Abstract
Receptor activator of nuclear factor (NF)-κB ligand (RANKL), a master cytokine that drives osteoclast differentiation, activation and survival, exists in both transmembrane and extracellular forms. To date, studies on physiological role of RANKL have been mainly carried out with extracellular RANKL probably due to difficulties in achieving high level expression of functional transmembrane RANKL (mRANKL). In the present study, we took advantage of codon optimization and response surface methodology to optimize the soluble expression of mRANKL in E. coli. We optimized the codon usage of mRANKL sequence to a preferred set of codons for E. coli changing its codon adaptation index from 0.64 to 0.76, tending to increase its expression level in E. coli. Further, we utilized central composite design to predict the optimum combination of variables (cell density before induction, lactose concentration, post-induction temperature and post-induction time) for the expression of mRANKL. Finally, we investigated the effects of various experimental parameters using response surface methodology. The best combination of response variables was 0.6 OD600, 7.5 mM lactose, 26°C post-induction temperature and 5 h post-induction time that produced 52.4 mg/L of fusion mRANKL. Prior to functional analysis of the protein, we purified mRANKL to homogeneity and confirmed the existence of trimeric form of mRANKL by native gel electrophoresis and gel filtration chromatography. Further, the biological activity of mRANKL to induce osteoclast formation on RAW264.7 cells was confirmed by tartrate resistant acid phosphatase assay and quantitative real-time polymerase chain reaction assays. Importantly, a new finding from this study was that the biological activity of mRANKL is higher than its extracellular counterpart. To the best of our knowledge, this is the first time to report heterologous expression of mRANKL in soluble form and to perform a comparative study of functional properties of both forms of RANKL.
Introduction
Receptor activator of nuclear factor (NF)-κB ligand (RANKL), a member of tumor necrosis factor (TNF) superfamily, is preferentially expressed on osteoblast and stromal lineage cells whereas its receptor RANK is preferentially expressed on osteoclast lineage cells [1]–[3]. RANKL is produced as a type II transmembrane protein on these cells, and cleaved into an extracellular soluble form by specific metalloproteinases [4]–[7]. The latter form has high similarity to TNF-related apoptosis inducing ligand (TRAIL), FasL (TNF-related ligand) and TNF itself [8]. RANKL and RANK are best known for their essential role in controlling osteoclastogenesis. The essential physiological roles of RANKL-RANK have been elucidated through several in vitro and in vivo studies. It is now known that RANKL-RANK system is required to trigger a network of intracellular signaling cascades that promote osteoclast differentiation, activation and survival [9]. Moreover, RANKL is essential to induce expression of genes including tartrate-resistant acid phosphatase (Trap), calcitonin receptor (CalcR), cathepsin K (CtsK), receptor for macrophage-colony-stimulating factor (Cfms) and nuclear factor of activated T-cells (Nfatc1) leading to the development of mature osteoclasts [9]–[11]. The biological activity of RANKL is balanced by its physiological decoy receptor, osteoprotegerin (OPG) that competes with RANK for RANKL and thus dictates the quantity of bone resorbed [12]–[14]. The elucidation of the signaling pathway mediated by OPG, RANK and RANKL provided a major breakthrough that clarified the role played by RANKL in osteoclast biology [15].
RANKL-RANK signaling is not only related to bone physiology but also has essential roles in maturation and activation of the immune system. RANKL-RANK system is also functionally involved in lymph node organogenesis, development of thymic medullary epithelial cells [16]–[20], central thermoregulation [21], development of a lactating mammary gland during pregnancy [22], promotes dendritic cell survival [2] and normal development of Peyer's patches. Recently, Knoop et al. showed that RANKL is the critical factor that controls the differentiation of M cells from RANK-expressing intestinal epithelial precursor cells. As a consequence, the RANKL null mice failed to develop M cells in their Peyer's patches which could be restored by systemic administration of exogenous RANKL [23].
Despite the diverse roles of RANKL in various cells, the physiological roles of RANKL are mainly studied using extracellular form due to its soluble nature. So, it is necessary to conduct experimental studies in order to shed light on the function of transmembrane RANKL. But, large quantities of functional proteins are required for biophysical characterization and other studies. Because the use of mammalian expression systems is usually expensive, time-consuming and difficult to scale up while providing optimal conditions for the expression of mammalian proteins themselves, E. coli can be used as a feasible alternative host to eukaryotic cells for the overexpression of mammalian proteins. The use of E. coli expression systems for heterologous production of protein by recombinant DNA technology has long been established. E. coli overexpression system provides the advantages of inexpensiveness, fast growth, straight forward genetics, a large number of mutant host strains and expression vectors, scalability and high expression [24]–[26].
However, bacteria and mammals prefer to use different codons and thus, biased codon usage is one of the major factors affecting the heterologous expression of mammalian protein in E. coli [27]. The problem of codon bias can be resolved by codon optimization, a genetic technique which involves the replacement of existing rare codons of a species with a set of more favorable host codons throughout the whole gene to achieve optimum expression of a foreign gene in a host's cellular system [27]–[29]. Accordingly, synthesis of the target gene is often faster and cheaper to get the codon-optimized gene. Gene synthesis offers the additional benefit that most gene optimization algorithms optimize not only rare codons but also mRNA secondary structure, the latter affects the translation efficiency [30].
Considerably, protein expression is influenced by multiple parameters including selection of vector with an appropriate promoter, fusion tag, expression host strain and the expression conditions such as temperature, concentration of inducer, induction time and composition of the culture medium. These parameters can be optimized to improve the yield of expressed proteins [31]. Optimization of protein expression and production can be achieved by a conventional one-factor-at-a-time approach [32]. In this method, optimization is usually done by varying a single factor, while keeping all the other factors fixed at a given set of conditions. But, this method is not only time consuming, but also incapable of achieving the true optimal conditions because the method ignores the interactions between the influencing factors [33]. Alternatively, response surface methodology (RSM) can be used to determine the individual role of each factor as well as their influences among the factors. RSM is a mathematical and statistical tool for designing experiments, building models, evaluating the effects of several factors, and achieving the optimum conditions for desirable responses with a limited number of experiments [34].
In this study, we utilized codon optimization strategy and response surface methodology to assess the heterologous production of both transmembrane and extracellular forms of RANKL as soluble proteins in E. coli. We optimized and evaluated the various important parameters for the production of these proteins. Finally, we analyzed and compared the functional properties of these proteins by tartrate resistant acid phosphatase (TRAP) assay and quantitative real-time polymerase chain reaction (qRT-PCR).
Materials and Methods
Reagents
Restriction enzymes were purchased from Takara (Shiga, Japan). All other chemicals used were purchased from Sigma- Aldrich (St. Louis, MO, USA) unless otherwise stated.
Bacterial strains, vectors and media
Escherichia coli (E. coli) DH5α (Invitrogen, USA) was used for DNA manipulation. For preliminary screening experiments, E. coli BL21 (DE3) (Stratagene, USA), E. coli BL21 (Stratagene, USA), TOP10F'E. coli (Invitrogen, USA) and SHuffle Express E.coli (New England Biolabs, UK) cells were tested for expression of proteins. To optimize the production of both forms of RANKL, SHuffle Express E. coli was used as expression host. All E. coli strains were grown at 37°C in Luria-Bertani (LB) Broth (Becton, Dickinson and Company, USA) or LB agar plate supplemented with ampicillin antibiotic (100 µg/ml) when required. While pGEM-T Easy vector (Promega, USA) was used for the cloning of PCR products, pMAL-c5X vector (New England Biolabs, UK), pET32a (+) vector (Novagen, USA), and pGEX-5X-1 (GE Healthcare, UK) were used as expression vectors.
Cell line and culture condition
The mouse macrophage cell line RAW 264.7, a well-established osteoclastogenic cell system that differentiate into TRAP-positive functional osteoclasts when co-cultured with RANKL [35], was purchased from American Type Culture Collection (ATCC, USA). RAW 264.7 cells were maintained in Dulbecco's modified Eagle's medium (Thermo Scientific HyClone, USA) supplemented with 10% fetal bovine serum (Thermo Scientific HyClone, USA) and antibiotics (100 U/ml penicillin G and 100 µg/ml streptomycin) at 37°C with 5% CO2.
Computational codon optimization, synthetic gene construction and amplification
The full-length mRNA sequence (951 bp) of transmembrane RANKL of Mus musculus (GenBank accession no. AF013170.1) was taken for codon optimization by DNAWorks (v3.2.2) software [36] (Table S1). Expected codon adaptation index (CAI) values of wild type and codon optimized mRANKL were calculated from E-CAI server (http://genomes.urv.es/CAIcal), using a predefined reference set of highly expressed E. coli genes. The codon optimized transmembrane RANKL gene (mRANKL), flanked by NdeI and SalI restriction sites, was synthesized and provided as an NdeI/SalI insert in pUCIDT vector by Mbiotech (Gyeonggi-Do, Korea). The synthetic mRANKL was excised by digesting with NdeI/SalI, purified with gel extraction kit (NucleoGen, Korea) and then ligated downstream of malE gene encoding maltose binding protein (MBP) in pMAL-c5X expression vector using T4 DNA ligase (Takara, Japan) to obtain recombinant plasmid pOmR-c5X. The ligation products were transformed into E. coli DH5α competent cells (Invitrogen, USA) for plasmid amplification and selected on LB agar plates with 100 µg/ml ampicillin. The plasmid was extracted using DNA purification kit (NucleoGen, Korea) and the ligation was confirmed by restriction enzyme digestion and DNA sequencing.
Cloning and construction of expression vectors
Polymerase chain reaction (PCR) was used to amplify mRANKL or optimized extracellular RANKL gene (RANKL-Ex) encoding for the extracellular domain (137–316 region) of full length mRANKL. PCR was performed in a Takara PCR thermal cycler (Takara, Japan) with a set of primers (Table 1) using AccuPower PCR PreMix containing Top DNA Polymerase (Bioneer, Korea) according to the manufacturer's instructions. PCR reactions were carried out in a total volume of 20 µl with 50 ng plasmid DNA and 10 pmole of each primer under the following conditions: 3 min denaturation at 94°C followed by 30 cycles of extension (30 s at 94°C, 30 s at 52°C, then 1 min at 72°C) and final extension of 7 min at 72°C. PCR products were purified with gel extraction kit and ligated into pGEM-T Easy vector using T4 DNA Ligase (Promega, USA). The ligation products were transformed into E. coli DH5α and selected on LB agar plates with 100 µg/ml ampicillin. The ligation was confirmed by plasmid DNA isolation, restriction enzyme digestion, and DNA sequencing.
Table 1. Primers used in this study.
| Gene | GenBank ID | Sequences |
| RANKL-Ex | - | Foreward: 5′GTGCATATGCAACGTTTCTCTGGTGCT 3′ Reverse:5′CCGGTCGACGTCGATGTCCTGAACTTT 3′ |
| mRANKL | - | Foreward:5′ TATCCCGGGATGCGTCGTGCGAGCCGC 3′ Reverse:5′CCGCTCGAGGTCGATGTCCTGAACTTT 3′ |
| Trap | NM_007388 | Foreward: 5′ GCGACCATTGTTAGCCACATACGG 3 Reverse: 5′ CGCCCAGGGAGTCCTCAGATCCAT 3′ |
| CalcR | NM_001042725 | Foreward: 5′ TGGTGCGGCGGGATCCTATAAGTT 3′ Reverse: 5′ GTGGATAATGGTTGGCACTATCGG 3′ |
| Cfms | NM_001037859 | Foreward: 5′ TGGATGCCTGTGAATGGCTCTGAT 3′ Reverse: 5′ ATGGGCAGCTGGCTCTGAATGATC 3′ |
| Ctsk | NM_007802 | Foreward:5′ GAAATCTCTCGGCGTTTAATTTGGGAG 3′ Reverse: 5′ GGGGTATAGAGAGTGTCATTACTGTAG 3′ |
| Nfatc1 | NM_001164109.1 | Foreward: 5′ TCCTGCTCCTCCTCCTGCTGCTCG 3′ Reverse: 5′ GCTGCTGGCAAGGCAGAGTGTGCT 3′ |
| Gapdh | XM_132897.1 | Foreward: 5′ AACTTTGGCATTGTGGAAGGGCTC 3′ Reverse: 5′ AAGGCCATGCCAGTGAGCTTC 3′ |
Further, mRANKL ligated into pGEM-T Easy vector was excised with XmaI-XhoI and inserted into pGEX-5X-1 to obtain pOmR-G5X. The plasmid pOmR-c5X harboring mRANKL was amplified, cut and ligated into NdeI- SalI sites of pET32a (+) to obtain pOmR-32. Likewise, RANKL-Ex was excised from its pGEM-T vector with NdeI/SalI, and then ligated into pMAL-c5X to generate pOsREx-c5X. The ligation products were transformed into E. coli DH5α for plasmid amplification.
Protein expression, isolation and analysis
To optimize the expression of protein in E. coli, different expression hosts were transformed with the recombinant expression plasmids (Table 2). The transformed single colony was inoculated in 4 ml of LB medium supplemented with 100 µg/ml of ampicillin and incubated overnight at 37°C. 500 µl of overnight culture was used to inoculate 100 ml of the same medium and incubated at 37°C. When the culture reached an OD600 of 0.5–0.7, expression of protein was induced by either 0.4 mM IPTG or 20 mM lactose and then incubated at 30 °C for 6 h. The cells were harvested by centrifugation at 3,000 rpm for 10 min, washed twice with ice-cold PBS buffer and resuspended in 2 ml of following buffers related to protein-tagged system such as column buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, pH 7.4) for MBP-tagged system, His6-binding buffer (20 mM Tris-HCl, 5 mM imidazole, 0.5 mM NaCl, pH 7.9) for His6-tagged system and GST-binding buffer (10 mM NaH2PO4, 1.8 mM KH2PO4, 2.7 mM KCl, 140 mM NaCl, pH 7.3) for GST-tagged system, followed by 10 min incubation on ice. The cells were then disrupted by sonication (Vibra Cell; Sonics & Materials, Newtown, USA) in a cycle of 9 s pulses and 4 s standbys for a total of 8 min in an ice bath. The lysate was cleared by centrifugation (12,000×g) for 30 min at 4°C. Expression of protein was monitored in 4–20% SDS gel (Komabiotech, Korea) using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Table 2. Bacterial strains and plasmids used in this study.
| E. coli host | Vector system | Recombinant plasmid | Expression system | Induction | Expression of mRANKL |
| E. coli BL21 (DE3) | pMAL-c5X | pOmR-c5X | E. coli BL21 (DE3)- pOmR-c5X | IPTG/Lactose | Too low |
| E. coli BL21 | pMAL-c5X | pOmR-c5X | E. coli BL21- pOmR-c5X | IPTG/Lactose | Too low |
| TOP10F'E. coli | pMAL-c5X | pOmR-c5X | TOP10F'E. coli-pOmR-c5X | IPTG/Lactose | Too low |
| Shuffle Express E. coli | pMAL-c5X | pOmR-c5X | Shuffle E. coli-pOmR-c5X | IPTG | Too low |
| Shuffle Express E. coli | pMAL-c5X | pOmR-c5X | Shuffle E. coli-pOmR-c5X | Lactose | Significant |
| E. coli BL21 (DE3) | pET32a(+) | pOmR-32 | E. coli BL21 (DE3)- pOmR-32 | IPTG/Lactose | ND |
| E. coli BL21 | pET32a(+) | pOmR-32 | E. coli BL21- pOmR-32 | IPTG/Lactose | ND |
| TOP10F'E. coli | pET32a(+) | pOmR-32 | TOP10F' E. coli-pOmR-32 | IPTG/Lactose | ND |
| Shuffle Express E. coli | pET32a(+) | pOmR-32 | Shuffle E. coli-pOmR-32 | IPTG/Lactose | ND |
| E. coli BL21 (DE3) | pGEX-5X-1 | pOmR-G5X | E. coli BL21 (DE3)- pOmR-G5X | IPTG/Lactose | ND |
| E. coli BL21 | pGEX-5X-1 | pOmR-G5X | E. coli BL21- pOmR-G5X | IPTG | Negligible |
| E. coli BL21 | pGEX-5X-1 | pOmR-G5X | E. coli BL21- pOmR-G5X | Lactose | ND |
| TOP10F'E. coli | pGEX-5X-1 | pOmR-G5X | TOP10F' E. coli-pOmR-G5X | IPTG/Lactose | ND |
| Shuffle Express E. coli | pGEX-5X-1 | pOmR-G5X | Shuffle E. coli-pOmR-G5X | IPTG/Lactose | ND |
| Shuffle Express E. coli | pMAL-c5X | pOsREx-c5X | Shuffle E. coli-pOsREx-c5X | Lactose | Significant |
Response surface methodology for optimization of protein expression
Central composite design (CCD) and response surface methodology (RSM) were employed to optimize the culture conditions for the production of maltose binding protein-tagged mRANKL in SHuffle Express E.coli. A 24 full factorial central composite rotary design for four independent variables with replicates at the center point and star points were used in this study. The variables; optical density before induction (OD600) (0.5–0.7), lactose concentration (5–10 mM), post-induction temperature (22–30°C) and post-induction time (4–6 h) were tested using a statistical analysis and RSM each at five coded levels (−α, −1, 0, +1, +α) as shown in Table 3. The central composite design at the given range of the above mentioned parameters in terms of codes is shown in Table 4. A total of 30 experimental trials including 16 trials for factorial design, eight trials for axial points and six trials for replication of the central points were performed. The experimental data obtained were analyzed statistically by regression analyses using the following second-order polynomial equation:
| (1) |
where Y is the predicted response (mRANKL (mg/L)) used as a dependent variable; n is the number of independent variables (factors), xi (i = 1, 2) is the input independent variable (factors); β0, a constant coefficient, is the value of the fixed response at the center point of the design, and βi, βij and βii are the coefficients of linear, interaction and quadratic regression terms, respectively. The statistical software package, Design-Expert 8.0.7.1 (Stat-Ease, Inc., Minneapolis, USA) was used for regression analysis of experimental data and to plot the 3D surface response models. The statistical analysis of the model was represented in the form of analysis of variance (ANOVA) and the optimal points of the variables were obtained by “point optimization” tool of Design Expert.
Table 3. The independent variables and coded values of variables used in central composite rotary design.
| Independent variables | Units | Levels | ||||
| -α | -1 | 0 | 1 | α | ||
| Cell density (OD600) | - | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 |
| Lactose | mM | 2.5 | 5 | 7.5 | 10 | 12.5 |
| Temperature | °C | 18 | 22 | 26 | 30 | 34 |
| Induction time | h | 3 | 4 | 5 | 6 | 7 |
Table 4. CCD with measured and predicted response of mRANKL production.
| Run | A OD600 | B Lactose (mM) | C Temperature (°C) | D Induction time (h) | Response mRANKL (mg/L) | |
| Predicted value | Actual value | |||||
| 1 | −1 | −1 | −1 | −1 | 16.7 | 15.9 |
| 2 | 1 | −1 | −1 | −1 | 17.3 | 16.3 |
| 3 | −1 | 1 | −1 | −1 | 16.8 | 17.5 |
| 4 | 1 | 1 | −1 | −1 | 26.8 | 27.9 |
| 5 | −1 | −1 | 1 | −1 | 26.1 | 26.8 |
| 6 | 1 | −1 | 1 | −1 | 16.1 | 19.2 |
| 7 | −1 | 1 | 1 | −1 | 24.7 | 23.1 |
| 8 | 1 | 1 | 1 | −1 | 23.9 | 23.9 |
| 9 | −1 | −1 | −1 | 1 | 39.7 | 40.5 |
| 10 | 1 | −1 | −1 | 1 | 23.7 | 27.4 |
| 11 | −1 | 1 | −1 | 1 | 37.8 | 36.8 |
| 12 | 1 | 1 | −1 | 1 | 31.1 | 31.3 |
| 13 | −1 | −1 | 1 | 1 | 41.8 | 42.8 |
| 14 | 1 | −1 | 1 | 1 | 15.1 | 15.3 |
| 15 | −1 | 1 | 1 | 1 | 38.3 | 40.2 |
| 16 | 1 | 1 | 1 | 1 | 20.9 | 23.8 |
| 17 | −2 | 0 | 0 | 0 | 33.5 | 34.3 |
| 18 | 2 | 0 | 0 | 0 | 16.8 | 13.3 |
| 19 | 0 | −2 | 0 | 0 | 39.1 | 36.9 |
| 20 | 0 | 2 | 0 | 0 | 45.1 | 44.6 |
| 21 | 0 | 0 | −2 | 0 | 11.5 | 11.3 |
| 22 | 0 | 0 | 2 | 0 | 10.8 | 8.3 |
| 23 | 0 | 0 | 0 | −2 | 15.7 | 16.2 |
| 24 | 0 | 0 | 0 | 2 | 35.8 | 32.6 |
| 25 | 0 | 0 | 0 | 0 | 49.5 | 49.7 |
| 26 | 0 | 0 | 0 | 0 | 49.5 | 50.6 |
| 27 | 0 | 0 | 0 | 0 | 49.5 | 48.7 |
| 28 | 0 | 0 | 0 | 0 | 49.5 | 47.8 |
| 29 | 0 | 0 | 0 | 0 | 49.5 | 48.9 |
| 30 | 0 | 0 | 0 | 0 | 49.5 | 51.6 |
Purification of soluble proteins and molecular weight determination
The soluble proteins were purified by amylose affinity chromatography according to the manufacturer's instructions. Briefly, crude protein extract in column buffer was loaded in 2 ml of amylose resin (New England Biolabs, UK) and washed with 12 column volumes of column buffer, and the proteins were eluted with 10 mM maltose in column buffer. Elution fractions were analyzed by SDS–PAGE followed by staining with Coomassie Brilliant Blue R-250. The purified MBP-tagged proteins were dialyzed against 20 mM Tris-HCl, 25 mM NaCl buffer (pH 7.4) at 4°C for 24 h with three buffer changes. Endotoxins were removed by Detoxi-gel endotoxin removing columns (Thermo Scientific Pierce, USA) according to the manufacturer's instructions. Protein concentrations were determined by measuring the absorbance at 280 nm using Nanophotometer (Implen GmbH, Germany).
Separation of RANKL from its MBP fusion partner was accomplished by proteolytic cleavage with Factor Xa (Amersham Biosciences, UK), and the reaction was performed overnight at 4°C in 50 mM Tris-HCl, 25 mM NaCl, 1 mM EDTA (pH 7.4). Uncleaved fusion protein and MBP were removed by second passage through amylose resin as described before. The protein was further purified by gel filtration chromatography using Superdex 20010/300 GL column (GE Healthcare, UK) equilibrated with buffer containing 20 mM Tris-HCl (pH 7.4). The column was connected to an AKTA explorer 100 fast protein liquid chromatography apparatus (Amersham Biosciences, UK) and eluted with the same buffer. Purified protein was run in gel electrophoresis under native condition using Tris-Glycine-PAG Pre-Cast Gel, non-SDS, 4–20% (Komabiotech, Korea) for mass estimation of native membrane protein. For molecular mass determination, 5 mg/ml of purified protein was applied to a Superdex 200 10/300 GL column. The column was calibrated with gel filtration molecular weight markers kit (MWGF200, Sigma). The gel-phase distribution coefficient (Kav) was calculated as
where Ve is elution volume, Vo is void volume and Vc is column volume.
Western blot analysis for confirmation of protein expression
Heterologous expression of the proteins was confirmed by western blot analysis. Briefly, the proteins were separated under reducing conditions in a 4–20% SDS gel using XCell SureLock Mini-Cell (Life Technologies, USA) at 130 V for 2 h. Precision plus protein dual-color standards (BioRad, USA) were used as molecular weight marker. After electrophoresis, the proteins were electro-transferred to nitrocellulose membranes (Protran nitrocellulose membrane, Whatman, UK) at 10 V for 60 min, using Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad, USA). The membrane was blocked with 5% skim milk in Tris-buffered saline-Tween (TBST) buffer for 60 min at room temperature and then washed three times with TBS-Tween. The membrane was then incubated with an antibody against RANKL (R&D Systems, USA) on a shaker for overnight at 4 °C, followed by washing with TBST buffer three times for 15 minutes each. The membrane was then incubated with goat IgG horseradish peroxidase (HRP)-conjugated antibody (R&D Systems, USA) in TBST buffer for 1 hour at room temperature. After three successive washing with TBST buffer for 15 minutes each, the proteins were detected by enhanced chemiluminescence (ECL) detection system (GE Healthcare, UK) and exposed to Gel Doc XR system (BioRad, USA) to capture chemiluminescent signal on the western blot.
Osteoclast differentiation assay
RAW 264.7 cells were seeded on 6-well plates at a density of 2×104 cells per well in the presence of different concentration of mRANKL (30–100 ng/ml) or RANKL-Ex (100 ng/ml) for 6 days. The culture medium was replaced with fresh medium containing above mentioned samples every 48 h over the course of 6 days. The cultured cells were then subjected to TRAP-staining (B-Bridge, USA) according to the manufacturer's instructions to confirm the generation of TRAP-positive osteoclast-like cells. Briefly, the cells were washed with PBS and fixed with the fixative reagent for 5 min at room temperature. The cells were washed three times with distilled water and then stained with the chromogenic substrate for 60 min at 37°C and finally washed with distilled water to stop the reaction when optimum color was achieved. TRAP-positive osteoclasts were visualized by light microscopy and photographed. Untreated cells were used as controls. Each osteoclast differentiation assay was performed at least 3 times. In order to quantify the TRAP activity, 30 µL of culture supernatant was incubated at 37°C for 3 h in the presence of 170 µL of the chromogenic substrate diluted in tartrate-containing buffer (3 mg/5 mL). The absorbance was measured at 540 nm using Infinite 200 PRO multimode reader (Tecan, Switzerland).
RNA extraction, reverse transcription and real-time PCR analyses
To determine the effect of RANKL in osteoclast differentiation, RAW 264.7 cells were treated with 100 ng/ml of mRANKL or RANKL-Ex. The level of transcription of the associated genes during osteoclast differentiation was then analyzed by isolating total RNAs from the RANKL-treated cells using Trizol reagent (Life Technologies, USA) following the manufacturer's instructions. Briefly, the cells were homogenized in 1 ml Trizol. Samples were mixed with 200 ml chloroform and then centrifuged 12,000×g for 15 minutes at 4°C. The upper aqueous phase was transferred carefully into fresh tube without disturbing the interphase and equal volume of isopropanol was added in the tube. Mixtures were thoroughly resuspended and centrifuged at 12,000×g for 10 minutes at 4°C. The precipitated RNA pellets were washed with 1 ml ethanol (75%, v/v). RNA pellets were recovered after centrifugation at 12,000×g for 5 min at 4°C. RNA samples were allowed to air-dry for 2–3 min and then resuspended in 50 µl diethyl pyrocarbonate-treated water (Life Technologies, USA). RNA was further purified using a Qiagen RNeasy mini kit and Rnase-free DNAse set (Qiagen, Germany) according to the manufacturer's specifications. RNA was quantified using Nanophotometer. The equal amount of total RNA (1 µg) from each sample was reverse-transcribed to cDNA at 42°C for 30 minutes in a final volume of 20 µl, using Quantitect reverse transcription kit (Qiagen, Germany) according to the manufacturer's protocol. Each cDNA was stored at −20°C until use.
qRT-PCR was conducted using TOPreal qPCR 2X PreMIX (SYBR Green) (Enzynomics, Korea) in a total reaction volume of 20 µl. cDNA templates and primers were added to SYBR Green containing nTaq-HOT DNA polymerase, dNTP mixture and SYBR-Green I. Real-time PCR was then performed using MyiQTM single color real-time PCR detection system (Bio-Rad) under following conditions: 30 s at 95°C, followed by 40 cycles at 95°C for 10 s, 60°C for 30 s; then 72°C for 1 min and followed by a dissociation stage (95°C for 15 s). A melting curve was plotted to ensure the specificity of amplification products. The 2−ΔΔCt method [37] was used to analyze the relative changes in the level of target gene transcription. The primers used for qRT-PCR are shown in Table 1.
Results
Codon-optimization and cloning of mRANKL
Different organisms use synonymous codons with different preferences. As a consequence, heterologous proteins, especially human proteins, fail to express in E. coli due to the presence of “rare” codons in the target mRNA that are infrequently used by E. coli. This includes the codons for arginine (AGA, AGG, CGA), isoleucine (AUA), leucine (CUA), and proline (CCC). One can improve the expression of heterologous proteins significantly in E. coli by selecting the synonymous codons that are favored by the E. coli host. The full-length sequence of mRANKL of Mus musculus possesses 19 rare codons including 11 codons for arginine (AGA, AGG, CGA), 3 codons for isoleucine (AUA), 2 codons for leucine (CUA), and 3 codons for proline (CCC) as predicted by the rare codon calculator, RaCC (http://nihserver.mbi.ucla.edu/RACC/). DNAWorks (v3.2.2) was used to modify 198 of the 316 codons with synonymous codons predicted to occur frequently in highly expressed genes in E. coli. Thus, mRANKL was codon optimized while maintaining the integrity of the native amino acid structure (Table S1). CAI was used to estimate the adaptation of codon optimized mRANKL to host codons. CAI is a measurement of the relative adaptiveness of the codon usage of a gene towards the codon usage of highly expressed genes in the host. The codon optimization of mRANKL increased the CAI value [38] from 0.64 (wild type) to 0.76 (codon optimized) indicating that the optimized gene was closer to the highly expressed genes in E. coli. Finally, mRANKL gene was synthesized and cloned into different expression vectors as described in the materials and method section.
Expression of mRANKL in E. coli
On the basis of ‘trial and error’ approach, three vector systems, pET32a(+), pGex-5X-1 and pMAL-c5X with three different solubility tags, i.e., hexahistidine (His6), glutathione S-transferase (GST) and maltose-binding protein (MBP), respectively, and four E. coli hosts including E. coli BL21 (DE3), E. coli BL21, TOP10F' E. coli and SHuffle Express E. coli were selected to test the heterologous expression of mRANKL. Accordingly, E. coli MBP-mRANKL, E. coli His6-mRANKL and E. coli GST-mRANKL expression systems were generated and recombinant RANKL was extracted as MBP-mRANKL, His6-mRANKL and GST-mRANKL from the respective system with IPTG or lactose as inducer. The expression of mRANKL in these systems was monitored by SDS-PAGE. There was no visible expression of mRANKL in SDS gel from all E. coli hosts with His6-tagged expression system and three E. coli hosts with GST-tagged system in all conditions tested. A negligible amount of soluble GST-mRANKL was obtained in E. coli BL21 when IPTG was used as inducer (data not shown). While three E. coli hosts with MBP-tagged system gave too low yield of mRANKL in all conditions tested, the significant amount of mRANKL was produced in SHuffle E.coli-pOmR-c5X where the high level expression of mRANKL was obtained in inclusion bodies when lactose was used as inducer (data not shown). These preliminary experiments showed that the highest protein yield was obtained from SHuffle E.coli-pOmR-c5X among the systems tested and thus, this system was further employed to optimize the production of soluble mRANKL in E. coli.
Optimization of mRANKL expression using response surface methodology
Besides appropriate expression vector, fusion tag and expression host, there are multiple parameters that can be varied when optimizing an expression of target protein in E. coli, with each parameter affecting the solubility and activity of the protein. Due to several parameters, RSM was applied to determine optimal conditions for the production of mRANKL from E. coli. RSM is a statistical method useful for analyzing the effect of several independent variables influencing the responses by varying them simultaneously with limited number of experiments. A central composite design (CCD), an efficient design tool for fitting second-order models under RSM, was used to investigate the effect of four most important variables such as cell density before induction, lactose concentration, post-induction temperature and post-induction time that influence the heterologous production of most recombinant proteins. In the optimization procedure, the response of the statistically designed combinations was determined, the coefficients by fitting the experimental data to the response functions were estimated, the response of the fitted model was predicted and the adequacy of the model was verified using software Design-Expert (version 8.0.1). A list of independent variables and coded factor levels are given in Table 3. A 24 full factorial CDD design matrix and the predicted and experimental results of mRANKL production are given in Table 4. A total of 30 experiments were conducted for four factors at five levels to optimize the production of mRANKL. The number of experiments required (N) is predicted by the expression: 2k (24 = 16; star points) +2 k (2×4 = 8; axial points) +6 (center points; 6 replications). Thirty observed responses were used to compute the model using the least square method. The response (mRANKL (mg/L) was correlated with the four different factors, cell density, lactose concentration, post-induction temperature and post-induction time, as a function of the second-order polynomial equation, as represented by Eq. (1). From the experimental data, following quadratic regression model was obtained in coded form:
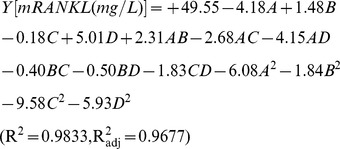 |
(2) |
where Y is the response (mRANKL production), and A, B, C and D are the coded terms for the four variables i.e. cell density before induction, lactose concentration, post-induction temperature and post-induction time respectively. The coefficients in front of A, B, C or D represent the effects of that particular factor, while the coefficients in front of AB, AC, AD, BC, BD or CD and those in front of A2, B2, C2 or D2 represent the interaction between the two factors and the quadratic effects, respectively. The positive sign indicates a synergistic effect, while the negative sign indicates an antagonistic effect.
Different combinations of four variables yielded MBP-tagged mRANKL production ranging from 8.3 to 51.6 mg/L as illustrated in Table 4. The maximum production of fusion mRANKL (51.6 mg/L) was obtained at central point values with OD600: 0.6, lactose concentration: 7.5 mM, post-induction temperature: 26°C and post-induction time: 5 h, respectively and the lowest production was found at OD600: 0.6, lactose concentration: 7.5 mM, post-induction temperature: 34°C and post-induction time: 5 h, respectively.
Validation of the model
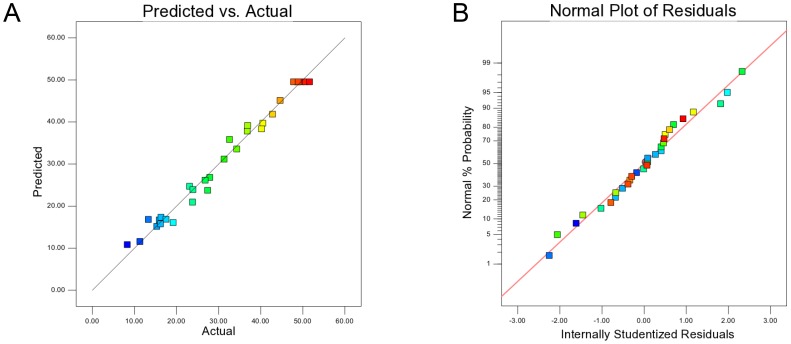
The results of analysis of variance (ANOVA) values for the quadratic regression model obtained from CCD employed in the optimization of mRANKL production are given in Table 5. The statistical significance of a quadratic model was tested through F- and p-values for analysis of variance. The large F-value indicates that most of the variation can be explained by a regression equation whereas a low p-value (<0.05) indicates that the model is considered to be statistically significant [34]. Thus, the high F-value (63.23) and very low probability of p > F value (0.0001) obtained revealed that the regression is statistically significant. Moreover, the results of the lack of fit test for the models showed that the lack of fit is not statistically significant at 95% confidence level. The adequacy of quadratic model can be confirmed by the coefficient of determination R2 along with an acceptable agreement with the adjusted determination coefficient R2 adj. The ANOVA analysis reported the high R2 value of 0.9833 and R2 adj value of 0.9677 for mRANKL production, both of which are close to 1, ensuring a high correlation between the experimental values and the predicted values. The diagnostic plots used for estimating the adequacy of the regression model are shown in Figure 1. From the correlation between the actual and the predicted values of mRANKL production, it is evident that there are tendencies in the linear regression fit, and the model adequately explains the experimental range studied (Figure 1A). The actual value is the result obtained for a specific run and the predicted value is obtained from the independent variables in the CCD model. The data points in the normal percentage probability and studentized residual plot indicated that neither response transformation was required nor there was any apparent problem with normality (Figure 1B). Thus, ANOVA results indicated the satisfactory adjustment of the quadratic models to the experimental data.
Table 5. ANOVA for response surface quadratic model for mRANKL production.
| Source | df | Sum of Squares | Mean Square | F- Value | p-value Prob > F |
| Model | 14 | 5218.066 | 372.719 | 63.23285 | <0.0001 |
| A-OD600 | 1 | 420.8438 | 420.8438 | 71.39735 | <0.0001 |
| B-Lactose | 1 | 53.10375 | 53.10375 | 9.009204 | 0.0089 |
| C-Temperature | 1 | 0.84375 | 0.84375 | 0.143145 | 0.7105 |
| D-Induction time | 1 | 603.0038 | 603.0038 | 102.3013 | <0.0001 |
| AB | 1 | 86.02563 | 86.02563 | 14.59449 | 0.0017 |
| AC | 1 | 115.0256 | 115.0256 | 19.51443 | 0.0005 |
| AD | 1 | 276.3906 | 276.3906 | 46.89046 | <0.0001 |
| BC | 1 | 2.640625 | 2.640625 | 0.44799 | 0.5135 |
| BD | 1 | 4.100625 | 4.100625 | 0.695683 | 0.4173 |
| CD | 1 | 53.65562 | 53.65562 | 9.102831 | 0.0087 |
| A∧2 | 1 | 1016.091 | 1016.091 | 172.3827 | <0.0001 |
| B∧2 | 1 | 93.7686 | 93.7686 | 15.90811 | 0.0012 |
| C∧2 | 1 | 2520.691 | 2520.691 | 427.6424 | <0.0001 |
| D∧2 | 1 | 966.625 | 966.625 | 163.9907 | <0.0001 |
| Residual | 15 | 88.41583 | 5.894389 | ||
| Lack of Fit | 10 | 78.88083 | 7.888083 | 4.136383 | 0.0653 |
| Pure Error | 5 | 9.535 | 1.907 |
A = OD600; B = Lactose concentration; C = Temperature; D = Induction time; df = Degrees of freedom.
Figure 1. Diagnostic plots for estimating the adequacy of the regression model.
Correlation between predicted and actual value for mRANKL production (A). The studentized and normal percentage probability plot of mRANKL production (B).
Interactive effect of process independent variables
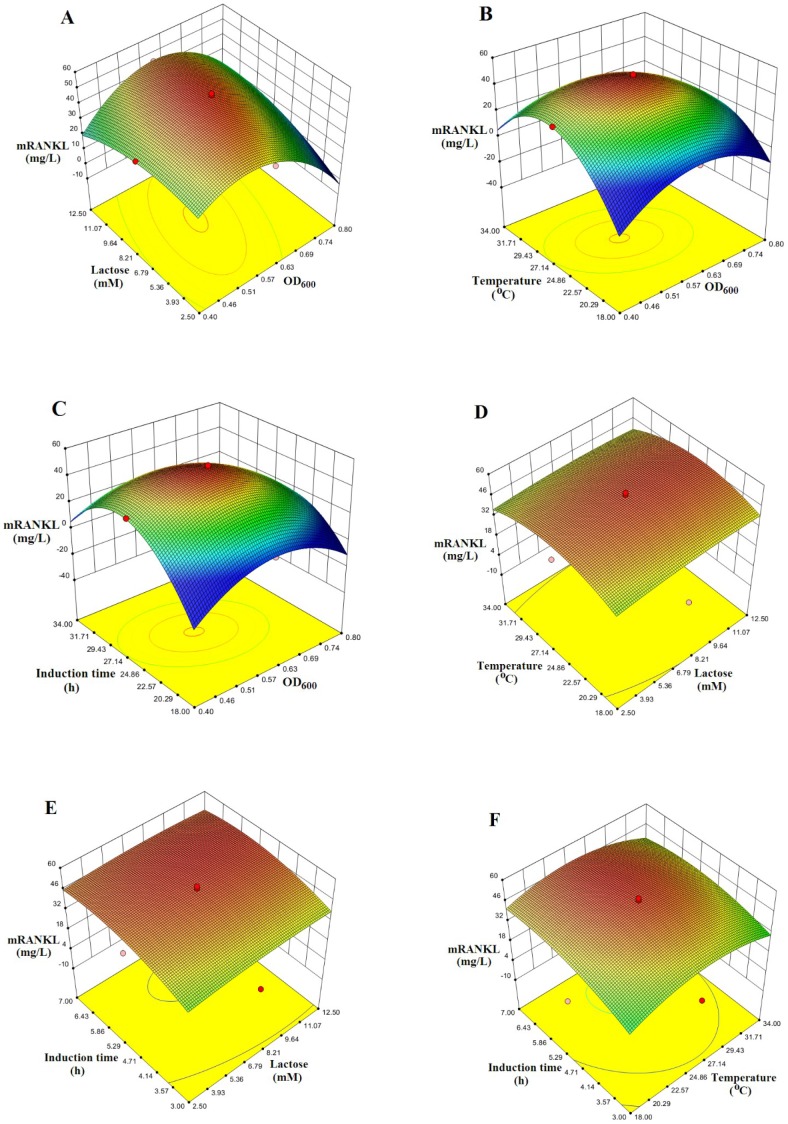
The interactive effects of variables on production of mRANKL were obtained in the form of three dimensional (3D) response surface plots as shown in Figure 2 (A-F). The response surface plots are the graphical representation of the regression equation used to visualize the relationship between the response and experimental levels of each factor. The integrated effect of lactose and cell density before induction on mRANKL production is shown in Figure 2A. The optimum conditions for mRANKL production was found to be at OD600 of 0.6 and lactose concentration of 7.5 mM. While, increased production of mRANKL was observed with increasing OD600, the lactose concentration showed the less effect on mRANKL production. However, an increase in OD600 beyond the optimum region resulted in a decreased production of mRANKL. Figure 2B indicates the production of mRANKL as a function of post-induction temperature and OD600. The production of mRANKL was increased with increasing temperature and OD600. At the temperatures higher than 30°C, the production of mRANKL began to decrease at all OD600. Figure 2C shows correlation of post-induction time and OD600 with mRANKL production. It can be observed that increase in mRANKL production occurred with both OD600 and post-induction time. An increase in both factors beyond the optimum region (OD600: 0.6 and induction time: 5 h) resulted in a decrease in mRANKL production. Figure 2D indicates the interactive influence of post-induction temperature and lactose concentration on the extent of mRANKL production. While the integrated impact of post-induction time and lactose concentration on mRANKL production is depicted in Figure 2E, the combined impact of induction time and post-induction temperature is shown in Figure 2F. Thus, these plots showed that OD600 lesser than 0.7, post-induction time of 5 h and lower post-induction temperature values were favorable for RANKL production, while concentration of lactose showed lower impact on mRANKL production.
Figure 2. Response surface plots to visualize the relationship between the response and experimental levels of each factor.
Three dimensional surface plots of combined effect of cell density (OD600) and lactose concentration (A), OD600 and post-induction temperature (B), OD600 and post-induction time (C), post-induction temperature and lactose concentration (D), post-induction time and lactose concentration (E) and post- induction temperature and post-induction time (F) on mRANKL production.
Confirmation experiments at optimum conditions
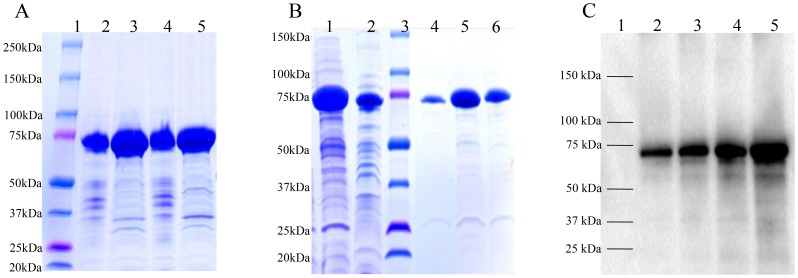
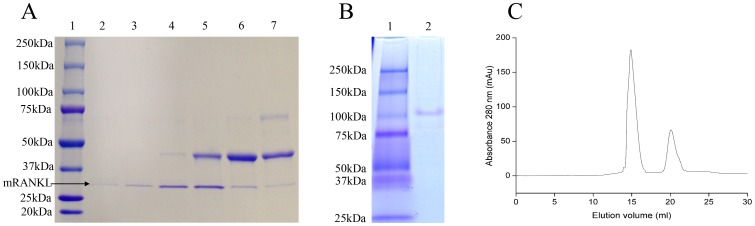
The agreements of the results obtained from the model and experiments were further confirmed by additional experiments by applying optimum conditions. The model predicted a maximum of 49.5 mg/L mRANKL production with a cell density before induction (OD600) of 0.6, lactose concentration of 7.5 mM, post-induction temperature of 26°C and post-induction time of 5 h. As shown in Table 6, mRANKL obtained from the additional experiments was found to be close to that predicted by the model. Experiments were performed in triplicate. The production of mRANKL was successfully scaled up from 100 ml to 1 L culture volume using these optimum conditions. Expression of fusion mRANKL (∼75 kDa) was assessed by SDS-PAGE (Figure 3A). The fusion protein was purified by amylose affinity chromatography (Figure 3B), and then confirmed by western blot analysis (Figure 3C). Finally, mRANKL was separated from MBP fusion partner and purified by affinity chromatography and gel filtration chromatography (Figure 4A).
Table 6. Confirmation experiments for mRANKL production.
| Expression condition | mRANKL (mg/L) | ||||
| OD600 | Lactose (mM) | Temperature (°C) | Induction time (h) | ||
| Predicted value | 0.6 | 7.5 | 26 | 5 | 49.5 |
| Experimental value | 0.6 | 7.5 | 26 | 5 | 52.4 |
Figure 3. SDS-PAGE and Western blot analyses of mRANKL fusion proteins.
A: Comassie blue stained SDS gel showing lysates from two SHuffle E. coli-pOmR-c5X colonies expressing the mRANKL fusion proteins (∼75 kDa). Lane 1: Protein standards, molecular masses are indicated in kilodaltons (kDa); Lanes 2 and 3: Insoluble and soluble fractions from clone 1, respectively; Lanes 4 and 5: Insoluble and soluble fractions from clone 2, respectively. B: Comassie blue stained SDS gel showing purification of mRANKL using amylose resin. Lane 1: crude fusion mRANKL; Lane 2: Flow through; Lane 3: Protein standards, molecular masses are indicated in kilodaltons (kDa); Lanes 4–6: Elution fractions. C: Western blot analysis of mRANKL. Different amounts of purified fusion mRANKL was run in 4–20% SDS gel and transferred onto a nitrocellulose membrane. Detection was performed with anti-RANKL primary antibody, goat IgG HRP-conjugated secondary antibody and chemiluminescent substrate. Lane 1: Protein marker; Lanes 2–5: 30, 50, 100, and 200 ng of purified fusion mRANKL protein respectively.
Figure 4. Purification of mRANKL by affinity chromatography and gel filtration chromatography.
A.SDS-PAGE analysis of purified mRANKL. The fusion mRANKL was cleaved by Factor Xa, purified by affinity chromatography followed by gel filtration chromatography. Eluted proteins fractions (Lane 2–7) were checked by SDS-PAGE gel. Lane 1: Protein standards, molecular masses are indicated in kilodaltons (kDa). B. Native PAGE analysis. PAGE analysis demonstrated the native state of mRANKL (molecular weight of approximately 100 kDa), consistent with the size of a homotrimer. Lane 1: Protein standard; Lane 2: mRANKL. C. Gel filtration chromatography profile. The elution peaks at 14.90 ml and 20.09 ml correspond to estimated molecular masses of ∼97 and ∼44 kDa, based on the elution volumes of known molecular standards. The estimated molecular masses are close to the predicted molecular weight of trimeric form (∼104 kDa) and monomeric form of the protein (∼35 kDa). The y axis indicates the absorption at 280 nm in milliabsorbance units (mAU) and the x axis indicates the volume in milliliters (ml) passed over the column.
Optimization of RANKL-Ex expression using response surface methodology
The successful expression of mRANKL in soluble form in SHuffle Express E. coli prompted us to overexpress RANKL-Ex. For this, RANKL-Ex was amplified and cloned to construct recombinant expression vector pOsREx-c5X, and transformed into SHuffle Express E.coli. CCD and RSM were employed to determine the optimum conditions for overexpression of RANKL-Ex in E. coli. Cell density before induction, lactose concentration, post-induction temperature and post-induction time were chosen as four independent variables. The experimental results were analyzed using Design Expert 8.0.7.1 and the regression model was proposed.
A list of independent variables and coded factor levels are given in Table 3. Accordingly, a total of 30 experiments were conducted for four factors at five levels to optimize the production of RANKL-Ex as shown in Table S2. The second-order polynomial model that characterizes the relationship between RANKL-Ex production and variables, is represented by Eq. (3).
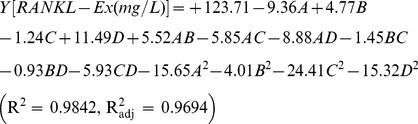 |
(3) |
where Y is the response (RANKL-Ex production), and A, B, C and D are the coded terms for the four variables, i.e., OD600 before induction, lactose concentration, post-induction temperature and post-induction time, respectively.
Different combinations of four variables yielded MBP-tagged RANKL-Ex production ranging from 19.9 to 128.7 mg/L as shown in Table S2. As in the case of fusion mRANKL, the maximum production of fusion RANKL-Ex (128.7 mg/L) was obtained at central point values with OD600: 0.6, lactose concentration: 7.5 mM, post-induction temperature: 26°C and post-induction time: 5 h, respectively, and the lowest production (19.9 mg/ml) was found at OD600: 0.6, lactose concentration: 7.5 mM, post-induction temperature: 34°C and post-induction time: 5 h, respectively, indicating that post-induction temperature above 30°C is not favorable for production of RANKL-Ex as well.
The results of analysis of variance (ANOVA) values for the quadratic regression model obtained from CCD employed in the optimization of RANKL-Ex production are given in Table S3. Thus, the high F-value (66.84), very low probability of p > F value (0.0001), R2 value of 0.9842 and R2 adj value of 0.9694 revealed the adequacy of quadratic model. The diagnostic plots used for estimating the adequacy of the regression model are shown in Figure S1 (A–B). The 3D response surface plots depicted in Figure S2 (A–F) represent the interactive effects of variables on production of RANKL-Ex within the experimental ranges. Additional confirmation experiments showed that the production of RANKL-Ex was found to be close to that predicted by the model under same conditions (Table S4). The production of RANKL-Ex was scaled up to 1 L culture media. Purified fusion RANKL-Ex (∼62 kDa) was assessed by SDS-PAGE and confirmed by western blot analysis as depicted in Figure S3 (A, B and C). Finally, RANKL-Ex was cleaved to remove its fusion partner and RANKL-Ex was further purified by affinity chromatography and gel filtration chromatography (data not shown).
mRANKL self-assembles into homotrimer
Based on the crystallographic studies that extracellular RANKL self-associates as a homotrimer (9), self-assembly of mRANKL was examined by native polyacrylamide gel electrophoresis (PAGE) and gel filtration chromatography. Proteins run on PAGE in the absence of SDS will separate on the basis of their native forms enabling for mass estimation of native membrane proteins. PAGE analysis of purified mRANKL under native conditions demonstrated that it migrated with a molecular weight of approximately 100 kDa, consistent with the size of a homotrimer (Figure 4B). Similarly, purified mRANKL was applied on to Superdex 200 10/300 GL column as described in materials and methods. The resulting chromatogram exhibited two major peaks at 14.9 ml and 20.0 ml of elution volume corresponding to calculated molecular weight of approximately 97 kDa and 44 kDa (Figure 4C), based on the elution volumes of known molecular standards. The calculated molecular weights are close to the predicted molecular weight of trimeric form (∼104 kDa) and monomeric form of mRANKL (∼35 kDa).
mRANKL induces osteoclast differentiation
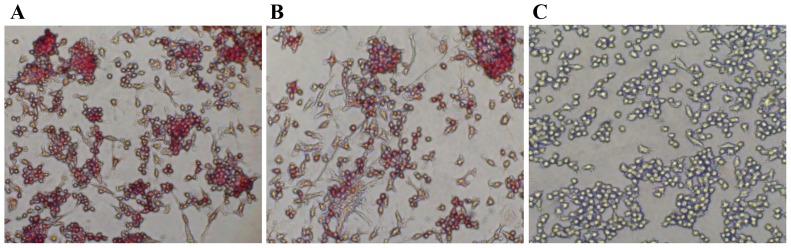
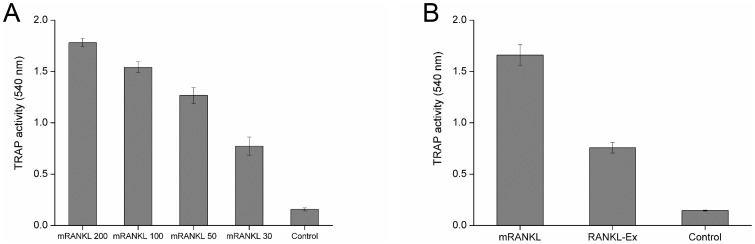
It has been well established that RANKL is the key cytokine that is essential for osteoclast differentiation/activation. RAW264.7 cell line is a model system that is being used extensively in in vitro osteoclast differentiation studies where osteoclasts are identified by TRAP assay. Since TRAP is expressed in high levels in the osteoclasts, it is used as a marker of osteoclast function [39]. Thus, to examine the potency of mRANKL to induce osteoclastogenesis in vitro, RAW264.7 cells were cultured at low cell density and treated with various concentrations of mRANKL for 6 days in order to promote cell differentiation. Cultures were then fixed and stained for TRAP activity as described in materials and method section. Untreated cultures are used as controls. After 6 days of culture, TRAP-positive osteoclasts were observed in the cultures treated with mRANKL when visualized under microscope (Figure 5). mRANKL at a concentration as low as 30 ng/ml was sufficient to induce TRAP release in RAW 264.7 cells. The TRAP activity increased significantly in the medium of RAW264.7 cells treated with mRANKL as compared to untreated cells. Moreover, mRANKL induced osteoclast differentiation in a concentration-dependent manner as evidenced by increasing TRAP formation in RAW264.7 cells (Figure 6A).
Figure 5. TRAP staining assay.
RAW 246.7 cells were treated with each protein at the concentration of 100/ml for 6 days and TRAP-positive osteoclasts were visualized under microscope. mRANKL (A); RANKL-Ex (B); Untreated RAW 246.7 cells (C).
Figure 6. Induction of osteoclastogenesis by RANKL in RAW264.7 cells.
TRAP activity was determined in the medium of RAW264.7 cells treated with mRANKL (30–200 ng/ml) for 6 days, numerical value with mRANKL denotes the concentration of mRANKL used (A); RAW264.7 cells were treated with 100 ng/ml of each mRANKL and RANKL-Ex for 6 days (B).
mRANKL upregulates osteoclast-associated genes
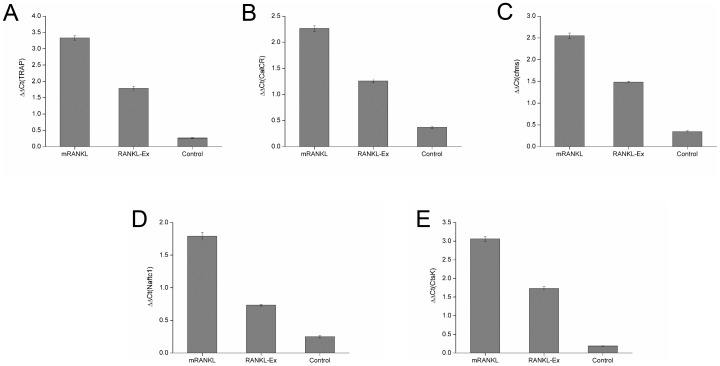
During osteoclastogenesis, osteoclasts express several marker genes, such as Trap, CalcR, Cfms, Nfatc1 and CtsK. These specific osteoclast genes are the key indicators of the differentiation of osteoclast precursor into osteoclasts. Therefore, the regulation of transcription of these genes, by the treatment of mRANKL in RAW 264.7 cells, were examined by qRT-PCR. When the transcription levels were quantitatively analyzed, Trap mRNA levels were approximately 15-fold higher in cells treated with mRANKL when compared to that of untreated cells (Figure 7A). Similarly, CalcR (Figure 7B) and Cfms (Figure 7C) mRNA expression levels were about 6-fold and 7.5-fold higher, respectively, in mRANKL treated cells. Likewise, Nfatc1 (Figure 7D) CtsK (Figure 7E) mRNA levels were approximately 6-fold and 14-fold higher, respectively, by the treatment of mRANKL as compared to that of untreated cells.
Figure 7. Quantitative RT-PCR analyses.
The expression of osteoclast differentiation (A–C) and functional markers (D, E) were studied at day 6 after mRANKL (100 ng/ml) and RANKL-Ex (100 ng/ml) exposure to RAW 246.7 cells. The results of the mRNA levels of genes of interest were normalized to GAPDH (control gene) and plotted. The data shown are representatives of three different experiments.
mRANKL is functionally superior to RANKL-Ex
To compare the functional difference between mRANKL and RANKL-Ex to induce osteoclastogenesis, RAW264.7 cells were treated with 100 ng/ml of mRANKL or RANKL-Ex for 6 days, and TRAP assay was performed as described in materials and method. It was found that secreted TRAP activity was approximately double in the medium of RAW264.7 cells treated with mRANKL as compared to that treated with RANKL-Ex (Figure 6B). Thus, the TRAP assay results showed that mRANKL is more efficient to induce osteoclast formation as compared to RANKL-Ex. Further, the effects of mRANKL and RANKL-Ex on mRNA levels of osteoclast phenotypic and functional markers were compared by qRT-PCR. Trap mRNA levels were approximately 2-fold higher in cells treated with mRANKL as compared to that of RANKL-Ex (Figure 7A). The mRNA levels of CalcR (Figure 7B) and Cfms (Figure 7C) were about 1.7-fold and 1.6-fold higher, respectively, in mRANKL treated cells. Similarly, the mRNA levels of Nfatc1 (Figure 7D) and CtsK (Figure 7E) were induced approximately 2.5-fold and 1.6-fold higher, respectively, by the treatment of mRANKL as compared to that of RANKL-Ex.
Discussion
In recent years, numerous studies have revealed the important roles of RANKL. But, most of the studies on physiological role of RANKL have been carried out with extracellular domain of RANKL. The extracellular RANKL is derived by proteolytic cleavage of mRANKL, a similar phenomenon observed in a transmembrane TNF-related Fas ligand (FasL) which undergoes metalloproteinase-mediated proteolytic cleavage to release the shed form of the ligand (sFasL). It was found that the apoptotic-inducing capacity of sFasL was reduced by 1,000-fold compared to transmembrane FasL [40]. Similarly, the distinct roles of transmembrane and shed forms of Syndecan-1 (Sdc1) in breast cancer progression were revealed [41]. Overexpression of wild type Sdc1 promoted cell proliferation, whereas its shed form inhibited proliferation. In contrast, the transmembrane Sdc1 inhibited invasiveness whereas soluble Sdc1 vastly promoted the invasion of MCF-7 in vitro [41].
On the basis of the previous comparative studies between transmembrane protein and its extracellular form, we speculated the functional difference between mRANKL and RANKL-Ex. For functional and structural studies, qualitative and quantitative production of proteins are essential. Yet, high level expression of stable and functional mammalian proteins, particularly membrane proteins, remains a challenging task. Moreover, there are several potential bottlenecks such as codon usage, expression systems and/or conditions that might affect both yield and solubility of membrane proteins in E. coli. In recent years, the significant divergent codon bias between the E. coli and mammalian genes are balanced by engineering the genes to preferentially used synonymous codons compatible with the host [27].
In this study, we explored the use of codon optimization and response surface methodology to achieve the high level expression of mRANKL in E. coli. Among different vector systems and E. coli hosts, SHuffle Express E. coli and pMAL-c5X possessing Ptac promoter were found to be most appropriate host and vector system, respectively for the expression of mRANKL in soluble form. We obtained significant amount of mRANKL from SHuffle E. coli-pOmR-c5X when induced with lactose. Among the three different solubility tags, i.e., MBP-, GST-, and His6-tag, MBP was found to be the most effective solubilizing agent for the expression of mRANKL in SHuffle Express E. coli. MBP is most commonly used as an N-terminal tag for cytosolic expression of the protein. As a fusion tag, it facilitates expression, solubility, and purification [42]. Initially, soluble mRANKL was not obtained from SHuffle E. coli-pOmR-c5X when IPTG was used as an inducer. As an alternative, lactose was selected because natural lactose-induction provides several advantages over the IPTG-induction such as low cost, metabolizable and non-toxicity [43].
After the selection of suitable expression system, we aimed to optimize several factors (cell density before induction, lactose concentration, post-induction temperature and post-induction time) for the high production of soluble proteins. Due to several parameters, we explored CCD and RSM to fix and evaluate the interactive effects of most influential parameters of culture condition. The five levels, four factors, CCD at the given range of the above mentioned parameters predicted a set of 30 combination of variables to optimize the expression of mRANKL in E. coli. The response (mRANKL (mg/L) was correlated with the four factors as a function of the second-order polynomial equation, given by Eq. 1, indicating that linear coefficients A (OD600 before induction), B (lactose concentration), C (post-induction temperature) and D (post-induction time) exhibited a significant impact on production of mRANKL. ANOVA analyses resulted high F-value, very low probability (p > F) value and statistically insignificant lack of fit test at 95% confidence level indicating that the model was adequate for representing the experimental data. The adequacy of quadratic model was confirmed by the high coefficient of determination, R2, and the adjusted determination coefficient, R2 adj, value both of which are close to 1, ensuring a high correlation between the experimental values and the predicted values.
Furthermore, in order to gain a better understanding of the four factors for optimal production of mRANKL, the models were presented as 3-D response surfaces. The optimum conditions for mRANKL production was found to be at OD600 of 0.6, lactose concentration of 7.5 mM, post-induction temperature of 26°C and post-induction time of 5 h. These plots showed that the production of mRANKL was found to be increased with increasing OD600 and post induction time. However, an increase in both factors beyond the optimum region (OD600: 0.6 and induction time: 5 h) resulted in a decrease in mRANKL production. While increased production of mRANKL was observed with increasing post-induction temperature, the lactose concentration showed less effect on the production of mRANKL. However, at the temperatures higher than 30°C, the production of mRANKL began to decrease at all OD600. While mRANKL as inclusion bodies was produced at high levels above 30°C, high yield of soluble mRANKL was produced when temperature was down-shifted to 26°C. Generally, protein expression processes operate a biphasic culture whereby cells are grown at 37°C to maximise biomass and then the culture is shifted to a lower temperature (25–30°C) while maintaining a longer and more viable stationary/production phase. It is known that the expression of the proteins at low temperatures usually improves both solubility and activity of proteins by increasing stability and correct folding patterns [44]. The results obtained from the experiment were found to be in good agreement with the values predicted by the model. In our laboratory condition, the maximum of 52.4 mg/L of purified MBP-tagged mRANKL was obtained under optimum conditions. Thus, OD600 lesser than 0.7, post-induction time of 5 h and lower post-induction temperature values are favorable for the production mRANKL while the concentration of lactose has less influence on the production of mRANKL.
Since we aimed to perform a comparative functional study between mRANKL and extracellular RANKL-Ex, the expression of RANKL-Ex was also optimized in SHuffle Express E. coli by CDD and RSM approach. We successfully produced about 130.8 mg/L of purified MBP-tagged RANKL-Ex by optimization of expression conditions whereas Papaneophytou et al., succeeded to produce 11.4 mg/L of extracellular RANKL in E. coli host using RSM [45]. Both mRANKL and RANKL-Ex were separated from their fusion partners prior to analysis of their biological activities.
It has been well established that extracellular RANKL self-aggregates into homotrimer and the trimeric form is essential for the activation of its cognate receptor RANK. Therefore, it is necessary to determine the native state of mRANKL prior to test its biological activity. The native form of mRANKL was determined by native gel electrophoresis and gel filtration chromatography. Both experiments showed that mRANKL exist as homotrimer by self-assembly. To distinguish between the roles of transmembrane and extracellular forms of RANKL, the efficiency of mRANKL and RANKL-Ex to induce osteoclastogenesis in RAW264.7 cells was tested and analyzed by TRAP staining and TRAP assay. It was observed that mRANKL is approximately 2-fold more potent than RANKL-Ex in inducing the osteoclastogenesis of RAW264.7 cells. In a similar experiment, the extracellular RANKL was oligomerized to mimic the function of transmembrane RANKL and tested its function in osteoclastogenesis [46]. The experiment demonstrated that oligomerized RANKL works more efficiently than extracellular RANKL in the generation of osteoclasts. Consistently, our results clearly demonstrated that the biological activity of mRANKL is comparatively higher than RANKL-Ex.
Osteoclast differentiation is associated with up-regulation of specific genes in response to RANKL. Osteoclasts express several markers, such as TRAP, CALCR and CFMS throughout the differentiation process which, along with multinucleation and resorption, characterize the osteoclast phenotype [47]. CALCR is expressed on the surface of mature osteoclast and it has been described as the best differentiation marker for the osteoclast [48]. CFMS triggers the proliferation and fusion of mononuclear cells to form multinucleated, mature osteoclasts [49]. NFATc1 is believed to be a master transcription factor for murine osteoclastogenesis [50]. RANKL signaling cascade plays a significant role in the regulation of cathepsin K expression. In fact RANKL stimulates the osteoclast to produce increased amounts of cathepsin K [51]. Our qRT-PCR analyses revealed that mRANKL significantly upregulates the genes that are induced during osteoclast differentiation. Not only the assay of TRAP activity showed that mRANKL is more efficient to induce osteoclastogenesis in vitro compared to RANKL-Ex, but quantitative RT-PCR assays also showed that mRANKL is approximately twice as much active as RANKL-Ex.
In conclusion, we successfully demonstrated the high level expression of both mRANKL and RANKL-Ex in soluble and active forms by codon optimization and response surface methodology. It demonstrates that the codon optimized synthetic gene products expressed in E. coli retain their functional properties. Therefore, it will offer great opportunity for structural and functional studies on transmembrane proteins. Importantly, our study also revealed that mRANKL is more potent than extracellular RANKL to induce osteoclastogenesis. We hope these findings will certainly contribute to broaden the insight of diverse roles of mRANKL, and to understand the RANKL/RANK system.
Supporting Information
Diagnostic plots for estimating the adequacy of the regression model. Correlation between predicted and actual value for RANKL-Ex production (A). The studentized and normal percentage probability plot of RANKL-Ex production (B).
(TIF)
Response surface plots to visualize the relationship between the response and experimental levels of each factor. Three dimensional surface plots of combined effect of cell density (OD600) and lactose concentration (A), OD600 and post-induction temperature (B), OD600 and post-induction time (C), post-induction temperature and lactose concentration (D), post-induction time and lactose concentration (E) and post- induction temperature and post-induction time (F) on RANKL-Ex production.
(TIF)
SDS PAGE and Western blot analyses of RANKL-Ex fusion proteins. A: Comassie blue stained SDS gel showing lysates from two SHuffle E. coli-pOsREx-c5x colonies expressing the RANKL-Ex fusion proteins (∼62 kDa). Lane 1: Protein standards, molecular masses are indicated in kilodaltons (kDa); Lanes 2 and 3: Insoluble and soluble fractions from clone 1, respectively; Lanes 4 and 5: Insoluble and soluble fractions from clone 2, respectively. B: Comassie blue stained SDS gel showing purification of crude RANKL-Ex using amylose resin. Lane 1: crude soluble RANKL-Ex; Lane 2: Flow through; Lane 3: Wash flow through; Lane 4: Protein standards, molecular masses are indicated in kilodaltons (kDa); Lanes 5–7: Elution fractions. C: Western blot analysis of purified RANKL-Ex. Different amount of purified RANKL-Ex was run in 4–20% SDS page and transferred onto a nitrocellulose membrane. Detection was performed with anti-RANKL primary antibody, goat IgG HRP-conjugated secondary antibody and chemiluminescent substrate. Lane 1: Protein marker; Lanes 2–5: 200, 100, 50 and 30 ng of purified RANKL-Ex protein respectively.
(TIF)
Codon usage table.
(DOCX)
CCD with measured and predicted response of RANKL-Ex production.
(DOCX)
ANOVA for response surface quadratic model for RANKL-Ex production.
(DOCX)
Confirmation experiments for RANKL-Ex production.
(DOCX)
Acknowledgments
We acknowledge the National Instrumentation Center for Environmental Management (NICEM), Seoul National University, Gwanak-gu, Seoul, Korea for analysis of gene sequence.
Funding Statement
This research is supported by National Research Foundation of Korea (NRF), Korea government (MEST) (Project No. 2011-0029416). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Anderson DM, Maraskovsky E, Billingsley WL, Dougall WC, Tometsko ME, et al. (1997) A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature 390: 175–179. [DOI] [PubMed] [Google Scholar]
- 2. Wong BR, Josien R, Lee SY, Sauter B, Li HL, et al. (1997) TRANCE (tumor necrosis factor [TNF]-related activation-induced cytokine), a new TNF family member predominantly expressed in T cells, is a dendritic cell-specific survival factor. J Exp Med 186: 2075–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, et al. (1998) Osteoclast differentiation factor is a ligand for osteoprotegerin osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. P Natl Acad Sci USA 95: 3597–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ikeda T, Kasai M, Utsuyama M, Hirokawa K (2001) Determination of three isoforms of the receptor activator of nuclear factor-kappaB ligand and their differential expression in bone and thymus. Endocrinology 142: 1419–1426. [DOI] [PubMed] [Google Scholar]
- 5. Lum L, Wong BR, Josien R, Becherer JD, Erdjument-Bromage H, et al. (1999) Evidence for a role of a tumor necrosis factor-alpha (TNF-alpha)-converting enzyme-like protease in shedding of TRANCE, a TNF family member involved in osteoclastogenesis and dendritic cell survival. J Biol Chem 274: 13613–13618. [DOI] [PubMed] [Google Scholar]
- 6. Hikita A, Tanaka S (2007) Ectodomain shedding of receptor activator of NF-kappa B ligand. Adv Exp Med Biol 602: 15–21. [DOI] [PubMed] [Google Scholar]
- 7. Hikita A, Tanaka N, Yamane S, Ikeda Y, Furukawa H, et al. (2009) Involvement of a disintegrin and metalloproteinase 10 and 17 in shedding of tumor necrosis factor-alpha. Biochem Cell Biol 87: 581–593. [DOI] [PubMed] [Google Scholar]
- 8. Wong BR, Rho JR, Arron J, Robinson E, Orlinick J, et al. (1997) TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. J Biol Chem 272: 25190–25194. [DOI] [PubMed] [Google Scholar]
- 9. Lacey DL, Boyle WJ, Simonet WS, Kostenuik PJ, Dougall WC, et al. (2012) Bench to bedside: elucidation of the OPG-RANK-RANKL pathway and the development of denosumab. Nature reviews Drug discovery 11: 401–419. [DOI] [PubMed] [Google Scholar]
- 10. Wong BR, Josien R, Lee SY, Vologodskaia M, Steinman RM, et al. (1998) The TRAF family of signal transducers mediates NF-kappaB activation by the TRANCE receptor. The Journal of biological chemistry 273: 28355–28359. [DOI] [PubMed] [Google Scholar]
- 11. Crockett JC, Mellis DJ, Scott DI, Helfrich MH (2011) New knowledge on critical osteoclast formation and activation pathways from study of rare genetic diseases of osteoclasts: focus on the RANK/RANKL axis. Osteoporosis international: a journal established as result of cooperation between the European Foundation for Osteoporosis and the National Osteoporosis Foundation of the USA 22: 1–20. [DOI] [PubMed] [Google Scholar]
- 12. Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, et al. (1997) Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell 89: 309–319. [DOI] [PubMed] [Google Scholar]
- 13. Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, et al. (1999) OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 397: 315–323. [DOI] [PubMed] [Google Scholar]
- 14. Teitelbaum SL (2000) Bone resorption by osteoclasts. Science 289: 1504–1508. [DOI] [PubMed] [Google Scholar]
- 15. Khosla S (2001) Minireview: the OPG/RANKL/RANK system. Endocrinology 142: 5050–5055. [DOI] [PubMed] [Google Scholar]
- 16. Dougall WC, Glaccum M, Charrier K, Rohrbach K, Brasel K, et al. (1999) RANK is essential for osteoclast and lymph node development. Gene Dev 13: 2412–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rossi SW, Kim MY, Leibbrandt A, Parnell SM, Jenkinson WE, et al. (2007) RANK signals from CD4(+)3(−) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp Med 204: 1267–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim N, Odgren PR, Kim DK, Marks SC Jr, Choi Y (2000) Diverse roles of the tumor necrosis factor family member TRANCE in skeletal physiology revealed by TRANCE deficiency and partial rescue by a lymphocyte-expressed TRANCE transgene. Proc Natl Acad Sci U S A 97: 10905–10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akiyama T, Shimo Y, Yanai H, Qin J, Ohshima D, et al. (2008) The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity 29: 423–437. [DOI] [PubMed] [Google Scholar]
- 20. Hikosaka Y, Nitta T, Ohigashi I, Yano K, Ishimaru N, et al. (2008) The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity 29: 438–450. [DOI] [PubMed] [Google Scholar]
- 21. Hanada R, Leibbrandt A, Hanada T, Kitaoka S, Furuyashiki T, et al. (2009) Central control of fever and female body temperature by RANKL/RANK. Nature 462: 505–509. [DOI] [PubMed] [Google Scholar]
- 22. Fata JE, Kong YY, Li J, Sasaki T, Irie-Sasaki J, et al. (2000) The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell 103: 41–50. [DOI] [PubMed] [Google Scholar]
- 23. Knoop KA, Kumar N, Butler BR, Sakthivel SK, Taylor RT, et al. (2009) RANKL Is Necessary and Sufficient to Initiate Development of Antigen-Sampling M Cells in the Intestinal Epithelium. J Immunol 183: 5738–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baneyx F, Mujacic M (2004) Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 22: 1399–1408. [DOI] [PubMed] [Google Scholar]
- 25. Sorensen HP, Mortensen KK (2005) Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol 115: 113–128. [DOI] [PubMed] [Google Scholar]
- 26. Makrides SC (1996) Strategies for achieving high-level expression of genes in Escherichia coli. Microbiological reviews 60: 512–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gustafsson C, Govindarajan S, Minshull J (2004) Codon bias and heterologous protein expression. Trends in biotechnology 22: 346–353. [DOI] [PubMed] [Google Scholar]
- 28. Gao W, Rzewski A, Sun H, Robbins PD, Gambotto A (2004) UpGene: Application of a Web-Based DNA Codon Optimization Algorithm. Biotechnology Progress 20: 443–448. [DOI] [PubMed] [Google Scholar]
- 29. Cai Y, Sun J, Wang J, Ding Y, Tian N, et al. (2008) Optimizing the codon usage of synthetic gene with QPSO algorithm. Journal of Theoretical Biology 254: 123–127. [DOI] [PubMed] [Google Scholar]
- 30. Hatfield GW, Roth DA (2007) Optimizing scaleup yield for protein production: Computationally Optimized DNA Assembly (CODA) and Translation Engineering. Biotechnology annual review 13: 27–42. [DOI] [PubMed] [Google Scholar]
- 31.Francis DM, Page R (2010) Strategies to optimize protein expression in E. coli. Current protocols in protein science/editorial board, John E Coligan [et al] Chapter 5: Unit 5 24 21–29. [DOI] [PMC free article] [PubMed]
- 32. Chambers SP, Swalley SE (2009) Designing experiments for high-throughput protein expression. Methods in molecular biology 498: 19–29. [DOI] [PubMed] [Google Scholar]
- 33.Mason RL, Gunst RF, Hess JL (2003) Statistical design and analysis of experiments with applications to engineering and science. 2nd ed. New York: John Wiley and Sons.
- 34.Myers RH, Montgomery DC, Anderson-Cook CM (2002) Response surface methodology: Process and product optimization using designed experiments. New York: John Wiley and Sons. 798 p. [Google Scholar]
- 35. Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, et al. (1999) Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci U S A 96: 3540–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoover DM, Lubkowski J (2002) DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic acids research 30: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 38. Henry I, Sharp PM (2007) Predicting gene expression level from codon usage bias. Mol Biol Evol 24: 10–12. [DOI] [PubMed] [Google Scholar]
- 39. Minkin C (1982) Bone acid phosphatase: tartrate-resistant acid phosphatase as a marker of osteoclast function. Calcified tissue international 34: 285–290. [DOI] [PubMed] [Google Scholar]
- 40. Schneider P, Holler N, Bodmer JL, Hahne M, Frei K, et al. (1998) Conversion of membrane-bound Fas(CD95) ligand to its soluble form is associated with downregulation of its proapoptotic activity and loss of liver toxicity. J Exp Med 187: 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nikolova V, Koo CY, Ibrahim SA, Wang ZH, Spillmann D, et al. (2009) Differential roles for membrane-bound and soluble syndecan-1 (CD138) in breast cancer progression. Carcinogenesis 30: 397–407. [DOI] [PubMed] [Google Scholar]
- 42. Kapust RB, Waugh DS (1999) Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci 8: 1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tian HS, Tang L, Wang Y, Wang XJ, Guan LL, et al. (2011) Lactose Induction Increases Production of Recombinant Keratinocyte Growth Factor-2 in Escherichia coli. Int J Pept Res Ther 17: 123–129. [Google Scholar]
- 44. Sorensen HP, Mortensen KK (2005) Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microbial cell factories 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Papaneophytou CP, Rinotas V, Douni E, Kontopidis G (2013) A statistical approach for optimization of RANKL overexpression in Escherichia coli: Purification and characterization of the protein. Protein Expres Purif 90: 9–19. [DOI] [PubMed] [Google Scholar]
- 46. Nakashima T, Kobayashi Y, Yamasaki S, Kawakami A, Eguchi K, et al. (2000) Protein Expression and Functional Difference of Membrane-Bound and Soluble Receptor Activator of NF-κB Ligand: Modulation of the Expression by Osteotropic Factors and Cytokines. Biochem Bioph Res Co 275: 768–775. [DOI] [PubMed] [Google Scholar]
- 47. Gallois A, Lachuer J, Yvert G, Wierinckx A, Brunet F, et al. (2010) Genome-wide expression analyses establish dendritic cells as a new osteoclast precursor able to generate bone-resorbing cells more efficiently than monocytes. Journal of Bone and Mineral Research 25: 661–672. [DOI] [PubMed] [Google Scholar]
- 48. Roodman GD (1996) Advances in bone biology: The osteoclast. Endocr Rev 17: 308–332. [DOI] [PubMed] [Google Scholar]
- 49. Boyle WJ, Simonet WS, Lacey DL (2003) Osteoclast differentiation and activation. Nature 423: 337–342. [DOI] [PubMed] [Google Scholar]
- 50. Takayanagi H (2007) Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol 7: 292–304. [DOI] [PubMed] [Google Scholar]
- 51. Troen BR (2006) The regulation of cathepsin K gene expression. Ann Ny Acad Sci 1068: 165–172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagnostic plots for estimating the adequacy of the regression model. Correlation between predicted and actual value for RANKL-Ex production (A). The studentized and normal percentage probability plot of RANKL-Ex production (B).
(TIF)
Response surface plots to visualize the relationship between the response and experimental levels of each factor. Three dimensional surface plots of combined effect of cell density (OD600) and lactose concentration (A), OD600 and post-induction temperature (B), OD600 and post-induction time (C), post-induction temperature and lactose concentration (D), post-induction time and lactose concentration (E) and post- induction temperature and post-induction time (F) on RANKL-Ex production.
(TIF)
SDS PAGE and Western blot analyses of RANKL-Ex fusion proteins. A: Comassie blue stained SDS gel showing lysates from two SHuffle E. coli-pOsREx-c5x colonies expressing the RANKL-Ex fusion proteins (∼62 kDa). Lane 1: Protein standards, molecular masses are indicated in kilodaltons (kDa); Lanes 2 and 3: Insoluble and soluble fractions from clone 1, respectively; Lanes 4 and 5: Insoluble and soluble fractions from clone 2, respectively. B: Comassie blue stained SDS gel showing purification of crude RANKL-Ex using amylose resin. Lane 1: crude soluble RANKL-Ex; Lane 2: Flow through; Lane 3: Wash flow through; Lane 4: Protein standards, molecular masses are indicated in kilodaltons (kDa); Lanes 5–7: Elution fractions. C: Western blot analysis of purified RANKL-Ex. Different amount of purified RANKL-Ex was run in 4–20% SDS page and transferred onto a nitrocellulose membrane. Detection was performed with anti-RANKL primary antibody, goat IgG HRP-conjugated secondary antibody and chemiluminescent substrate. Lane 1: Protein marker; Lanes 2–5: 200, 100, 50 and 30 ng of purified RANKL-Ex protein respectively.
(TIF)
Codon usage table.
(DOCX)
CCD with measured and predicted response of RANKL-Ex production.
(DOCX)
ANOVA for response surface quadratic model for RANKL-Ex production.
(DOCX)
Confirmation experiments for RANKL-Ex production.
(DOCX)