 81.3 | 90.9 | 77.8 | 100 | 93.8 | 100 |
| GBNV | 82.6 | 96.9 | 88.2 | 100 | 87.0 | 100 | 84.9 | 100 | 85.0 | 100 | 81.3 | 100 |
| MYSV | 79.4 | 93.7 | 80.4 | 100 | 89.9 | 100 | 81.3 | 100 | 79.6 | 100 | 81.8 | 100 |
| TSWV | 69.0 | 66.7 | 80.0 | 76.5 | 78.3 | 100 | 84.9 | 81.8 | 78.8 | 88.9 | 83.9 | 90.9 |
| INSV | 66.4 | 72.6 | 75.5 | 82.4 | 81.2 | 100 | 78.1 | 81.8 | 83.0 | 94.4 | 86.2 | 90.9 |
Sequences of tospoviral L RNAs for the comparison were obtained from GenBank: WSMoV, NC_003832; Tomato spotted wilt virus (TSWV), NC_002052; Impatiens necrotic spot virus (INSV), NC_003625; Groundnut bud necrosis virus (GBNV), NC_003614; Capsicum chlorosis virus (CaCV), NC_008302; and Melon yellow spot virus (MYSV), NC_008306.
Figure 2. Comparison of tospoviral L gene region encoding RNA-dependent RNA polymerase (RdRp) conserved motifs of L protein.
(A) Alignment of L gene sequences corresponding to the RdRp motifs, A, B, C, D and E of L proteins of Watermelon silver mottle virus (WSMoV), Capsicum chlorosis virus (CaCV), Groundnut bud necrosis virus (GBNV), Melon yellow spot virus (MYSV), Tomato spotted wilt virus (TSWV) and Impatiens necrotic spot virus (INSV). (B) The recognizability of the L RNAs of WSMoV, TSWV and INSV by the 32P-labeled probe prepared using the cloned L gene conserved region (WLm), as analyzed by northern hybridization. Genomic RNAs of Zucchini yellow mosaic virus (ZYMV), Turnip mosaic virus (TuMV), Papaya ringspot virus (PRSV) and Cucumber mosaic virus (CMV) were also used in the hybridization analysis. RNA from mock inoculated plant was used as the negative control. Ribosomal RNAs indicate the loading controls.
To confirm the relationship among the conserved region of L genes of tospoviruses, the construct WLm was used as a template for preparation of 32P-labeled DNA probes to hybridize with total RNAs extracted from leaf tissues of plants infected with various plant viruses, including WSMoV, TSWV and INSV belonging to the genus Tospovirus; TuMV, ZYMV and PRSV belonging to the genus Potyvirus; and CMV belonging to the genus Cucumovirus. Strong hybridization signals were observed from RNA prepared from WSMoV-infected plants. Comparably strong signals were also observed from RNA prepared from plants infected by TSWV or INSV that are serologically unrelated to WSMoV, suggesting that WLm-derived probes derived from the construct WLm are able to react with the conserved regions of L genes of TSWV and INSV. Corroboratively, no hybridization signals were detected from the RNAs prepared from TuMV, ZYMV, PRSV, or CMV-infected plant and the negative control of mock-inoculated plant (Figure 2B). Based on the results of sequence analysis (Figure 2A) and hybridization analysis (Figure 2B), the region spanning nt 3975 to 4928 of the vc strand of WSMoV L RNA (denoted WLm) was used for construction of various transgenes targeting the conserved RdRp motifs of tospovirus L genes to trigger transgenic resistance.
Resistance to Homologous WSMoV in Transgenic Tobacco Lines Triggered by Various Transgene Constructs
N. benthamiana plants were transformed using the five constructs, pBGWLm, pBGWLmt, pBGWLmts, pBGWLmA and pBWLmhp (Figure 1), carrying transgenes derived from the conserved region of WSMoV L gene. The GUS leader sequence of PB1121 was retained also at the 5′-terminal region of the untranslatable transgenes in the constructs pBGWLmt, pBGWLmts and pBGWLmA to maintain the translation capability similar to that of the translatable pBGWLm. Different categories of transgenic N. benthamiana lines carrying different transgenes were denoted by the names of the transgenes they carried (i.e., WLm, WLmt, WLmts, WLmA or WLmhp) and individual lines/plants from each category were identified by numeral(s) following their category name.
After kanamycin selection and PCR detection, the putative transgenic N. benthamiana R0 lines, 30 lines for each construct, were first evaluated for resistance to WSMoV and the results are summarized in Table 2. The transgenic lines which exhibited delay in symptom development for more than 7 days relative to the non-transgenic control, and also ELISA positive for WSMoV N protein were classified as moderately resistant (MR) lines; while those that did not show any symptoms one month after inoculation, and were ELISA negative were classified as highly resistant (HR) lines. Fourteen WLm lines (46.7%), 19 WLmt lines (63.3%), 19 WLmts lines (63.3%), 20 WLmA lines (66.7%) and 21 WLmhp lines (70.0%) exhibited resistance to WSMoV. Among them, 30.0–40.0% (9–12 lines) of resistant lines were HR type to WSMoV infection and 13.3–40.0% (4–12 lines) were MR type (Table 2). The results indicated that each of the five transgene constructs was able to trigger high levels of resistance to WSMoV in N. benthamiana transgenic lines. However, the non-translatable and inverted repeat transgene constructs WLmt, WLmts, WLmA and WLmhp rendered higher protection efficiencies (63.3% to 70.0%) compared to the translatable transgene construct WLm (46.7%) (Table 2), indicating that RNA-mediated resistance plays a key role.
Table 2. Evaluation of transgenic Nicotiana benthamiana R0 lines carrying individual constructs containing L gene conserved region for resistance to the homologous Watermelon silver mottle virus (WSMoV) under greenhouse conditions.
| Transgene | Total assayed line No. | Challenged withWSMoV | |||
| HRa | MRb | Sc | Total R lines (%)d | ||
| WLm | 30 | 10 | 4 | 16 | 14 (46.7)A * |
| WLmt | 30 | 12 | 7 | 11 | 19 (63.3)B |
| WLmts | 30 | 12 | 7 | 11 | 19 (63.3)B |
| WLmA | 30 | 11 | 9 | 10 | 20 (66.7)B |
| WLmhp | 30 | 9 | 12 | 9 | 21 (70.0)B |
Transgenic N. benthamiana lines that remained asymptomatic and ELISA negative for WSMoV NP beyond a month after inoculation were considered highly resistant (HR) lines.
Transgenic N. benthamiana lines that showed more than 7 days delay in symptom development in comparison to the non-transformed plants were classified as moderately resistant (MR) lines.
Transgenic N. benthamiana lines that exhibited typical systemic symptoms similar to non-transformed N. benthamiana plants, at the same time or less than 7 days delay after inoculation with WSMoV, were regarded as susceptible (S) lines.
Both HR and MR lines were considered as resistant (R) lines. The percentages of resistance were expressed as number of R lines/total number of total assayed lines.
*A & B: adjacent values that are superscribed by same alphabet are not significantly different as analyzed by chi-square using software JMP 5.0.1 (SAS Institute Inc.) (p<0.05).
The L Gene Conserved Region of WSMoV Confers Broad-spectrum Resistance Against Serologically Unrelated Tospoviruses in Transgenic Tobacco Plants
Subsequently, the WSMoV-resistant transgenic N. benthamiana lines were challenged with serologically unrelated tospovirus species of TSWV, GRSV, INSV and GCFSV, to further evaluate the spectrum of the resistance. The results are presented in Table 3. The virus infection of symptomatic transgenic plants was confirmed by indirect ELISA using the antiserum against the N protein of TSWV, GRSV, INSV or GCFSV. If no significant accumulation of the virus was detected by ELISA one month after inoculation, the resistant transgenic plants were considered immune to virus infection. Of the 14 WSMoV-resistant WLm transgenic lines, 35.7–71.4% exhibited different levels of broad-spectrum resistance. Most WSMoV-resistant lines showed different levels of resistance against 2 to 5 different challenged tospovirus species, except WLm-16 that was resistant only to WSMoV. Especially, the lines WLm-11 and WLm-30 were resistant to all the tested tospovirus species. WLm-11 was immune to INSV, but developed delayed symptoms when challenged with TSWV, GRSV and GCFSV. WLm-30 was immune to GRSV and GCFSV, but developed delayed symptoms when challenged with TSWV and INSV.
Table 3. Evaluation of the Watermelon silver mottle virus (WSMoV)-resistant transgenic Nicotiana benthamiana lines against other serologically unrelated tospovirus species, Tomato spotted wilt virus (TSWV), Groundnut ringspot virus (GRSV), Impatiens necrotic spot virus (INSV) and Groundnut chlorotic fan-spot virus (GCFSV) under greenhouse conditions.
| Transgene | Total assayed line No. | TSWV | GRSV | INSV | GCFSV | ||||||||
| HRa | MRb | Total R lines (%)c | HR | MR | Total R lines (%) | HR | MR | Total R lines (%) | HR | MR | Total R lines (%) | ||
| WLm | 14 | 0 | 6 | 6 (42.9) | 2 | 5 | 7 (50.0) | 2 | 3 | 5 (35.7) | 4 | 6 | 10 (71.4) |
| WLmt | 19 | 4 | 6 | 10 (52.6) | 4 | 6 | 10 (52.6) | 5 | 6 | 11 (57.9) | 7 | 7 | 14 (73.7) |
| WLmts | 19 | 7 | 4 | 11 (57.9) | 4 | 9 | 13 (68.4) | 7 | 6 | 13 (68.4) | 9 | 10 | 19 (100.0) |
| WLmA | 20 | 5 | 5 | 10 (50.0) | 6 | 5 | 11 (55.0) | 2 | 9 | 11 (55.0) | 5 | 10 | 15 (75.0) |
| WLmhp | 21 | 6 | 9 | 15 (71.4) | 2 | 13 | 15 (71.4) | 9 | 7 | 16 (76.2) | 6 | 10 | 16 (76.2) |
The lines that remained asymptomatic and ELISA negative for the N proteins beyond a month after inoculation were considered highly resistant (HR) lines.
The lines that showed more than 7 days delay in symptom development, in comparison to the non-transformed plants were classified as moderately resistant (MR) lines.
Both HR and MR lines were regarded as resistant (R) lines. The percentages of resistance were expressed as number of R lines/total number of assayed WSMoV-resistant lines.
Among 19 WSMoV-resistant WLmt transgenic lines, 52.6–73.7% of them showed different levels of resistance to TSWV, GRSV, INSV and GCFSV. Six lines, WLmt-4, 5, 7, 9, 13 and 17, were resistant, at various levels, to all the five challenged tospoviruses. Indeed, two of them, WLmt-4 and WLmt-13, were immune to all challenged tospoviruses; from the asymptomatic plants of these lines, virus accumulation was not detectable by indirect ELISA (Figure 3).
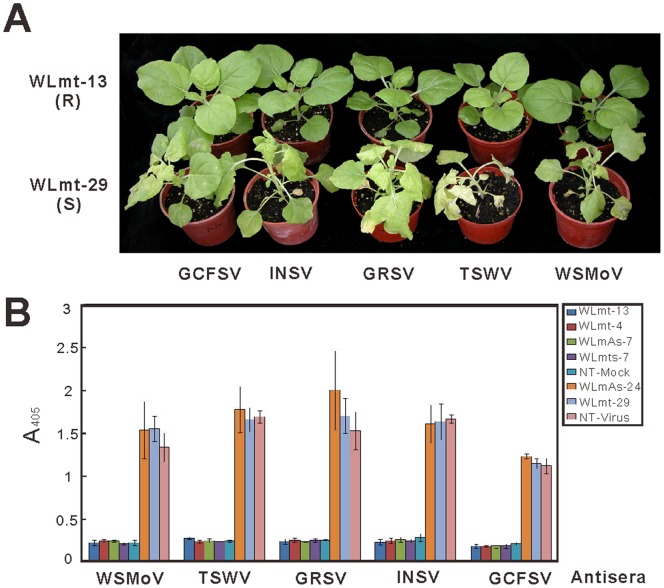
Figure 3. Evaluation of the transgenic Nicotiana benthamiana lines for resistance to different tospovirus species under greenhouse conditions.
The lines with individual constructs of the conserved region of L gene of Watermelon silver mottle virus (WSMoV) were tested. (A) Resistant (R) line WLmt-13 exhibited broad-spectrum resistance to five tospovirus species, WSMoV, Tomato spotted wilt virus (TSWV), Groundnut ringspot virus (GRSV), Impatiens necrotic spot virus (INSV) and Groundnut chlorotic fan-spot virus (GCFSV). Susceptible (S) line WLmt-29 showed typical systemic symptoms similar to the non-transformed control after inoculation with individual tospoviruses was used for comparison. (B) Virus infections were confirmed by indirect enzyme-linked immunosorbent assay (ELISA) using the antiserum against the nucleocapsid protein of WSMoV, TSWV, GRSV, INSV or GCFSV. ELISA readings were recorded from the average of three independent experiments. Fourteen days post inoculation, the symptoms were photographed and ELISA was conducted.
Among the 19 WSMoV-resistant WLmts transgenic lines, 57.9–100% of them exhibited different levels of resistance to TSWV, GRSV, INSV and GCFSV. Seven lines, WLmts-2, 5, 7, 10, 13, 16 and 24, were resistant to all challenged tospoviruses with different levels of resistance. Particularly, WLmts-7 was immune to all challenged tospoviruses, similar to WLmt-13 (Figure 3). Among the 20 WSMoV-resistant WLmA transgenic lines, 50–75% of them exhibited different degrees of resistance to TSWV, GRSV, INSV and GCFSV. Five lines, WLmA-7, 8, 19, 20 and 21, showed different levels of resistance to the challenged tospoviruses. Particularly, WLmA-7 was immune to these challenged viruses, similar to WLmt-13 (Figure 3).
Moreover, in 21 WSMoV-resistant WLmhp transgenic lines, 71.4–76.2% of the plants showed different levels of broad-spectrum resistance to TSWV, GRSV, INSV and GCFSV. Seven lines, WLmhp-2, 4, 10, 15, 16, 22 and 24, exhibited different levels of resistance to all the challenged tospoviruses. However, no line was immune to any of those viruses.
Taken together, our results demonstrated that the L gene conserved region of WSMoV is able to trigger resistance not only against the homologous WSMoV, but also against other serologically unrelated tospoviruses, such as TSWV, GRSV, INSV and GCFSV, in transgenic N. benthamiana plants. Four particular lines, WLmt-4, WLmt-13, WLmts-7 and WLmA-7, showed complete resistance to the four heterologous tospovirus species. In further tests, these four tobacco lines also provided complete resistance against WBNV, MYSV, TYRV, IYSV and CaCV (data not shown). The 10 tospoviruses species used for challenge inoculation belong to different clades of tospoviruses: CaCV, WBNV, WSMoV and MYSV from WSMoV clade; TSWV and GRSV from TSWV clade; IYSV and TYRV from IYSV clade; GCFSV from GYSV clade; and INSV (Figure 4).
Figure 4. Phylogenetic relationship of tospoviruses based on the sequences of nucleocapsid (N) proteins.
The dendrogram was generated using the Neghbour-Joining algorithm based on 1000 bootstrap replicates. Viruses sharing close serological relationship in their N proteins are highlighted and designated as a clade. The viruses used to evaluate the transgenic resistance in this investigation are asterisked. The sequences of N genes were retrieved from GenBank of National Center for Biotechnology Information. ANSV, Alstroemeria necrotic streak virus (GQ478668); BeNMV, Bean necrosis mosaic virus (NC_018071); CaCV, Capsicum chlorosis virus (NC_008301); CCSV, Calla lily chlorotic spot virus (AY867502); CSNV, Chrysanthemum stem necrosis virus (AB600873); GBNV, Groundnut bud necrosis virus (U27809); GCFSV, Groundnut chlorotic fan-spot virus (AF080526); GRSV, Groundnut ringspot virus (L12048); GYSV, Groundnut yellow spot virus (AF013994); HCRV, Hippeastrum chlorotic ringspot virus (KC290943); INSV, Impatiens necrotic spot virus (NC_003624); IYSV, Iris yellow spot virus (AF001387); MeSMV, Melon severe mosaic virus (EU275149); MYSV, Melon yellow spot virus (AB038343); PNSV, Pepper necrotic spot virus (HE584762); PolRSV, Polygonum ringspot virus (EF445397); SVNaV, Soybean vein necrosis-associated virus (JF808214); TCSV, Tomato chlorotic spot virus (S54325); TNRV, Tomato necrotic ringspot virus (FJ489600); TSWV, Tomato spotted wilt virus (D13926); TYRV, Tomato yellow ring virus (AY686718); TZSV, Tomato zonate spot virus (NC_010489); WBNV, Watermelon bud necrosis virus (EU249351); WSMoV, Watermelon silver mottle virus (U78734); and ZLCV, Zucchini lethal chlorosis virus (AF067069).
When RNA viruses not belonging to the genus Tospovirus such as TuMV (Potyvirus) and CMV (Cucumovirus) were used to challenge the four lines immune to tospoviruses, the plants showed full susceptibility similar to the non-transgenic controls. Thus, our results suggest that the broad-spectrum resistance occurs at the genus level specifically to Tospovirus.
The L Gene Conserved Region Provides Broad-spectrum Resistance to WSMoV, TSWV and GRSV in Transgenic Tomato Plants
Nearly 80–90 independent lines of transgenic tomato plants were regenerated after transformation with each construct of pBGWLm, pBGWLmt, pBGWLmts, pBGWLmA and pBWLmhp. Following the selection on the kanamycin medium, plants of individual R0 lines were transferred to the greenhouse and challenged with a virulent TSWV isolate. A remarkably high number of 38–40% resistance of all WLmhp and WLmA lines showed high or moderate resistance to TSWV infection (Table 4). From almost all cases in our previous results of transgenic tobacco, we noticed that when a tomato line was resistant to the heterologous unrelated TSWV, it was also resistant to the homologous WSMoV and GRSV. The results for the WLm, WLmt and WLmts lines were less consistent. The resistance conferred by these three constructs was lower for TSWV and other tospoviruses (Table 4).
Table 4. Evaluation of transgenic tomato (Solanum lycopersicum L. cv. Feminini Beauty) lines, which carry individual constructs containing the conserved region of L gene of Watermelon silver mottle virus (WSMoV), for resistance to the homologous virus and two serologically unrelated tospovirus species of Tomato spotted wilt virus (TSWV) and Groundnut ringspot virus (GRSV) under greenhouse conditions.
| Transgene | No. of line assayed | Challenged with | TSWV-HR lines challenged with | ||||
| TSWV | WSMoV | GRSV | |||||
| HRa | MRb | Sc | HR | HR | MR | ||
| WLm | 94 | 5 (5.3%) | 25 (26.6%) | 64 | 5 (100%) | 2 (40%) | 2 (40%) |
| WLmt | 82 | 7 (8.5%) | 23 (28%) | 52 | 7 (100%) | 3 (42.9%) | 3 (42.9%) |
| WLmts | 88 | 7 (8%) | 23 (26.1%) | 58 | 7 (100%) | 3 (42.9%) | 3 (42.9%) |
| WLmA | 76 | 9 (11.8%) | 21 (27.6%) | 36 | 9 (100%) | 5 (55.6%) | 3 (33.3%) |
| WLmhp | 78 | 12 (15.4%) | 18 (23.1%) | 38 | 12 (100%) | 7 (58.3%) | 3 (25%) |
Transgenic tomato lines exhibited no symptoms; lines that were ELISA negative for TSWV NP were regarded as highly resistant (HR) lines.
Transgenic tomato lines exhibited delayed systemic symptoms for more than 7 days as compared with non-transgenic control were classified as moderately resistant (MR) lines.
Transgenic tomato lines that exhibited typical systemic symptoms similar to non-transformed plants after inoculation with TSWV at the same time or less than 7-day delay as compared to the non-transgenic control were classified as susceptible (S) lines.
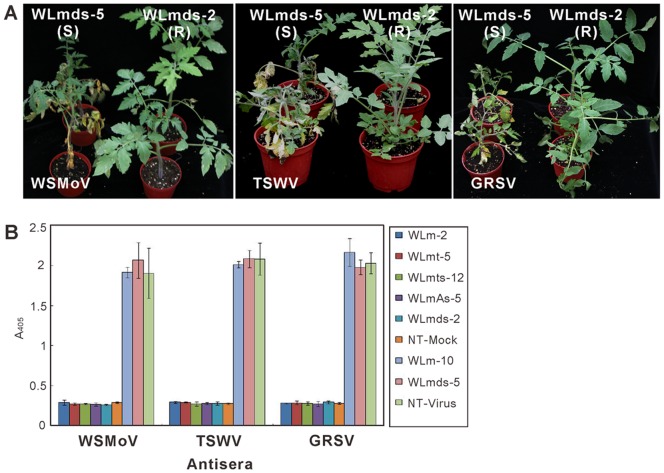
The virus infection of symptomatic transgenic tomato plants (Figure 5) was confirmed by indirect ELISA using the antiserum against the N protein of WSMoV, TSWV or GRSV. No significant accumulation of the virus was detected in the systemic leaves of symptomless plants, indicating that the resistant transgenic plants confer complete resistance to virus infection (Figure 5). Taken together, our results demonstrated that the L gene conserved region of WSMoV is able to trigger broad-spectrum resistance not only against the homologous WSMoV, but also against other serologically unrelated tospoviruses, such as TSWV and GRSV, in transgenic tomato plants.
Figure 5. Evaluation of the transgenic tomato lines for resistance to distinct tospovirus species under greenhouse conditions.
All transgenic lines carried the double-stranded inverted repeat of conserved region of L gene (WLmhp) of Watermelon silver mottle virus (WSMoV). The photograph was taken and the indirect enzyme-linked immunosorbent assay (ELISA) was conducted 14 days post-inoculation. (A) Resistant (R) line WLmhp-2 exhibited broad-spectrum resistance to three tospovirus species, including WSMoV, Tomato spotted wilt virus (TSWV) and Groundnut ringspot virus (GRSV). Susceptible (S) line WLmhp-5, which was included as a control, showed typical systemic symptoms similar to the non-transgenic control after inoculation with these three tospoviruses. (B) Virus infections were confirmed by indirect ELISA using the antiserum against the nucleocapsid protein of WSMoV, TSWV or GRSV. ELISA readings were recorded from the average of three independent experiments.
Broad-spectrum Resistant Transgenic Tobacco Lines Carrying One to Multiple Copies of Transgenes
The genomic DNAs isolated from the leaves of transgenic tobacco plants exhibiting broad-spectrum resistance against five different tospovirus species, including WLm-30, WLmA-7, 8, 19, 20 and 21, WLmt-4, 5, 9 and 13, WLmts-2, 16 and 24, and WLmhp-2 and 4, were digested with SspI. Results of hybridization revealed that these resistant transgenic tobacco lines carried one to seven copies of transgenes as shown in Figure 6. The similar patterns observed for WLmA-19 and WLmA-20 suggested that they might have originated from a same transformed cell. The particular lines WLmA-7, WLmt-4 and WLmt-13, which exhibited immunity to 10 different tospoviruses, contain 1–3 inserts of transgenes. Hence, our result indicated that the broad-spectrum resistance is not correlated to the insert number of transgenes.
Figure 6. Detection of transgene insert numbers in the transgenic Nicotiana benthamiana R0 lines.
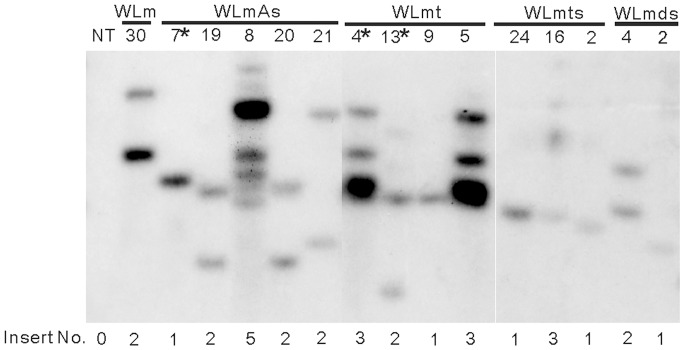
(A) The insert numbers of individual transgenes are indicated. A non-transgenic (NT) N. benthamiana plant was used as a negative control.
Broad-spectrum Resistance is Mediated by RNA Silencing
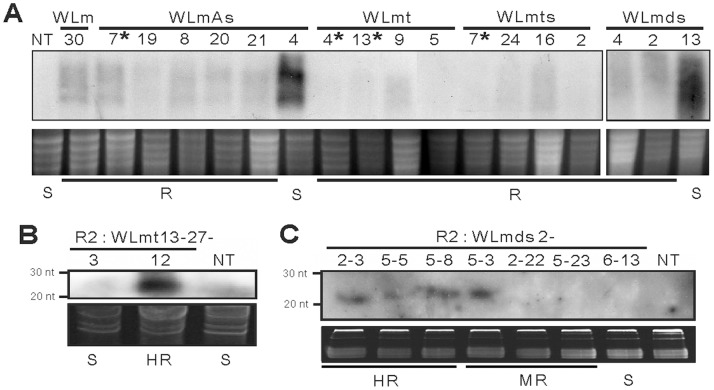
Significant amounts of transgene transcript were detected from samples from N. benthamiana plants of two randomly selected susceptible transgenic lines, WLmA-4 and WLmhp-13 (Figure 7A). Steady-state levels of transgene transcripts were not detected in all the assayed transgenic N. benthamiana samples conferring broad-spectrum resistance to five different tospovirus species, including WLm-30, WLmA-7, 8, 19, and 21, WLmt-4, 5, 9 and 13, WLmts-2, 7, 16 and 24, and WLmhp-2 and 4 (Figure 7A).
Figure 7. Detection of transgene transcripts in R0 lines of transgenic Nicotiana benthamiana and siRNA accumulation in R2 progeny of N. bentamiana and tomato lines resistant to Watermelon silver mottle virus (WSMoV).
(A) All the analyzed resistant (R) lines showed lower levels of transgene transcripts than the susceptible (S) lines. The transgenic N. benthamiana lines completely resistant to 10 distinct tospovirus species are indicated by asterisks. Transgene transcripts were not detected in the NT plant of N. benthamiana. Ribosomal RNAs were used as loading controls. (B) siRNA accumulation of a susceptible (S) R2 individual (WLmt 13-27-3) and a highly resistant (HR) R2 individual (WLmt 13-27-12) of a tobacco line immune to WSMoV, in comparison to a non-transgenic (NT) plant, was analyzed by 32P-labeled WLm-specific probe. (C) siRNA accumulation of R2 progeny of a HR tomato line WLmhp 2. Accumulation of siRNA in R2 HR individuals (WLmhp 2-2-3, 2-5-5 and 2-5-8), moderately resistant (MR) individuals (WLmhp 2-5-3, 2-2-22 and 2-5-23), an S individual (WLmhp 2-6-13), and a NT plant were analyzed by 32P-labeled WLmhp-specific probe.
Since transgenic tobacco and tomato lines were obtained at different times, siRNA analyses were conducted on the R2 population of selected HR tobacco or tomato lines. High accumulation of siRNA was observed in the progeny of the highly resistant tobacco line WLmt13, the 21 nt siRNA was found in the resistant individual WLmt-13-27-12, but not present in the susceptible individual WLmt-13-27-3 (Figure 7B). Also, when the R2 progeny of the HR tomato line WLmhp-2 were analyzed, the 21 nt siRNA was found present in the HR individuals of WLmhp-2-2-3, 2-5-5, and 2-5-8, and WR individual of WLmhp-2-5-3, whereas no siRNA accumulation was detected in the two WR and one susceptible individuals (Figure 7C). Taken together, the results of transcript silencing and siRNA accumulation were correlated to the degrees of the transgenic resistance, indicating that the broad-spectrum resistance conferred by the L gene conserved region-transgenic tobacco plants is mediated by PTGS.
Discussion
The effectiveness of PDR in terms of strength, spectrum and durability is largely depends upon the selection of appropriate segment of pathogen genome and its engineering into an effective transgene. In the present study, the tospovirus L gene region containing the highly conserved five RdRp motifs [16], [43], [46], [47] was amplified from WSMoV L gene (nt 3975 to 4928 of vc strand of WSMoV L gene) and designed into various transgenes that can trigger PTGS in the transformants of the model plant N. benthamiana and the real crop tomato. When the resistance levels of transgenic N. benthamiana plants were compared, the transgenic lines carrying individual transgenes were able to provide complete resistance to the homologous virus WSMoV and also to serologically unrelated 9 heterologous tospovirus species of different phylogenetic clades of tospoviruses. However, the resistance is not extended to unrelated viruses like TuMV and CMV that do not belong to the genus Tospovirus, indicating that the broad-spectrum resistance is Tospovirus genus-specific. The broad-spectrum resistance was further evidenced in transgenic tomato lines which provide complete resistance to the homologous and serologically unrelated heterologous tospovirus species.
The transgene transcripts were not detected in all the tested resistant transgenic N. benthamiana lines, while they were detectable in the susceptible lines. Furthermore, siRNA accumulation was detected in the resistant R2 individuals of the selected tobacco and tomato HR lines, but not in the susceptible R2 individuals. These results demonstrated that PTGS is the underlying the mechanism for resistance. The non-translatable transgenes of WLmt, WLmts, WLmA and WLmhp effectively triggered PTGS, as described previously for RNA-mediated transgenic resistance [67], [68].
Some studies reported that translatability of transgenes was necessary for replicase-mediated resistance to TMV, Pea early browning virus and CMV [69], [70], [71]. The CMV-resistant tobacco lines containing translatable 2a replicase transgenes exhibited either delay in symptom development or complete resistance [71]. It was considered that replication of CMV was interfered and the cell-to-cell and/or long-distance movement of the virus was restricted through unidentified cellular processes [72], [73], [74], [75]. However, the effectiveness of transgene-induced silencing could not be ruled out in these previous studies, because some resistant lines containing translatable transgenes also generated RNA-mediated resistance correlating with low levels of accumulation of transgene mRNA [71]. In the present study, similar replicase-mediated effect from the translatable construct WLm could not be excluded due to the lack of specific antibody for verifying translation from WLm. However, lower transgene transcript levels observed with the translatable WLm construct (Figure 7A) suggest that PTGS is the main underlying resistance mechanism.
PTGS is sequence homology-dependent. The WLm region within L genes of TSWV and INSV show 69% and 66.4% nucleotide identity, respectively, to that of WSMoV. This level of identity (66–69%) is lower than the level of identity (∼88%) considered being required for effective targeting for PTGS [76]. However, the contexts of L genes of TSWV and INSV corresponding to the highly conserved five RdRp motifs of L protein share higher identities of 78–85% and 76–86%, respectively, with those of WSMoV L gene. This level of identity in combination with the relatively less mutable nature of RdRp motifs may explain the observed PTGS-mediated resistance of WLm transgenic plants to the serologically unrelated tospoviruses. The DNA probe corresponding to the conserved region of WSMoV L gene was able to hybridize with total RNAs extracted from plant tissues infected with TSWV and INSV, suggesting that the WLm transgenes are also able to interact with the L genes of tospoviruses not belonging to the WSMoV clade (Figure 2B). Corroboratively, hybridization signals were not detected for TuMV (Potyvirus) and CMV (Cucumovirus). Challenge assay with TuMV and CMV validated the results of hybridization. Taken together, the results of hybridization analysis and challenge assay suggest that the broad-spectrum transgenic resistance mediated by WSMoV L gene RdRp motifs is specific against the genus Tospovirus.
Although some of the full-length L RNAs of the other challenged tospoviruses have not been determined yet, the transgenic resistance against these viruses logically demonstrates that the L RNAs of GRSV and GCFSV share homology with those of WSMoV, TSWV and INSV, as described in our previous reports [43], [46]. The conserved region of the L RNAs of MYSV, GBNV and CaCV share high identities of 79.6% to 89.9%, 81.3% to 88.2% and 77.8% to 93.8%, respectively, with the five RdRp motifs of WSMoV (Table 1). Thus, it is not surprising that the L gene conserved region-transgenic tobacco plants also provide resistance against these three economically important Asia-type tospoviruses, and other Asia-type tospovirus species of WBNV, TYRV and IYSV, which belong to WSMoV and IYSV clades [16], [47], [77], [78]. The tospovirus N protein dendrogram (Figure 4) highlighting different clades and tested viruses illustrates the Tospovirus genus level broad spectrum resistance, though we are yet to test the members of Bean necrosis mosaic virus (BeNMV)-Soybean vein necrosis-associated virus (SVNaV) clade, which was discerned recently by phylogenetic analyses with tospoviral protein sequences [79]. SVNaV, one of the known two members of the BeNMV-SVNaV clade [79], was shown to be a distinct serotype by analyzing its serological relationship with GRSV, INSV, TCSV, TSWV and TYRV [80].
The negative sense viral (v) L RNA of tospoviruses is the template for L RNA replication and transcription. The transcribed positive sense viral complementary (vc) L RNA is the messenger RNA for expressing replicase protein [4]. Both v and vc L RNAs, believed to be present simultaneously in the infected cells, are possibly targeted by the siRNAs processed from the transcripts, resulting in inhibition of L RNA replication and translation.
The members of the genus Tospovirus are distributed worldwide and they cause severe damage to many economically important crops. Wide genetic variations of tospoviruses and their persistent transmission by thrips make the diseases caused by tospoviruses difficult to be controlled. Sources of natural resistant genes to Tospovirus spp. suitable for commercial breeding are very limited and extensive studies have been made to create engineered resistance against these viruses. Several studies have shown that transgenic plants can confer broad-spectrum resistance against distinct tospovirus species. For instance, transgenic N. benthamiana plants expressing the N protein-interacting peptide derived from the N ORF of TSWV generated high levels of resistance to TSWV, GRSV, TCSV and CSNV [35] belonging to TSWV clade.
A strategy using a single transgene by combining small RNA fragments from N genes of WSMoV, TSWV, GRSV and TCSV in hairpin construct to trigger RNA silencing for generating a high frequency broad-spectrum resistance against all the four tospoviruses was reported [34]. However, this approach is N gene homology-dependent and difficult to be applied for controlling the different tospoviral strains or newly emergent species in the genus Tospovirus. Hassani-Mehraban et al. [81] generated transgenic N. benthamiana plants resistant to five different tospoviruses, i.e., WSMoV, TSWV, GRSV, TCSV and TYRV-t (tomato-infecting strain of TYRV) using an inverted repeat construct containing partial N gene sequences from these viruses. However, the transgenic resistance against TYRV-t does not hold against the soybean-infecting strain TYRV-s; the resistance against TYRV-s was broken down by TYRV-t, when it was co-inoculated with TYRV-s [81].
We have first reported that an alternative approach that the 21 nt viral sequences of the conserved motif of a viral PTGS suppressor gene constructed as amiRNAs is able to confer complete resistance to virus infection in transgenic Arabidopsis plants [65]. Our further effort revealed that amiRNAs precisely targeting at the conserved RdRp motifs of WSMoV L gene, individually or in different combination, confer high degrees of transgenic resistance against the homologous virus, but not against different tospovirus species with 2 or 3 mismatches in the targeted RdRp motifs in the same WSMoV clade [32].
Differing from all the aforementioned approaches for generating transgenic resistance to tospoviruses, here we have successfully developed a novel transgenic approach using a single fragment from the L RNA of WSMoV, which encompasses all five highly conserved RdRp motifs in their native contiguous context, as transgenes to generate transgenic tobacco and tomato plants conferring broad-spectrum resistance against distinct tospovirus species at the genus level. In earlier reported tospovirus N gene transgenic plants, the N gene-mediated resistance was restricted to homologous or closely related viruses [25], [27], [30], [31], owing to relatively less conserved nature of N gene sequences. The Tospovirus genus level resistance of the present WLm transgenic N. benthamiana and tomato plants can be attributed to higher degree of conservation of L gene and relatively non-mutable nature of L gene sequence contexts corresponding to the essential RdRp motifs of L protein. The superior performance of WLmhp transgene to other transgenes may partly be due to the hairpin structure of the transcribed RNA and the presence of the eight conserved terminal nucleotides of tospoviral RNAs as the linker connecting the two arms of the hairpin. However, the possible contribution of the eight terminal nucleotides of tospoviral RNAs to the genus level resistance conferred by WLmhp transgene was not assessable because of the lack of an appropriate control. Our transgenic approach should provide a more practical and durable measure for controlling the devastating diseases caused by the prevailing and newly emerging tospoviral strains or species in different regions of the world.
Funding Statement
The authors thank the funding supports of the National Science Council (NSC92-2317-B-005-024, NSC-101-2911-I-005-301 and NSC-102-2911-I-005-301), and the Ministry of Education, Taiwan, R.O.C. under the ATU plan for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Prins M, Goldbach R (1998) The emerging problem of tospovirus infection and nonconventional methods of control. Trends Microbiol 6: 31–35. [DOI] [PubMed] [Google Scholar]
- 2. Parrella G, Gognalons P, Gebre-Selassiè K, Vovlas C, Marchoux G (2003) An update of the host range of Tomato spotted wilt virus . J Plant Pathol 85: 227–264. [Google Scholar]
- 3. Lebas BSM, Ochoa-Corona FM (2007) Impatiens necrotic spot virus . In: Vol Rao GP, Bragard C, Lebas BSM, editors. Characterization, diagnosis and management of plant viruses. 4: 221–243. [Google Scholar]
- 4.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (2012) Ninth Report of the International Committee on Taxonomy of Viruses. United States of America: Elsevier. 1338 p.
- 5. de Haan P, Wagemakers L, Peters D, Goldbach R (1990) The s-rna segment of Tomato spotted wilt virus has an ambisense character. J Gen Virol 71: 1001–1007. [DOI] [PubMed] [Google Scholar]
- 6. Takeda A, Sugiyama K, Nagano H, Mori M, Kaido M, et al. (2002) Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus . Febs Letters 532: 75–79. [DOI] [PubMed] [Google Scholar]
- 7. Kormelink R, Dehaan P, Meurs C, Peters D, Goldbach R (1992) The nucleotide-sequence of the messenger-rna segment of Tomato spotted wilt virus, a bunyavirus with 2 ambisense rna segments. Journal of General Virology 73: 2795–2804. [DOI] [PubMed] [Google Scholar]
- 8. Storms MMH, van der Schoot C, Prins M, Kormelink R, van Lent JWM, et al. (1998) A comparison of two methods of microinjection for assessing altered plasmodesmal gating in tissues expressing viral movement proteins. Plant Journal 13: 131–140. [Google Scholar]
- 9. de Haan P, Kormelink R, Resende RD, Vanpoelwijk F, Peters D, et al. (1991) Tomato spotted wilt virus-l rna encodes a putative rna-polymerase. Journal of General Virology 72: 2207–2216. [DOI] [PubMed] [Google Scholar]
- 10. de Ávila AC, de Haan P, Kormelink R, Resende RdeO, Goldbach RW, et al. (1993) Classification of tospoviruses based on phylogeny of nucleoprotein gene Sequences. J Gen Virol 74: 153–159. [DOI] [PubMed] [Google Scholar]
- 11.Tsompana M, Moyer JW (2008) Tospovirus. Encyclopedia of Virology (B.W.J. Mahy and M.H.V. Van Regenmortel, Editors), 157–163 Oxford: Elsevier.
- 12.Jan FJ, Chen TC, Yeh SD (2003) Occurrence, importance, taxonomy, and control of thrips-borne tospoviruses. In: Huang HC, Acharya SN, editors. Advances in Plant Disease Management. Research Signpost, India. 399–421.
- 13. Lin YH, Chen TC, Hsu HT, Liu FL, Chu FH, et al. (2005) Serological comparison and molecular characterization for verification of Calla lily chlorotic spot virus as a new tospovirus species belonging to Watermelon silver mottle virus serogroup. Phytopathology 95: 1482–1488. [DOI] [PubMed] [Google Scholar]
- 14. Hassani-Mehraban A, Saaijer J, Peters D, Goldbach R, Kormelink R (2007) Molecular and biological comparison of two Tomato yellow ring virus (TYRV) isolates: challenging the Tospovirus species concept. Arch Virol 152: 85–96. [DOI] [PubMed] [Google Scholar]
- 15. Dong JH, Cheng XF, Yin YY, Fang Q, Ding M, et al. (2008) Characterization of Tomato zonate spot virus, a new tospovirus in China. Arch Virol 153: 855–864. [DOI] [PubMed] [Google Scholar]
- 16. Li JT, Yeh YC, Yeh SD, Raja JAJ, Rajagopalan PA, et al. (2011) Complete genomic sequence of Watermelon bud necrosis virus. Arch Virol 156: 359–362. [DOI] [PubMed] [Google Scholar]
- 17. Chen TC, Lu YY, Cheng YH, Li JT, Yeh YC, et al. (2010) Serological relationship between Melon yellow spot virus and Watermelon silver mottle virus and differential detection of the two viruses in cucurbits. Arch Virol 155: 1085–1095. [DOI] [PubMed] [Google Scholar]
- 18. Stevens MR, Scott SJ, Gergerich RC (1992) Inheritance of a gene for resistance to Tomato spotted wilt virus (TSWV) from Lycopersicon esculentum Mill. Euphytica 59: 9–17. [Google Scholar]
- 19. Rosello S, Diez Maria J, Nuez F (1998) Genetics of Tomato spotted wilt virus resistance coming from Lycopersicon peruvianum . Eur J Plant Pathol 104: 499–509. [Google Scholar]
- 20. Jahn M, Paran I, Hoffmann K, Radwanski ER, Livingstone KD, et al. (2000) Genetic mapping of the Tsw locus for resistance to the Tospovirus Tomato spotted wilt virus in Capsicum spp. and its relationship to the Sw-5 gene for resistance to the same pathogen in tomato. Mol Plant-Microbe Interact 13: 673–682. [DOI] [PubMed] [Google Scholar]
- 21. Margaria P, Ciuffo M, Pacifico D, Turina M (2007) Evidence that the nonstructural protein of Tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the Tsw gene. Mol Plant-Microbe Interact 20: 547–558. [DOI] [PubMed] [Google Scholar]
- 22. López C, Aramburu J, Galipienso L, Soler S, Nuez F, et al. (2011) Evolutionary analysis of tomato Sw-5 resistance-breaking isolates of Tomato spotted wilt virus . J Gen Virol 92: 210–215. [DOI] [PubMed] [Google Scholar]
- 23. Sanford JC, Johnston SA (1985) The concept of parasite-derived resistance-deriving resistance genes from the parasite’s own genome. J Theor Biol 113: 395–405. [Google Scholar]
- 24. Powell-Abel P, Nelson RS, De B, Hoffmann N, Rogers SG, et al. (1986) Delay of disease development in transgenic plants that express the Tobacco mosaic virus coat protein gene. Science 232: 738–743. [DOI] [PubMed] [Google Scholar]
- 25. Pang SZ, Bock JH, Gonsalves C, Slightom JL, Gonsalves D (1994) Resistance of transgenic Nicotiana benthamiana plants to tomato spotted wilt and impatiens necrotic spot tospoviruses: Evidence of involvement of the N protein and N gene RNA in resistance. Phytopathology 84: 243–249. [Google Scholar]
- 26. Schwach F, Adam G, Heinze C (2004) Expression of a modified nucleocapsid-protein of Tomato spotted wilt virus (TSWV) confers resistance against TSWV and Groundnut ringspot virus (GRSV) by blocking systemic spread. Mol Plant Pathol 5: 309–316. [DOI] [PubMed] [Google Scholar]
- 27. Prins M, Goldbach R (1996) RNA-mediated virus resistance in transgenic plants. Arch Virol 141: 2259–2276. [DOI] [PubMed] [Google Scholar]
- 28. Pang SZ, Jan FJ, Carney K, Stout J, Tricoli DM, et al. (1996) Post-transcriptional transgene silencing and consequent tospovirus resistance in transgenic lettuce are affected by transgene dosage and plant development. Plant J 9: 899–909. [Google Scholar]
- 29. Prins M, Kikkert M, Ismayadi C, de Graauw W, de Haan P, et al. (1997) Characterization of RNA-mediated resistance to Tomato spotted wilt virus in transgenic tobacco plants expressing NSm gene sequence. Plant Mol Biol 33: 235–243. [DOI] [PubMed] [Google Scholar]
- 30. Pang SZ, Nagpala P, Wang M, Slightom JL, Gonsalves D (1992) Resistance to heterologous isolates of Tomato spotted wilt virus in transgenic tobacco expressing its nucleocapsid protein gene. Mol Plant Pathol 82: 1223–1229. [Google Scholar]
- 31. Pang SZ, Slightom JL, Gonsalves D (1993) Different mechanisms protect transgenic tobacco against tomato spotted wilt and impatiens necrotic spot tospoviruses. Nat Biotechnol 11: 819–824. [DOI] [PubMed] [Google Scholar]
- 32. Kung YJ, Lin SS, Huang YL, Chen TC, Harish SS, et al. (2012) Multiple artificial microRNAs targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded RNA plant virus. Mol Plant Pathol 13: 303–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prins M, de Haan P, Luyten R, van Veller M, van Grinsven MQ, et al. (1995) Broad resistance to tospoviruses in transgenic tobacco plants expressing three tospoviral nucleoprotein gene sequences. Mol Plant-Microbe Interact 8: 85–91. [DOI] [PubMed] [Google Scholar]
- 34. Bucher E, Lohuis D, Pieter M, van Poppel JA, Geerts-Dimitriadou C, et al. (2006) Multiple virus resistance at a high frequency using a single transgene construct. J Gen Virol 87: 3697–3701. [DOI] [PubMed] [Google Scholar]
- 35. Rudolph C, Schreier PH, Uhrig JF (2003) Peptide-mediated broad-spectrum plant resistance to tospoviruses. Proc Natl Acad Sci USA 100: 4429–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yeh SD, Lin YC, Cheng YH, Jih CL, Chen MJ, et al. (1992) Identification of tomato spotted wilt-like virus infecting watermelon in Taiwan. Plant Dis 76: 835–840. [Google Scholar]
- 37. Iwaki M, Honda Y, Hanada K, Tochihara H, Yonaha T, et al. (1984) Silver mottle disease of watermelon caused by Tomato spotted wilt virus . Plant Dis 68: 1006–1008. [Google Scholar]
- 38.Singh SJ, Krishnareddy M (1995) Diagnosis and epidemiology of a tospovirus infecting watermelon in India. Abstracts of the International Symposium on Tospoviruses and Thrips of Floral and Vegetable Crops, Taiwan, p.42.
- 39. Chiemsombat P, Gajanandana O, Warin N, Hongprayoon R, Bhunchoth A, et al. (2008) Biological and molecular characterization of tospoviruses in Thailand. Arch Virol 153: 571–577. [DOI] [PubMed] [Google Scholar]
- 40. Yeh SD, Chang TF (1995) Nucleotide sequence of the N gene of Watermelon silver mottle virus, a proposed new member of the genus Tospovirus . Phytopathology 85: 58–64. [Google Scholar]
- 41. Yeh SD, Sun IJ, Ho HM, Chang TF (1996) Molecular cloning and nucleotide sequence analysis of the S RNA of watermelon silver mottle tospovirus. Acta Hortic 431: 244–260. [Google Scholar]
- 42. Chu FH, Yeh SD (1998) Comparison of replication forms and ambisense M RNA of Watermelon silver mottle virus with other tospoviruses. Phytopathology 88: 351–358. [DOI] [PubMed] [Google Scholar]
- 43. Chu FH, Chao CH, Chung MH, Chen CC, Yeh SD (2001) Completion of the genome sequence of Watermelon silver mottle virus and utilization of degenerate primers for detecting tospoviruses in five serogroups. Phytopathology 91: 361–368. [DOI] [PubMed] [Google Scholar]
- 44. Kunkalikar SR, Poojari S, Arun BM, Rajagopalan PA, Chen TC, et al. (2011) Importance and genetic diversity of vegetable-infecting tospoviruses in India. Phytopathology 101: 367–376. [DOI] [PubMed] [Google Scholar]
- 45. Shimomoto Y, Kobayashi K, Okuda M (2014) Identification and characterization of Lisianthus necrotic ringspot virus, a novel distinct tospovirus species causing necrotic disease of lisianthus (Eustoma grandiflorum). J Gen Plant Pathol 80: 169–175. [Google Scholar]
- 46. Chen CC, Chen TC, Lin YH, Yeh SD, Hsu HT (2005) A chlorotic spot disease on calla lilies (Zantedeschia spp.) is caused by a tospovirus serologically but distantly related to Watermelon silver mottle virus . Plant Dis 89: 440–445. [DOI] [PubMed] [Google Scholar]
- 47. Chen TC, Li JT, Fan YS, Yeh YC, Yeh SD, et al. (2013) Molecular characterization of the full-length L and M RNAs of Tomato yellow ring virus, a member of the genus Tospovirus . Virus Genes 46: 487–495. [DOI] [PubMed] [Google Scholar]
- 48. Chen TC, Li JT, Lin YP, Yeh YC, Kang YC, et al. (2012) Genomic characterization of Calla lily chlorotic spot virus and design of broad-spectrum primers for detection of tospoviruses. Plant Pathol 61: 183–194. [Google Scholar]
- 49. Chen TC, Lu YY, Cheng YH, Chang CA, Yeh SD (2008) Melon yellow spot virus in watermelon: a first record from Taiwan. Plant Pathol 57: 765. [Google Scholar]
- 50. Chu FH, Chao CH, Peng YC, Lin SS, Chen CC, et al. (2001) Serological and molecular characterization of Peanut chlorotic fanspot virus, a new species of the genus Tospovirus . Phytopathology 91: 856–863. [DOI] [PubMed] [Google Scholar]
- 51. Hsu HT, Ueng PP, Chu FH, Ye Z, Yeh SD (2000) Serological and molecular characterization of a high temperature-recovered virus belonging to Tospovirus serogroup IV. J Gen Plant Pathol 66: 167–175. [Google Scholar]
- 52. Pang SZ, Slightom JL, Gonsalves D (1993) The biological properties of a distinct tospovirus and sequence analysis of its S RNA. Phytopathology 83: 728–733. [Google Scholar]
- 53. Law MD, Moyer JW (1990) A tomato spotted wilt-like virus with a serologically distinct N protein. J Gen Virol 71: 933–938. [Google Scholar]
- 54. Cortes I, Livieratos IC, Derks A, Peters D, Kormelink R (1998) Molecular and serological characterization of Iris yellow spot virus, a new and distinct tospovirus species. Phytopathology 88: 1276–1282. [DOI] [PubMed] [Google Scholar]
- 55. Hassani-Mehraban A, Saaijer J, Peters D, Goldbach R, Kormelink R (2005) A new tomato-infecting tospovirus from Iran. Phytopathology 95: 852–858. [DOI] [PubMed] [Google Scholar]
- 56. Lin SS, Hou RF, Huang CH, Yeh SD (1998) Characterization of Zucchini yellow mosaic virus (ZYMV) isolates collected from Taiwan by host reactions, serology, and RT-PCR. Plant Prot Bull 40: 163–176. [Google Scholar]
- 57. Yeh SD, Jan FJ, Chiang CH, Doong TJ, Chen MC, et al. (1992) Complete nucleotide sequence and genetic organization of Papaya ringspot virus RNA. J Gen Virol 73: 2531–2541. [DOI] [PubMed] [Google Scholar]
- 58. Chen CC, Chao CH, Chen CC, Yeh SD, Tsai HT, et al. (2003) Identification of Turnip mosaic virus isolates causing yellow stripe and spot on calla lily. Plant Dis 87: 901–905. [DOI] [PubMed] [Google Scholar]
- 59. Cheng YH, Yeh SD (2000) Construction and evaluation of transgenic tobacco plants expressing the coat protein gene of Papaya ringspot virus with different translation leaders. Bot Bull Acad Sin 41: 1–10. [Google Scholar]
- 60. Chen H, Nelson RS, Sherwood JL (1994) Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. BioTechniques 16: 664–670. [PubMed] [Google Scholar]
- 61. Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, et al. (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231. [DOI] [PubMed] [Google Scholar]
- 62. Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497. [Google Scholar]
- 63. McCormick S, Niedermeyer J, Fry J, Barnason A, Horseh R, et al. (1986) Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. . Plant Cell Reports 5: 81–84. [DOI] [PubMed] [Google Scholar]
- 64. Yeh SD, Gonsalves D (1984) Purification and immunological analysis of cylindrical-inclusion protein induced by Papaya ringspot virus and Watermelon mosaic virus I. Phytopathology. 74: 1273–1278. [Google Scholar]
- 65. Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, et al. (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24: 1420–1428. [DOI] [PubMed] [Google Scholar]
- 66. Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, et al. (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20: 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Prins M (2003) Broad virus resistance in transgenic plants. Trends Biotechnol 21: 373–375. [DOI] [PubMed] [Google Scholar]
- 68. Lindbo JA, Dougherty WG (2005) Plant pathology and RNAi: a brief history. Annu Rev Phytopathol 43: 191–204. [DOI] [PubMed] [Google Scholar]
- 69. Carr JP, Marsh LE, Lomonossoff GP, Sekiya ME, Zaitlin M (1992) Resistance to Tobacco mosaic virus induced by the 54-kDa gene sequence requires expression of the 54-kDa protein. Mol Plant-Microbe Interact 5: 397–404. [DOI] [PubMed] [Google Scholar]
- 70. MacFarlane SA, Davies JW (1992) Plants transformed with a region of the 201-kilodalton replicase gene from pea early browning virus RNA 1 are resistant to virus infection. Proc Natl Acad Sci USA 89: 5829–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wintermantel WM, Zaitlin M (2000) Transgene translatability increase effectiveness of replicase-mediated resistance to Cucumber mosaic virus . J Gen Virol 81: 587–595. [DOI] [PubMed] [Google Scholar]
- 72. Carr JP, Gal-On A, Palukaitis P, Zaitlin M (1994) Replicase-mediated resistance to Cucumber mosaic virus in transgenic plants involves suppression of both virus replication in the inoculated leaves and long-distance movement. Virology 199: 439–447. [DOI] [PubMed] [Google Scholar]
- 73. Nguyen L, Lucas WJ, Ding B, Zaitlin M (1996) Replicase-mediated resistance in transgenic tobacco plants involves inhibition of viral cell-to-cell movement. Proc Natl Acad Sci USA 93: 12643–12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wintermantel WM, Banerjee N, Oliver JC, Paolillo DJ, Zaitlin M (1997) Cucumber mosaic virus is restricted from entering minor veins in transgenic tobacco exhibiting replicase-mediated resistance. Virology 231: 248–257. [DOI] [PubMed] [Google Scholar]
- 75. Canto T, Palukaitis P (1999) Replicase-mediated resistance to Cucumber mosaic virus does not inhibit localization and/or trafficking of the viral movement protein. Mol Plant-Microbe Interact 12: 743–747. [Google Scholar]
- 76. Mueller E, Gilbert J, Davenport G, Brigneti G, Baulcome DC (1995) Homology-dependent resistance: Transgenic virus resistance in plants related to homology-dependent gene silencing. Plant J 7: 1001–1013. [Google Scholar]
- 77. Knierim D, Blawid R, Maiss E (2006) The complete nucleotide sequence of a Capsicum chlorosis virus isolate from Lycopersicum esculentum in Thailand. Arch Virol 151: 1761–1782. [DOI] [PubMed] [Google Scholar]
- 78. Bag S, Druffel KL, Pappu HR (2010) Structure and genome organization of the large RNA of Iris yellow spot virus (genus Tospovirus, family Bunyaviridae). Arch Virol 155: 275–279. [DOI] [PubMed] [Google Scholar]
- 79. de Oliveira AS, Melo FL, Inoue-Nagata AK, Tatsuya Nagata T, Kitajima EW, et al. (2012) Characterization of Bean necrotic mosaic virus: A member of a novel evolutionary lineage within the genus Tospovirus . PloS One 7: e38634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Khatabi B, Wen RH, Hershman DE, Kennedy BS, Newman MA, et al. (2012) Generation of polyclonal antibodies and serological analyses of nucleocapsid protein of Soybean vein necrosis-associated virus: A distinct soybean infecting tospovirus serotype. Eur J Plant Pathol 133: 783–790. [Google Scholar]
- 81. Hassani-Mehraban A, Brenkman AB, van den Broek NJF, Goldbach R, Kormelink R (2009) RNAi-mediated transgenic tospovirus resistance broken by intraspecies silencing suppressor protein complementation. Mol Plant-Microbe Interact 22: 1250–1257. [DOI] [PubMed] [Google Scholar]