Abstract
The sodium -pumping NADH: ubiquinone oxidoreductase (Na+-NQR) is the main ion pump and the primary entry site for electrons into the respiratory chain of many different types of pathogenic bacteria. This enzymatic complex creates a transmembrane gradient of sodium that is used by the cell to sustain ionic homeostasis, nutrient transport, ATP synthesis, flagellum rotation and other essential processes. Comparative genomics data demonstrate that the nqr operon, which encodes all Na+-NQR subunits, is found in a large variety of bacterial lineages with different habitats and metabolic strategies. Here we studied the distribution, origin and evolution of this enzymatic complex. The molecular phylogenetic analyses and the organizations of the nqr operon indicate that Na+-NQR evolved within the Chlorobi/Bacteroidetes group, after the duplication and subsequent neofunctionalization of the operon that encodes the homolog RNF complex. Subsequently, the nqr operon dispersed through multiple horizontal transfer events to other bacterial lineages such as Chlamydiae, Planctomyces and α, β, γ and δ -proteobacteria. Considering the biochemical properties of the Na+-NQR complex and its physiological role in different bacteria, we propose a detailed scenario to explain the molecular mechanisms that gave rise to its novel redox- dependent sodium -pumping activity. Our model postulates that the evolution of the Na+-NQR complex involved a functional divergence from its RNF homolog, following the duplication of the rnf operon, the loss of the rnfB gene and the recruitment of the reductase subunit of an aromatic monooxygenase.
Introduction
The sodium -dependent NADH: ubiquinone oxidoreductase (Na+-NQR) is a bacterial respiratory complex that catalyzes the transfer of electrons from NADH to ubiquinone, and thus it is the gateway by which the redox equivalents, produced by the primary and intermediary metabolism, enter into the respiratory chain [1]–[3] (Figure 1). Na+-NQR is not only the first enzyme in the respiratory chain but also one of the main ion transporters in the cell, since the energy released by the redox activity is coupled to the pumping of sodium to the periplasmic space. Thus, Na+-NQR coordinates the operation of the entire metabolism with the mechanisms used for osmotic regulation. Na+-NQR is commonly associated with the aerobic respiratory metabolism of pathogenic bacteria, such as Vibrio cholerae [4], Vibrio harveyi [5], Klebsiella pneumoniae [6], Haemophilus influenzae [7], among others, in which it plays essential roles in different homeostatic functions [8], [9]. Although, a comprehensive study of the role that this enzyme plays in pathogens has not been carried out, the evidence indicates that Na+-NQR has an important role in infection, regulating the production of virulence factors [10], and the survival of the cells in the internal host environment. The enormous amount of information that has been obtained with the sequencing of prokaryotic genomes has shown that the genes encoding the subunits of this respiratory enzyme are also found in the genomes of diverse types of bacteria, including aerobic, microaerophilic, obligate anaerobes, photosynthetic, chemolithotrophic, halophilic, alkalophilic and predatory microorganisms (Table S1).
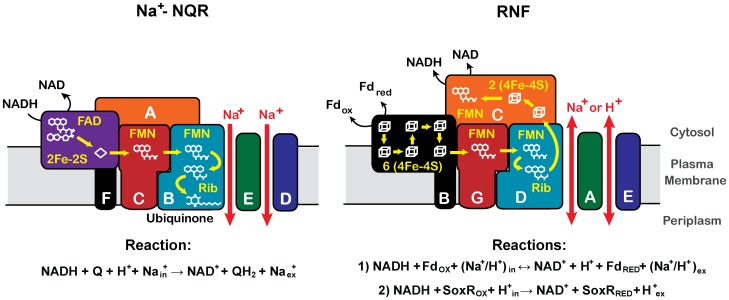
Figure 1. Physiological activities, topology predictions, subunit and cofactor composition and electron transfer pathways of Na+-NQR and RNF.
Na+-NQR catalyzes the transfer of electrons from NADH to ubiquinone, which under physiological conditions is irreversible, and uses the free energy released by the redox reaction to pump sodium [1]–[3]. On the other hand, in most bacterial phyla RNF catalyzes the transfer of electrons between NADH and ferredoxin, which can be reversible depending on the metabolic adaptations of the bacteria [24], [25]. However, the evidence indicates that RNF can also catalyze the reduction the transcriptional regulatory protein SoxR [63], which controls the expression of proteins involved in reactive oxygen species handling. In contrast with Na+-NQR, which is specific for sodium [11], the RNF complex might use sodium and protons [25]. The pathway for electron transport in Na+-NQR is shown in the yellow lines [1], as well as the proposed pathway for RNF [62]. Ubiquinone is a hydrophobic molecule located in the plasma membrane of bacteria. It has been demonstrated previously that the reduction of this compound occurs in the cytosolic face of the membrane [2], but for illustrative purposes it is shown on the bottom of the figure.
Na+-NQR is a sodium -specific ionic pump [11], composed of six subunits (NqrA-F) (Figure 1, Table 1) encoded in the nqr operon, that contains five redox cofactors: FAD and a 2Fe-2S center, located in the NqrF subunit, two covalently -bound FMN molecules, located in the NqrB and NqrC subunits, and riboflavin (Table 1) [1]. The binding sites for NADH and ubiquinone are located in the NqrF [12], [13] and NqrB subunits [14], respectively, and although there are some indications suggesting that the riboflavin binding site might be located in NqrB subunit [15], the exact localization of this cofactor remains elusive. On the other hand, the subunits NqrB, NqrD and NqrE contain several transmembrane helices [16], with a large number of acid residues that could be involved in sodium binding and transport [17]–[19]. In contrast with other subunits, NqrA is cytosolic, does not contain either transmembrane segments or cofactor binding sites [1], [20], and although there is controversial evidence about its function [14], [21], [22], it has been considered to fulfill structural roles, since its elimination prevents the association of the complex [15]. Na+-NQR is evolutionary related to a membrane -bound NADH: ferredoxin dehydrogenase that was originally described in Rhodobacter capsulatus, in which it might have roles in Nitrogen fixation, and thus it is called RNF (Rhodobacter Nitrogen Fixing) [23], [24]. Although in most cases RNF catalyzes the reversible electron transfer between NADH and ferredoxin [25]–[30], coupled to the sodium or proton transport, its function can vary enormously depending on the metabolic adaptations of the bacteria in which it is found (see below). The RNF complex is also composed of six subunits, encoded in the rnf operon, most of which are homologous to the subunits of Na+-NQR (Figure 1, Table 1, Table S2) [20], [24], [25].
Table 1. Structural and catalytic motifs of Na+-NQR and RNF subunits.
| Subunit/ | TH | Cofactors | Substrate binding sites | Function |
| homologue | ||||
| NqrA/RnfC | None (NqrA)/ | None (NqrA)/ | Structural? (NqrA)/ | |
| 2(4Fe-4S) and FMN (RnfC) | NAD+ (RnfC) | NAD+ reduction (RnfC) | ||
| NqrB/RnfD | 9/8 | Covalently -bound FMN and Riboflavin (?) | Ubiquinone (NqrB) and Sodium | Ubiquinone reduction (NqrB) and Sodium transport |
| NqrC/RnfG | 2 | Covalently -bound FMN | ||
| NqrD/RnfE | 6 | Sodium | Sodium transport | |
| NqrE/RnfA | 6 | Sodium | Sodium transport | |
| NqrF | 1 | FAD and 2Fe-2S | NADH | NADH oxidation |
| RnfB | 1–2 | 6(4Fe-4S) and 2Fe-2S? | Ferredoxin | Ferredoxin oxidation |
Na+-NQR has many characteristics that are rather uncommon among respiratory enzymes. For instance, it is the only enzyme reported that can use riboflavin (vitamin B2) directly as a redox cofactor [31]. Until recently, it was generally accepted that riboflavin acts exclusively as precursor in the biosynthesis of FAD and FMN. Hence, the role of riboflavin as a cofactor in Na+-NQR is completely novel. Moreover, Na+-NQR produces several flavin radicals during the catalytic cycle, and surprisingly also contains stable flavin semiquinone radical species in different inactive forms of the enzyme [32], an extremely unusual characteristic. Indeed, riboflavin is actually found as a neutral semiquinone radical [31], [32], which is stable in the presence of oxygen and other strong oxidizing agents, indicating that Na+-NQR presents a mechanism, like no other reported so far, to stabilize the unpaired electron of the flavin. Furthermore, the binding of the two FMN molecules to NqrB and NqrC subunits involves a phosphoester bond [33], which is also very unusual and can only be found in NqrB and NqrC, their RNF homologues (Table 1) [34] and in the regulatory proteins of N2O reductase expression, NOsR and NirI [35]. Although the catalytic mechanisms of the respiratory and photosynthetic enzymes are diverse, the ion pumping strategies of these enzymes can be grouped into two categories: redox loop mechanisms, such as the Q -cycle (i.e., bc 1 complex and photosystem II) [36], [37] and the localized or “direct” coupling (used by cytochrome c oxidase and complex I) [38], [39]. Under these two mechanisms the reduction of a molecule or cofactor in a hydrophobic environment is involved in the electroneutral ion capture, and its release on the other side of the membrane, upon oxidation. According to our data, Na+-NQR does not follow these ion -pumping strategies. Instead, ion translocation by this complex involves a series of conformational states that allow the capture of sodium in the internal side of the membrane and its release on the opposite side [1], [40]. Given all of these characteristics, it is evident that Na+-NQR has evolved separately from other redox enzymes and ion transporters, which has led to the arising of novel enzymatic strategies. In order to gain insights into the evolution and phylogenetic history of Na+-NQR, we analyzed the distribution and organization of the nqr operon in diverse pathogenic and non -pathogenic bacteria. Our results allow us to hypothesize and discuss a series of molecular events that may have been important in the origin, evolution and spreading of Na+-NQR, as well as to postulate some ecological factors that could have been important in its spreading and evolution. The data indicate that Na+-NQR appeared originally in the Chlorobi/Bacteroidetes (C/B) phyla, after a duplication of the rnf operon. The subsequent loss on the rnfB gene prompted significant changes in the mechanism of the enzyme, which together with the recruitment of the reductase subunit of the aromatic (phenol, toluene and bencene) monooxygenases (AMOr protein) underlie the origin of Na+-NQR. The current distribution of this complex in diverse bacterial lineages can be explained by at least two ancestral independent horizontal gene transfer (HGT) events, to proteobacteria, and Chlamydiae and Planctomyces, and at least two posterior HGT events, from Chlorobi to δ-proteobacteria and from Pasteurellales (γ-proteobacteria) to Neisseriales (β-proteobacteria).
Materials and Methods
Homolog Identification, Multiple Sequence Alignment and Phylogenetic Analyses
Using the amino acid sequences of the Na+-NQR subunits of Vibrio cholerae (O1 El Tor strain N16961) orthologous sequences were identified in the RefSeq GenBank database. After an exhaustive similarity search (blastp; cutoff E-value ≤1e-15), protein sequences were retrieved, covering all major bacterial groups where Na+-NQR subunits were identified (Table S1). The nqr operon was not found in the bacterial groups Acidobacteria, Actinobacteria, ε-proteobacteria, Aquificae, Chloroflexi, Cyanobacteria, Deinococcus/Thermus and Thermotogae, neither in Archaea or Eukaryotes (Table 2). Our final protein data set comprised sequences of the six Na+-NQR subunits, from 90 bacterial taxa (Table S1). The six protein sets were aligned with MAFFT v7 [41], using the FFT-NS-I method, with the BLOSUM 62 substitution matrix. After visual inspection to discard incomplete protein models, fused or chimeric sequences, the raw multiple alignments were processed with Gblocks v0.91b [42], to discard poorly aligned positions and ambiguous regions. In all cases the alignments obtained after Gblocks processing contained all the catalytically and structurally relevant sections of the six subunits. Interestingly, the alignment of the NqrA subunits showed that the sequence of this subunit is highly variable. This variability is possibly related to relaxed functional constraints, given the lack of catalytic function of NqrA, reflected in a relatively high amino acid substitution rate. To test the hypothesis of homology (i.e., common origin) of the Na+-NQR and RNF subunits we used the global alignment software Needdle (EMBOSS) to estimate the pairwise percentage of sequence identity (PID) between Na+-NQR and RNF putative homolog pairs, and NqrF with the AMOr protein. We carried out two independent intraspecific sequence comparisons using the Na+-NQR and RNF proteins from both Chlorobium phaeobacteroides (Chlorobi) and Anaerophaga thermohalophila (Bacteroidetes), respectively. A single randomly selected AMOr sequence (Cupriavidus metallidurans CH34) was aligned independently against the Chlorobium and Anaerophaga nqrF subunit.
Table 2. Distribution of Na+-NQR and RNF in Archaea and Bacteria.
| Domain | Group | RNF | Na+-NQR | ||||||||||
| C | D | G | E | A | B | A | B | C | D | E | F | ||
| Bacteria | Acidobacteria | ||||||||||||
| Actinobacteria | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |||||||
| α -proteobacteria | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |
| β-proteobacteria | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |
| γ-proteobacteria | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |
| δ-proteobacteria | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |
| ε -proteobacteria | |||||||||||||
| Aquificae | |||||||||||||
| Chlamy/Verrua | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |||||||
| Planctomycetes | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |||||||
| Chlorobi | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |
| Bacteroidetes | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |
| Chloroflexi | |||||||||||||
| Cyanobacteria | |||||||||||||
| Deinococcus/Thermus | |||||||||||||
| Firmicutes | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |||||||
| Fusobacteria | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |||||||
| Spirochaetes | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |||||||
| Tenericutes | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |||||||
| Thermotogae | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |||||||
| Archaea | Methanosarcinales | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ||||||
Please note that although the nqr and rnf operons were found in these phyla, there is a large variability among the bacterial orders and some of them might not contain any of the two complexes. Moreover, the presence of the genes might not indicate that the complexes are functional and further biochemical characterizations are necessary to clarify these and other aspects.
The intraspecific sequence similarity surveys revealed normalized PIDs of at least 30% in four out of six tested homolog pairs (see PID2-PID4 values in Table S2). According with typically used PID thresholds (30%) [43]–[45], the identity values between RnfE/NqrD, RnfA/NqrE, RnfD/NqrB and AMOr/NqrF (Table S2) indicate that these pairs are homologous, supporting previous works [2], [25]. The PID values in the case of the RnfC/NqrA and RnfG/NqrC pairs were between 22 and 33% depending on the normalization strategy. Even though the PID in these last two Na+-NQR/RNF pairs are in the “twilight zone” [46] to clearly define families of homologous proteins, the hypothesis of common origin is substantially reinforced by structural similarities and shared protein domains observed between these protein duos, in particular in the case of NqrC/RnfG, which present a similar topology and conserved motifs for the interactions with the covalently -bound FMN cofactor (Table 1) [25], [34]. In the case of NqrA/RnfC, the evolutionary relationship of these subunits has been difficult to establish, due to the borderline PID values (22–27%). To offer further evidence supporting the homology of these subunits, secondary structure prediction analysis were performed and illustrated together with the pairwise sequence alignment of the NqrA and RnfC subunits of A. thermohalophila (Fig. S1). Both subunits show strong similarities in the amino terminus, containing a barrel sandwich hybrid domain and remnants of the NADH, FMN and Fe-S binding domains. The secondary structure predictions show that these two proteins have a very similar (>80%) structure (and probably a similar folding pattern), which together with their relative positions in the operon (see below), and the PID values, support the homology hypothesis.
To gain additional insights into the evolution of the Na+-NQR complex, we generated multiple alignments of each of the Na+-NQR subunits, together with the homologous proteins that constitute the RNF complex (see Table 1 for Na+-NQR/RNF homolog-pair details) [24], [25]. NqrF is the only Na+-NQR subunit with no homologous counterpart in the RNF complex. Our PSI blast similarity analysis showed that the AMOr subunit is the closest homolog to NqrF, sharing the FAD, NADH and 2Fe-2center binding sites [47]. Thereafter, we were able to generate multiple alignments of the homolog pairs: NqrB/RnfD, NqrD/RnfF, NqrE/RnfA and NqrF/AMOr. The high divergence in the amino acid sequence of the homolog pairs NqrA/RnfC and NqrC/RnfG precluded the estimation of reliable multiple alignments for phylogenetic analysis.
The best amino acid substitution model for each Na+-NQR subunit alignment was selected according to ProtTest. The Le and Gascuel (LG) substitution model [48], considering a gamma distribution of substitution rates (+G) and a proportion of invariable sites (+I), was selected in all cases. ML trees using the LG+G+I substitution model were estimated with RAxML HPC-MPI 7.2.8 [49] with 500 bootstrap pseudoreplicates. Bayesian posterior probabilities were calculated with MrBayes 3.2.1 [50], considering the WAG+G+I substitution model (LG model is not implemented in MrBayes) running a Metropolis-coupled Markov Chain Monte Carlo (MC3) for 2 million generations. Two independent MC3 runs were performed simultaneously, with 1 “cold” and 3 “heated” chains starting from different random trees. Chain convergence was evaluated every 250,000 generations, until the average standard deviation of split frequencies dropped below 0.01. The pool of trees was sampled every 100th generation. Final posterior probabilities were estimated after discarding the trees of the first 500,000 generations. Visual inspection of trees was carried out with Archaeopteryx 0.997 beta version [51].
Results and Discussion
Distribution and Metabolic Roles of Na+-NQR
The RNF and Na+-NQR complexes are among the few respiratory enzymes that couple their redox activities with the ability to create an electrochemical gradient of sodium (Figure 1) [1], [25]. These two enzymes share numerous characteristics that include a similar cofactor composition, sodium translocation mechanisms and structural motifs (Table 1) [25], [27], [28]. Nonetheless, one of the main differences between RNF and Na+-NQR is the mechanism used to incorporate electrons into the complex. In the case of Na+-NQR, electrons are introduced into the transport chain through NqrF [12], [52], and in the case of RNF, it has been proposed that the redox equivalents are introduced from ferredoxin, using the RnfB subunit [25]. NqrF and RnfB subunits are complex-specific and have no homologous counterpart in RNF or Na+-NQR.
The rnf operon is present in numerous bacterial lineages and can also be found in the Methanosarcinales order from the domain Archaea (Table 2). In contrast, the nqr operon is exclusively found in bacteria, and it is absent in both eukaryotes and archaea. Na+-NQR has been extensively studied in marine/opportunistic pathogens, especially in γ -proteobacteria, such as V. cholerae [4], [9], in which it forms part of the aerobic respiratory chain. However, our results now show that the nqr operon is found in bacteria with diverse metabolic capabilities and life-styles, including photosynthetic, chemolithotrophic and anaerobic bacteria (Table S1). In spite that the Na+-NQR complex is found in distantly related bacterial groups, such as α, β, γ and δ -proteobacteria, Planctomyces, Chlamydiae, Chlorobi and Bacteroidetes, it is apparently absent in many major lineages (Table 2). Na+-NQR patchy distribution suggests that this enzymatic complex was not present in the most recent common bacterial ancestor. Overall, the rnf operon presents a wider distribution among major bacterial lineages than Na+-NQR. Interestingly, in some bacterial lineages, such as Chlorobi/Bacteroidetes and most proteobacteria, both homologous complexes coexist (Table 2).
Na+-NQR -containing bacteria can be classified according to their habitat into marine/halophilic and endo/pathogenic bacteria. Pathogenic bacteria harboring Na+-NQR include β and γ -proteobacteria (Enterobacteriales, Vibrionalles, Pasteurellales, Aeromonadales, Pseudomonadales, Neisserales), Bacteroidetes and Chlamydiae, the latter are obligate intracellular parasites [8]. Most free -living organisms (as well as mitochondria and plastids) produce a transmembrane gradient of protons to convert the free energy released by different chemical reactions, into electrochemical energy that can be used to sustain a variety of processes, such as ATP synthesis. In contrast, pathogenic bacteria might not be able to use protons as energy currency, mostly because the extracellular pH is more alkaline (plasma pH = 7.2–7.4), compared to the internal pH of the cell (pH 6.0–6.5) [53], [54], and thus they are actually facing an “inverted” proton gradient [55]. Instead of using protons, pathogenic bacteria would produce a gradient of sodium, which is the most common cation in the plasma. It has been proposed that the sodium gradient is essential for survival of bacterial parasites, because it is used to support a large variety of processes, including pH regulation, ATP synthesis, cell motility, uptake of nutrients, toxin extrusion, as well as the efflux of drugs in antibiotic- resistant strains [8], [55]. Indeed, the evidence indicates that Na+-NQR plays an essential role in the physiology of many pathogens. In fact, the expression of Na+-NQR increases greatly during the infection of the small intestine by V. cholerae [56], and random insertion mutagenesis has demonstrated that the inactivation of nqr operon reduces drastically the infectivity of these cells [57]. Moreover, Na+-NQR activity regulates the expression of the V. cholerae virulence factors, such as the cholera toxin (CTX) and the major subunit of the toxin co-regulated pillus (TcpA), through the modulation of the ToxR, TcpP, and ToxT regulatory cascade [10].
Marine/halophilic bacteria containing Na+-NQR include Plactomycetes, Chlorobi, as well as α, δ, free- living β and “early diverging” γ-proteobacteria. All these types of bacteria have different metabolic strategies, including photoautotrophic or chemolitotrophic anaerobes, chemorganothrophs, as well as strict aerobic bacteria. In these organisms, Na+-NQR pumping activity could be used both to generate a gradient of sodium, coupled to the activity of the oxidative phosphorylation, and to maintain the intracellular ionic composition. Indeed, in the extreme haloalkalophile Desulfurivibrio alkaliphilus (which is a sulfate reducing bacteria -see below- found in soda lakes [58]), the activity of Na+-NQR should be an essential part of the mechanisms used for ionic homeostasis. Interestingly, Na+-NQR is also found in the predatory δ -proteobacteria Bacteriovorax marinus, in which its activity might be employed to form an electrochemical gradient, driving flagellum rotation. Ion -driven cell motility is essential for predatory bacteria, since is the main strategy to find prey bacteria [59].
The nqr operon is also found in sulfate reducing (SR) bacteria, such as δ -proteobacteria, especially in marine species. In these bacteria Na+-NQR might participate in osmotic protection, but may have other roles in the metabolism, especially in menaquinone reduction. Menaquinol is the main electron donor in the first stage of sulfate -based respiration. The Qmo complex catalyzes the menaquinol -mediated reduction of the activated sulfate (APS), which is converted to sulfite. Further reduction of sulfite to sulfuric acid requires electrons provided by periplasmic hydrogenases [60]. The full reduction of sulfate to sulfur requires a large amount of redox equivalents (8 electron pairs) and 2ATP molecules, thus it has been postulated that several respiratory enzymes such as RNF [61] or Na+-NQR could have an important role in this process.
Horizontal Gene Transfer in the Evolution of nqr Operon
One of the distinguishing aspects of RNF is that its function can vary enormously, depending on the metabolic adaptations of the bacteria in which is found. For instance, in Acetobacterium woodii (anaerobic acetogenic), RNF catalyzes the reduction of NAD (−320 mV), using the redox equivalents of Ferredoxin (−500 mV) [25], [62]. This can be considered as the “forward reaction”, since the electron transfer is downhill, and the energy released is used by the enzyme to pump sodium (Figure 1). In Rhodobacter capsulatus (purple non -sulfur photosynthetic), and probably in other nitrogen fixing bacteria, RNF uses the electrochemical gradient to drive the backwards reaction, reducing ferredoxin, using NADH as electron donor [27]. On the other hand, in E. coli RNF transfers the electrons from NADH, to the low molecular weight SoxR protein, which is a transcription factor that controls the expression of proteins involved in oxidative stress protection [63]. In addition, indirect evidence suggest that in some cases RNF could function as a proton pump [64]. Thus, it seems that the ability of RNF to sustain major catalytic innovations, conserving the basic structural and catalytic mechanisms, may have been a key factor in the evolution of Na+-NQR.
The fact that the nqr operon is found only in certain bacterial lineages, while the rnf operon is widely distributed (Table 2), suggests that RNF might have been the enzymatic ancestor of Na+-NQR. The common ancestry of 5/6 of the Na+-NQR and RNF subunits posits a unique scenario to investigate the origin and evolution of these enzymatic complexes from a common ancestral gene cluster. In order to investigate the origin of the Na+-NQR components, we estimated ML trees from multiple alignments generated for each Na+-NQR/RNF homolog pairs (NqrB/RnfD, NqrD/RnfF, NqrE/RnfA), defining the RNF clades as outgroups (see Table 1 and Methods section for Na+-NQR/RNF homolog-pair details). The NqrF and RnfB subunits are complex-specific and they have no homologous counterpart in RNF or Na+-NQR, respectively. Thereafter, a multiple alignment NqrF/AMOr was generated as well. Each of the four rooted ML trees (Figs. S2–S5) consistently depict C/B and some SR δ -proteobacteria (of the family Desulfobacteraceae) as the earliest diverging sequences within the Na+-NQR subunits. Given the consistent early branching position of the C/B and the “anomalous” position of the SR Desulfobacteraceae nested within them, we hypothesize that the Na+-NQR evolved first within the Chlorobi/Bacteroidetes group, after an ancestral duplication and posterior neofunctionalization of some elements the rnf operon (see discussion below). The phylogenetic analysis suggests that other bacterial lineages acquired the Na+-NQR complex via multiple HGT events (see below).
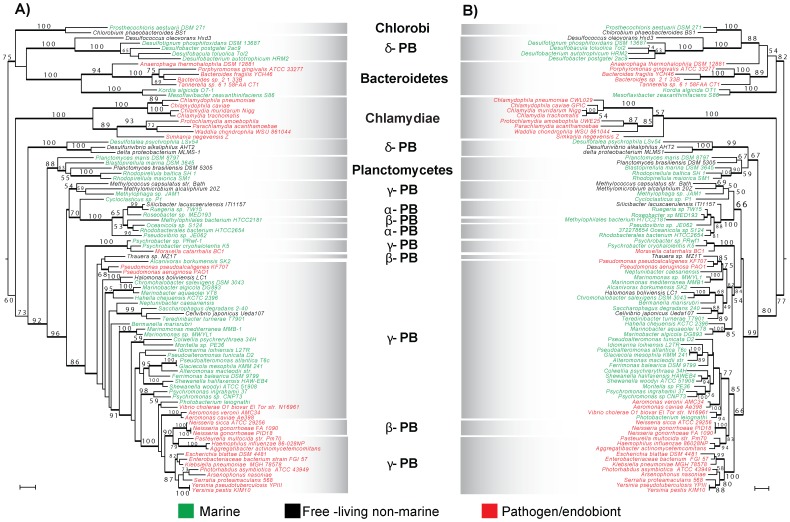
The ML trees shown in figure 2 were estimated with the sequences of subunits NqrB (Fig. 2A) and NqrF (Fig. 2B), respectively, and rooted in the C/B and SR Desulfobacteraceae node. Overall, the topologies of both trees show important consistencies. The Chlamydiae group branches immediately after the defined rooting node. However this position has to be taken with caution given that the Chlamydiae nqr operon is split in three fragments [65], [66] and it is evident the Chlamydiae clade has evolved through a high amino acid replacement rate. The main branch of SR δ -proteobacteria and Planctomyces are found immediately after the Chlamydiae cluster, these two groups are united in a single branch (67% bootstrap support; BS) in the NqrF tree (Fig. 2B), whereas branch separated in NqrB (Fig. 2A). Both ML trees depict a strongly supported clade (92% and 99% BS, respectively) comprising the diverse proteobacterial lineages. Overall, the NqrB (414 residues long in Vibrio cholerae) and NqrF (404 residues) ML trees show considerable resolution in different nodes and it is likely that the catalytic roles of NqrB and NqrF have restrained the substitution rate in the catalytic regions, but allow higher variability in non-essential regions, producing a relatively consistent phylogenetic signal. The topologies of the ML trees of the short subunits NqrD (Fig. S6; 210 residues), NqrE (Fig. S7; 198 residues), NqrC (256 residues; Fig. S9) and the longer NqrA (414 residues; Fig. S8) show some important differences with respect the positions of the Chlamydiae long-branch and the “anomalous” SR δ-proteobacteria branches (i.e., C/B are not branching in the same clade with Chlamydiae and/or SR δ-proteobacteria intermingled; Fig. S6 and S7), therefore in these cases rooting in the Chlorobi/Bacteroidetes clade was not attainable. Even though the resolution and branch support values are relatively low in the NqrD and NqrE trees, similar topological patterns within the proteobacterial clades are consistent with the NqrB and NqrF results. Overall the ML tree topologies suggest a common history underlying the evolution of the whole Na+-NQR complex.
Figure 2. Maximum Likelihood phylogenetic analyses of NqrB (A) and NqrF (B) subunits of the Na+-NQR complex.
Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%) and Bayesian posterior probabilities ≥0.95. Branch lengths are proportional to the number of substitutions per site (scale bars corresponds to 0.1 substitutions per site, respectively). Major bacterial groups are highlighted with shaded boxes. Taxa in red characters are endobionts or pathogens. Organisms in green type set have been found in marine habitats and those in black characters are free-living non-marine bacteria.
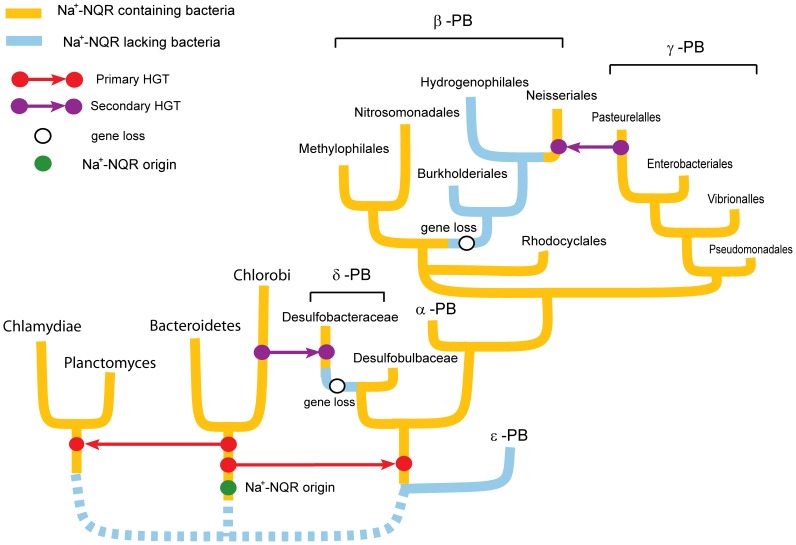
We hypothesize that after the emergence of the Na+-NQR in C/B, it was subsequently dispersed to other bacterial lineages by, at least, three independent HGT events (Figure 3): to the Chlamydiae and Planctomyces ancestor and in two different occasions to proteobacteria. The results suggest that the common ancestor of α, β, γ and δ -proteobacteria originally recruited Na+-NQR via HGT, since it is not found in ε -proteobacteria. Indeed, the topology of the proteobacterial branch resembles the consensus tree of this bacterial lineage, with α, β and δ -proteobacteria in the base of the tree [67]. Also, our trees display a pattern that is similar to γ-proteobacteria phylogeny, with Pasteurelalles and Enterobacteriales in the top branches [68]. It must be pointed out that the minor differences in the proteobacterial clade found in our single-locus trees, in comparison to evolutionary patterns found elsewhere [67], are not unexpected, since those trees were estimated by consensus analysis of dozens of different proteins. The phylogenetic analyses of NqrB (Figs. 2A) and NqrF (Figs. 2B) show that the evolution of Na+-NQR has followed a convoluted pattern inside the proteobacteria lineage, particularly in the case of β and δ -proteobacteria, which appear branching in different locations in the trees. Our data demonstrate that “basal” β-proteobacteria, such as Rhodocyclales (Thauera sp. MX1T) and other species, including Candidatus Accumulibacter phosphatis (not shown), as well as Nitrosomonadales (Nitrosomonas europaea ATCC 19718, not shown) and Methylophilales (Methylophilales bacterium HTCC2181) contain Na+-NQR, which is closely related to α -proteobacteria and “basal” γ -proteobacteria, matching relatively well their phylogenetic history [67]. Additionally, another β-proteobacteria group, comprising exclusively Neisseriales, is found branching with pathogenic γ-proteobacteria, specifically with Pasteurellales (90% and 83% BS in NqrB and NqrF trees, respectively) and is separated from the main β-proteobacteria branch. Interestingly, the sister groups of the Neisseriales, including Hydrogenophilales and Burkholderiales, lack the nqr operon, indicating that the common ancestor of these groups lost the genes coding for Na+-NQR. Thus, it is possible that Neisseriales, which are pathogenic, reacquired the nqr operon from pathogenic γ-proteobacteria (Figure 3).
Figure 3. Multiple horizontal gene transfer (HGT) events involved in the dispersal of the nqr operon.
Our data indicate that Na+-NQR appeared in the common ancestor of the Chlorobi and Bacteroidetes groups. The operon was transferred by HGT to Chlamydiae and Planctomyces, and to the ancestor of α, β, γ and δ–proteobacteria, after this lineage split from ε -proteobacteria. The evidence indicates that both Desulfobacteraceae (δ–proteobacteria) and the ancestor of Burkholderales, Hydrogenophilales and Neisseriales (β–proteobacteria), lost the nqr operon and was reacquired later. In the case of Desulfobacterales, from Chlorobi, and in the case of Neisseriales, from pathogenic Pasteurelalles (γ–proteobacteria).
As mentioned before, a group of SR δ-proteobacteria (Desulfobacteraceae) branch consistently with Chlorobi/Bacteroidetes (Figs. 2A, 2B). This topology does not reflect the evolutionary history of these non-related bacterial major groups, but instead may indicate a more recent HGT event from C/B into Desulfobacteraceae. This transference may have included other genes, not only Na+-NQR, as shown by Biegel et al, in the evolutionary analysis of RnfD, which shows almost exactly the same topology [25]. Moreover, SR δ-proteobacteria and Chlorobi contain other metabolic pathways that are not common among bacteria, such as the reductive Tricarboxylic acid cycle [69], [70]. Thus, it appears that these two distantly related bacterial lineages have exchanged a significant amount of genetic information. It should be pointed out that several proteobacteria have lost Na+-NQR, most conspicuously Escherichia coli. This effect probably represents adaptations of different organisms to their environmental conditions. Surprisingly, the nqr operon can also found in the closely related enteric bacteria Escherichia blattae.
The ML trees of NqrB and NqrF also show that Na+-NQR was likely spread to Chlamydiae and Planctomyces, through one or two lateral ancient HGT events. Phylogenetic trees obtained with 16S rRNA have located Chlamydia and Planctomyces as distantly related groups [71]. Furthermore, peptidoglycan, which is a universal component of bacterial cell walls, is absent in the cell walls of both Chlamydiae [72] and Plantomyces [73], [74]. Interestingly, Chlamydiae contains most of the genes involved in peptidoglycan synthesis [65], [75] and the addition of β-lactam antibiotics blocks its normal development [76], but their cell walls do not contain detectable amounts of peptidoglycans. Thus, it is possible that Na+-NQR may have been transferred from C/B to the common ancestor of Chlamydiae and Planctomyces, and that Chlamydiae Na+-NQR would have evolved faster, including a fragmentation of the nqr operon, than the Planctomyces homologs, explaining the Chlamydiae long branch and the separation of both lineages in the trees.
An important ecological aspect that may have contributed to the evolution and spread of Na+-NQR to different bacterial lineages could have been a limited iron availability, which is now reflected on the particular cofactor composition of the “modern” Na+-NQR complex. One of the most intriguing aspects of this enzyme is the preferential use of flavin molecules compared to iron containing cofactors [1]. While most respiratory enzymes contain a large number of cofactors that contain iron, including complex I that has up to nine Fe-S centers [77], Na+-NQR contains only one 2Fe-2S center and four flavin molecules. Iron has been considered one of the key elements involved in the origin of life, probably forming part of the first cofactors in enzymatic reactions [78], [79]. The ability to use minimal amounts of iron could represent an early adaptation of primordial anaerobic cells to the rising levels of oxygen. While ferrous iron is “life -compatible” and easy accessible under anaerobic conditions, due to its high solubility, under aerobic conditions ferric iron has a very low solubility and can react easily with oxygen, producing superoxide that can damage different cell components [78], [80], [81]. On the other hand, in the case of parasitic organisms, where Na+-NQR is well represented [8], [55], the ability of operate with minimal amounts of iron would be advantageous, since an important part of the immunity mechanisms of the body include the limitation of the availability of iron or its complete depletion in the infection site [81], [82].
The Organizations of the nqr Operon and the Origin of the Na+-NQR Complex
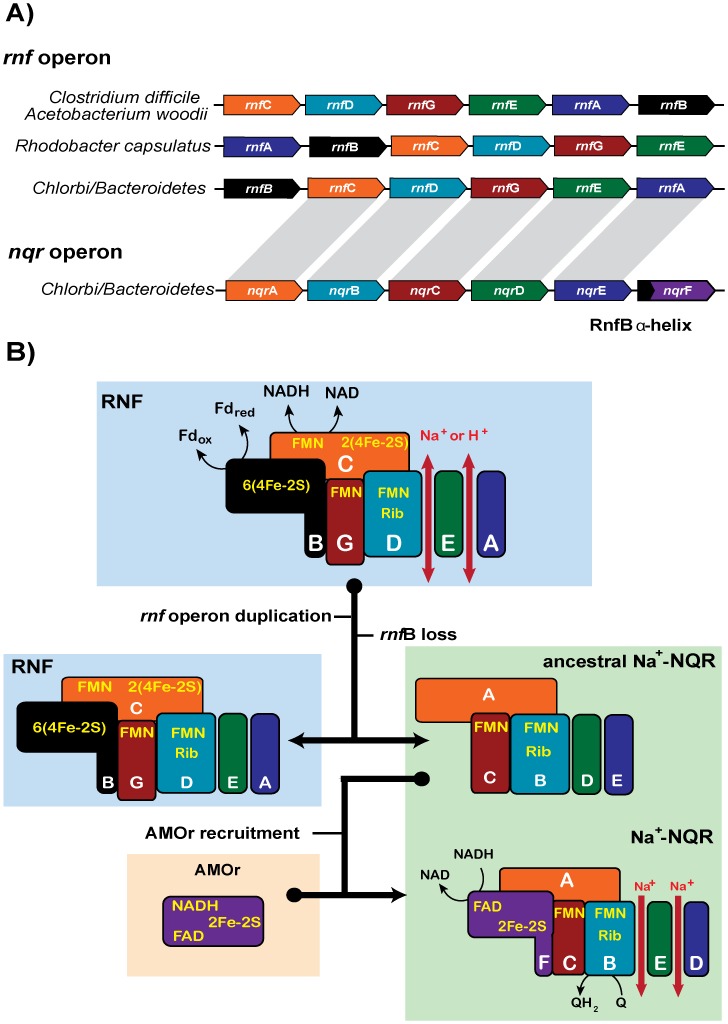
The phylogenetic results and the distributions of the rnf and nqr operons in the major bacterial lineages suggest that Na+-NQR appeared from the duplication of the rnf operon in the C/B common ancestor. After its putative duplication, the original rnf operon kept its function, as sodium -pumping ferredoxin: NADH oxidoreductase, while the paralogue copy lost a key gene (Fig. 4A and 4B). Under our proposal, the gene encoding the subunit RnfB, which is probably involved in the electron uptake from reduced ferredoxin [25], was lost in the nqr-predecessor operon after the duplication event, although the core organization of the operon elements (rnfCDGEA/nqrABCDE) remained essentially identical. Examination of the rnf operon organization in different bacteria offers further evidence to support this hypothesis (Fig. 4A). In organisms such as R. capsulatus, and most proteobacteria, the genes in the rnf operon are assembled in the order rnfABCDGE [83]. The origin of nqr operon in these types of bacteria would have not only involved the duplication of the rnf operon, but major gene rearrangements, which makes this scenario unlikely. On the other hand, in the C/B lineage, the organization of the rnf operon is rnfBCDGEA, very similar to the organization found in the typical nqr operon [25]. Thus, it is conceivable that the current organization of the nqr operon would have appeared first in the C/B rnf operon, fact reflected in the conserved order of the homologous genes. There are other bacteria in which the organization of the rnf operon could have also led to the evolution of Na+-NQR, such as in Firmicutes, including different species of the Clostridium genus [84], [85] and A. woodii [62], where the organization of the genes is rnfCDGEB, but in contrast to the case of C/B, they do not harbor the nqr operon (Table 2). It must be pointed out that some authors have proposed that the RNF complex might include other subunits, such as RnfH [83], RnfX, RnfY [86] or endonuclease III [87]. However, these subunits are not found in the rnf operons in other bacteria and their role in the function of RNF is still obscure.
Figure 4. Model of Na+-NQR complex origin after the ancestral duplication of the rnf operon.
A) Gene organization of the rnf operon in diverse bacterial lineages. The order rnfBCDGEA of the Chlorobi/Bacteroidetes rnf operon resembles the gene organization nqrABCDEF of the homologues genes that constitute the rnf operon of most bacteria. B) Schematic model for the functional evolution of the Na+-NQR complex from a duplicated RNF complex encoded by an rnf operon which likely lost the rnfB gene. According to our hypothesis subunit NqrF complex evolved from the reductase subunit of aromatic monoxygenase (AMOr) family that recruited the α -helix region of the former RnfB subunit.
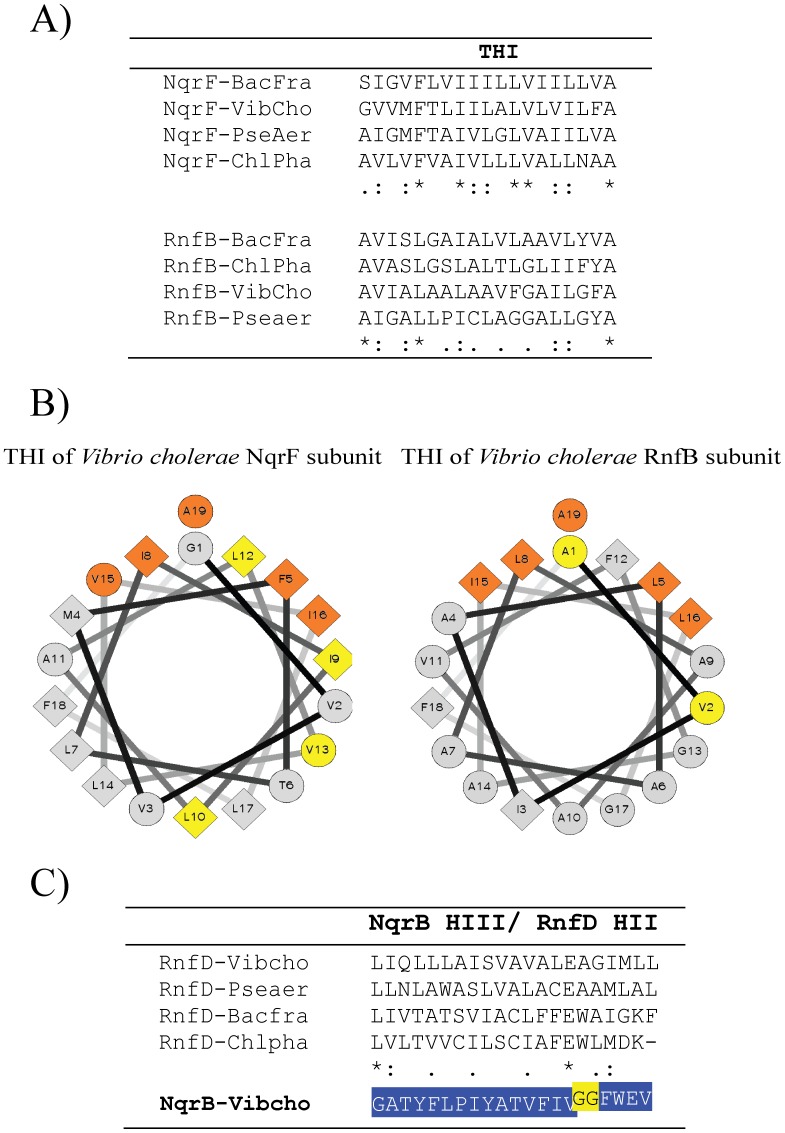
We propose that the loss of the rnfB gene in the duplicated rnf operon was the key event that elicited the evolution of Na+-NQR, triggering the evolutionary process underlying the change in the function of RNF into Na+-NQR, which included the switch in electron acceptor and donor. In Na+-NQR, the electrons are accepted by the FAD molecule in subunit NqrF and are transferred according to the pathway shown in Figure 1 [14]. On the other hand, Biegel et al [25] postulated that in A. woodi RNF, the electrons may enter the enzyme in RnfB subunit, which contains a polyferredoxin domain, with a number of Fe-S centers, and would follow a similar electron pathway as found in Na+-NQR (Figure 1). In the scenario depicted in Fig. 4, the loss of RnfB would have rendered the RnfC subunit non -functional, which eventually might have led to the loss of the binding sites for NAD and FMN, as well as the cysteines involved in the binding of the two 4Fe-4S clusters, but nonetheless this subunit retained its structural role, thus giving rise to the NqrA subunit. Indeed, NqrA contains reminiscences of the substrate and cofactor binding sites of RnfC, but the critical residues involved in the binding have been lost (Figure S1). After the RnfB loss, the next step in Na+-NQR evolution might have been the recruitment of a “new” electron-accepting module, which is the NADH dehydrogenase module in NqrF. As mentioned before, none of the RNF complex subunits are evolutionary related to NqrF and the closest known homologue is the AMOr subunit. We postulate that a gene encoding an AMOr was recruited by the primordial C/B nqrABCDE operon, establishing its function as the reductase module of Na+-NQR complex (Fig. 4B). However, a complete integration of this subunit into the complex required the acquisition of a membrane attachment domain, since typical AMOr are hydrophilic proteins with no membrane attachment sites. Although the transmembrane segment is the most variable part of NqrF, it contains the conserved general structure: X4-F-X2-I-X-(I/V/L/M)-X-L-X3-(L/I)-X3-(A/S), implying that it is not only an membrane anchor site, but also should be able to interact with the transmembrane segments of other subunits. Remarkably, helix I of the RnfB subunit resembles the transmembrane helix of NqrF, sharing a very similar structure A-(V/I)-X2-L-X2-(L/I)-X6-(V/I/L)-(I/L)-X2-(A). The wheel projections of both helices show that positions 5, 8, 15, 16 and 19 have similar residues and are conserved in both transmembrane segments and are concentrated in a relatively small face of the helical cylinder (Figure 5B). Thus, it is likely that the NqrF subunit evolved as the result of the gene fusion of AMOr and the first transmembrane segment of RnfB or that the acquisition of the membrane domain could have followed a convergent evolution pattern, producing a structure similar to the RnfB transmembrane helix that was able to establish the same structural contacts. Under both of these hypotheses, the integration of NqrF into the complex would have been facilitated through the hydrophobic interactions of the transmembrane segment with other subunits.
Figure 5. Comparative analysis of Na+-NQR and RNF transmembrane helices.
A) Multiple sequence alignment of the transmembrane helix I of NqrF and RnfB subunits. B) Helical wheel projection of Vibrio cholerae NqrF and RnfB transmembrane helices. Conserved residues within NqrF and RnfB subunits are highlighted in yellow, and residues that are conserved in both NqrF and RnfB are found in orange. C) Multiple sequence alignment of the transmembrane helix II of RnfD subunit compared to the homologous helix III of NqrB subunit. The position of the two conserved glycine residues 140 and 141 in NqrB is highlighted in yellow. In this panel the conserved and semiconserved residues correspond exclusively to the sequences of RnfD. BacFra; Bacillus fragilis, VibCho; Vibrio cholerae, PseAer; Pseudomonas aeruginosa, ChlPha; Chlorobium phaeobacteroides.
Moreover, another important part of Na+-NQR redox partner switching is the use of ubiquinone as electron acceptor. Clues about this molecular event can be found by examining the topology and sequence of the subunits NqrB and RnfD. Figure 5C shows the sequence alignment of transmembrane helix III of V. cholerae NqrB subunit (shown highlighted in blue as reference on the bottom), compared to the homologous region of RnfD (transmembrane helix II) from different bacteria. Helix III in NqrB subunit contains two conserved glycine residues that are involved in the formation of the ubiquinone binding site and whose mutation, even to other small residues, such as alanine or valine, disrupts completely the interaction with ubiquinone [14]. Although these residues might not interact directly with the substrate, they probably create a cavity for ubiquinone. The analysis of the homologous transmembrane segment in RnfD shows that this helix does not contain the two conserved glycine residues. Thus, it is evident that the acquisition of the ubiquinone reductase activity involved the opening of the ubiquinone binding site, through the loss of bulky residues in transmembrane helix III and their substitution by glycine. In addition, our data indicate that Na+-NQR originally evolved as a NADH: menaquinone reductase, since ubiquinone (Em = +90 mV) is absent in both Chlorobi [88]–[90] and Bacteroidetes [90], [91], and instead they present the low potential analog, menaquinone (Em = −80 mV). The ability to use menaquinone might be one of the key elements that determined the spread to other types of bacteria, for instance to δ-proteobacteria and planctomyces, which also contain menaquinone [90]. The ability of Na+-NQR to use ubiquinone appeared later on evolution, probably first in aerobic α-proteobacteria and was inherited to β and γ-proteobacteria, all of which contain exclusively ubiquinone [90]. It is important to highlight that the mechanism of sodium translocation by Na+-NQR should be essentially the same with the two redox electron acceptors, since the reduction of the quinone molecule is not involved directly in sodium translocation [40]. The role played by riboflavin is another aspect to consider during the transition of the catalytic mechanism of RNF, to become Na+-NQR. Our group described that riboflavin is a functional cofactor in Na+-NQR, delivering the electrons to the final substrate [31], [52], a very unusual and probably unique case in nature. Little inferences can be done at this point about the evolutionary mechanisms involved in the acquisition of riboflavin, since the location of this cofactor remains elusive. However, W. Buckel’s group found evidence indicating that RNF also contains riboflavin [92]. Thus, it seems that the evolution of Na+-NQR did not require the acquisition of riboflavin, but merely an adaptation in the function of RNF.
In addition to the subunit acquisition and motif evolution, the adjustment in the midpoint potentials of the redox centers should have been one of the most important aspects necessary to complete the transition from RNF into Na+-NQR. The forward reaction catalyzed by RNF involves the electron transfer from Ferredoxin (Em = −500 mV) to NADH (Em = −320 mM) [62]. In Na+-NQR, the electrons move downhill from NADH to ubiquinone (Em = +90mV), following a pathway in which most redox centers have midpoint potentials higher than NADH [93], [94]. Hence, in order for the covalently -bound FMN molecules (and presumably for riboflavin too) to become functional in Na+-NQR, a significant change in the redox potential should have occurred, increasing the potentials probably by 50–490 mV, depending on the specific cofactor. In most proteins, the control of the redox potential of the centers is achieved by modifying the environment surrounding the cofactors. For instance, classic studies on Myoglobin have shown that the mutation of valine 68, which is close to the heme group, to aspartate or asparagine decreases the midpoint potential by 200 or 80 mV, respectively [95]. In flavin proteins, the dielectric constant of the immediate environment [96] and the interactions with aromatic residues have been reported to affect strongly the midpoint potential of the flavin cofactors [97], [98]. Thus, it is possible that in the evolution of Na+-NQR the environment of the flavins was modified, replacing negatively charged residues and decreasing the polarity of the flavin environment, which will increase the midpoint potential of the three flavin cofactors.
Conclusion
The distribution and the phylogenetic history of the Na+-NQR subunits in different bacterial lineages suggest that the nqr operon, and subsequently the Na+-NQR complex, is the outcome of an ancestral duplication of the homologous rnf operon. Here, we present evidence that suggest a series of molecular events, comprising a specific partial gene loss, a gene fusion and enzymatic recruitment, that occurred during the divergence and neo-fuctionalization of the ancestral versatile RNF complex into the specialized sodium pump that became the modern Na+-NQR assemblage. Moreover, our phylogenic survey indicates that the putative duplication of the rnf operon occurred in the C/B group, and later the ancestral nqr operon dispersed via independent HGT events into different proteobacterial groups, and Chlamydiae and Planctomyces. This model is consistent with the idea that genes with related functions and organized in clusters (i.e., operons) are prone material for successful HGT, given that the encoded products, as an effective functional unit, are potentially able to confer novel metabolic traits, such as sodium pumping coupled to electron transport. Our analyses highlight the major relevance that duplication of genes, or clusters of genes, has in the emergence and consolidation of evolutionary innovations. In this case underlying the diversification and expansion of the bacterial metabolic capabilities under particular ecologic scenarios, such as the use of sodium gradients across membranes to generate electromotive forces when proton “inverted” (i.e., alkaline outside) gradients prevail in the surroundings. Our model posits that the emergence, and later independent recruitments, of the Na+-NQR allowed different types of bacteria with diverse metabolic adaptations to take advantage of the abundance of Na+ ions in particular habitats, such as marine, alkaline and intracellular environments. Finally, our work invites to thoroughly investigate the presence of paralogous operons in prokaryotic genomes to study the magnitude that duplication of gene clusters have had in the diversification and expansion of the microbial primary metabolism.
Supporting Information
Alignment of NqrA and RnfC subunits. Secondary structure prediction of Anaerophaga thermohalophila (Bacteroidetes) RnfC and NqrA. Figure shows the secondary structure prediction consensus of eight algorithms, using the Network Protein Sequence Analysis software. The conserved residues of RnfC involved in NADH and FAD binding sites, as well as the cysteine residues involved in the 4Fe-4S centers attachement are highlighted in black.
(PDF)
Rooted phylogenetic tree of NqrB subunit. Maximum Likelihood phylogenetic analysis of the NqrB subunit of the Na+-NQR complex and the homolog RnfD subunit from the RNF complex. The tree was rooted using the RnfD clade as outgroup. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site. NCBI GI numbers precede the taxa identification.
(PDF)
Rooted phylogenetic tree of NqrF subunit. Maximum Likelihood phylogenetic analysis of the NqrF subunit of the Na+-NQR complex and sequences of the reductase subunit of aromatic monooxigenases (AMOr), which are the closes know homologues of NqrF. The tree was rooted using the AMOr (benC/ToL) proteins as outgroup. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site. NCBI GI numbers precede the taxa identification.
(PDF)
Rooted phylogenetic tree of NqrD subunit. Maximum Likelihood phylogenetic analysis of the NqrD subunit of the Na+-NQR complex and the homolog RnfE subunit from the RNF complex. The RnfE branch was defined as outgroup. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site. NCBI GI numbers precede the taxa identification.
(PDF)
Rooted phylogenetic tree of NqrE subunit. Maximum Likelihood phylogenetic analysis of the NqrE subunit of the Na+-NQR complex and the homolog RnfA subunit from the RNF complex. The tree was rooted using the RnfA branch (100% BS) as outgroup. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site. NCBI GI numbers precede the taxa identification.
(PDF)
Unrooted phylogenetic tree of NqrD subunit. Maximum Likelihood tree of the subunit D (NqrD) of the Na+-NQR complex. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site.
(PDF)
Unrooted phylogenetic tree of NqrE subunit. Maximum Likelihood tree of the subunit NqrE of the Na+-NQR complex. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site.
(PDF)
Unrooted phylogenetic tree of NqrA subunit. Maximum Likelihood tree of the subunit A (NqrA) of the Na+-NQR complex. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site.
(PDF)
Unrooted phylogenetic tree of NqrC subunit. Maximum Likelihood tree of the subunit NqrC of the Na+-NQR complex. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site.
(PDF)
List of the analyzed bacterial taxa harboring Na+-NQR complex.
(PDF)
Similarity profiles of the Na+-NQR subunits with the closest homologues in RNF and AMOr.
(PDF)
Acknowledgments
Authors would like to thank Dr. R.K. Thauer and Dr. W. Buckel for helpful discussions.
Funding Statement
This work was partially supported by NSF Grant MCB 1052234 (OJ and BB), and NSERC Discovery 402421-2011 CFI-LOF 28276 and NBIF RIF2012-006 (ARP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Juárez O, Barquera B (2012) Insights into the mechanism of electron transfer and sodium translocation of the Na(+)-pumping NADH:quinone oxidoreductase. Biochim Biophys Acta 1817: 1823–1832 10.1016/j.bbabio.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bogachev AV, Verkhovsky MI (2005) Na(+)-Translocating NADH:quinone oxidoreductase: progress achieved and prospects of investigations. Biochem Biokhimiia 70: 143–149. [DOI] [PubMed] [Google Scholar]
- 3. Hayashi M, Nakayama Y, Unemoto T (2001) Recent progress in the Na(+)-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. Biochim Biophys Acta 1505: 37–44. [DOI] [PubMed] [Google Scholar]
- 4. Barquera B, Hellwig P, Zhou W, Morgan JE, Häse CC, et al. (2002) Purification and characterization of the recombinant Na(+)-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry (Mosc) 41: 3781–3789. [DOI] [PubMed] [Google Scholar]
- 5. Zhou W, Bertsova YV, Feng B, Tsatsos P, Verkhovskaya ML, et al. (1999) Sequencing and preliminary characterization of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio harveyi. Biochemistry (Mosc) 38: 16246–16252. [DOI] [PubMed] [Google Scholar]
- 6. Bertsova YV, Bogachev AV (2004) The origin of the sodium-dependent NADH oxidation by the respiratory chain of Klebsiella pneumoniae. FEBS Lett 563: 207–212 10.1016/S0014-5793(04)00312-6 [DOI] [PubMed] [Google Scholar]
- 7. Hayashi M, Nakayama Y, Unemoto T (1996) Existence of Na+-translocating NADH-quinone reductase in Haemophilus influenzae. FEBS Lett 381: 174–176. [DOI] [PubMed] [Google Scholar]
- 8.Häse CC, Fedorova ND, Galperin MY, Dibrov PA (2001) Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol Mol Biol Rev MMBR 65: 353–370, table of contents. doi:10.1128/MMBR.65.3.353-370.2001. [DOI] [PMC free article] [PubMed]
- 9. Häse CC, Barquera B (2001) Role of sodium bioenergetics in Vibrio cholerae. Biochim Biophys Acta 1505: 169–178. [DOI] [PubMed] [Google Scholar]
- 10. Häse CC, Mekalanos JJ (1998) TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A 95: 730–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Juárez O, Shea ME, Makhatadze GI, Barquera B (2011) The Role and Specificity of the Catalytic and Regulatory Cation-binding Sites of the Na+-pumping NADH:Quinone Oxidoreductase from Vibrio cholerae. J Biol Chem 286: 26383–26390 10.1074/jbc.M111.257873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rich PR, Meunier B, Ward FB (1995) Predicted structure and possible ionmotive mechanism of the sodium-linked NADH-ubiquinone oxidoreductase of Vibrio alginolyticus. FEBS Lett 375: 5–10. [DOI] [PubMed] [Google Scholar]
- 13. Barquera B, Nilges MJ, Morgan JE, Ramirez-Silva L, Zhou W, et al. (2004) Mutagenesis study of the 2Fe-2S center and the FAD binding site of the Na(+)-translocating NADH:ubiquinone oxidoreductase from Vibrio cholerae. Biochemistry (Mosc) 43: 12322–12330 10.1021/bi048689y [DOI] [PubMed] [Google Scholar]
- 14. Juárez O, Neehaul Y, Turk E, Chahboun N, DeMicco JM, et al. (2012) The role of glycine residues 140 and 141 of subunit B in the functional ubiquinone binding site of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J Biol Chem 287: 25678–25685 10.1074/jbc.M112.366088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Casutt MS, Huber T, Brunisholz R, Tao M, Fritz G, et al. (2010) Localization and function of the membrane-bound riboflavin in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J Biol Chem 285: 27088–27099 10.1074/jbc.M109.071126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duffy EB, Barquera B (2006) Membrane topology mapping of the Na+-pumping NADH: quinone oxidoreductase from Vibrio cholerae by PhoA-green fluorescent protein fusion analysis. J Bacteriol 188: 8343–8351 10.1128/JB.01383-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Juárez O, Athearn K, Gillespie P, Barquera B (2009) Acid residues in the transmembrane helices of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae involved in sodium translocation. Biochemistry (Mosc) 48: 9516–9524 10.1021/bi900845y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neehaul Y, Juárez O, Barquera B, Hellwig P (2013) Infrared spectroscopic evidence of a redox-dependent conformational change involving ion binding residue NqrB-D397 in the Na(+)-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry (Mosc) 52: 3085–3093 10.1021/bi4000386 [DOI] [PubMed] [Google Scholar]
- 19. Shea ME, Juárez O, Cho J, Barquera B (2013) Aspartic acid 397 in subunit B of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae forms part of a sodium-binding site, is involved in cation selectivity, and affects cation-binding site cooperativity. J Biol Chem 288: 31241–31249 10.1074/jbc.M113.510776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verkhovsky MI, Bogachev AV (2010) Sodium-translocating NADH:quinone oxidoreductase as a redox-driven ion pump. Biochim Biophys Acta 1797: 738–746 10.1016/j.bbabio.2009.12.020 [DOI] [PubMed] [Google Scholar]
- 21. Casutt MS, Nedielkov R, Wendelspiess S, Vossler S, Gerken U, et al. (2011) Localization of ubiquinone-8 in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J Biol Chem 286: 40075–40082 10.1074/jbc.M111.224980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nedielkov R, Steffen W, Steuber J, Möller HM (2013) NMR reveals double occupancy of quinone-type ligands in the catalytic quinone binding site of the Na+-translocating NADH:Quinone oxidoreductase from Vibrio cholerae. J Biol Chem 288: 30597–30606 10.1074/jbc.M112.435750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, et al. (1993) Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet MGG 241: 602–615. [DOI] [PubMed] [Google Scholar]
- 24. Kumagai H, Fujiwara T, Matsubara H, Saeki K (1997) Membrane localization, topology, and mutual stabilization of the rnfABC gene products in Rhodobacter capsulatus and implications for a new family of energy-coupling NADH oxidoreductases. Biochemistry (Mosc) 36: 5509–5521 10.1021/bi970014q [DOI] [PubMed] [Google Scholar]
- 25. Biegel E, Schmidt S, González JM, Müller V (2011) Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci CMLS 68: 613–634 10.1007/s00018-010-0555-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeong HS, Jouanneau Y (2000) Enhanced nitrogenase activity in strains of Rhodobacter capsulatus that overexpress the rnf genes. J Bacteriol 182: 1208–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hess V, Schuchmann K, Müller V (2013) The ferredoxin:NAD+ oxidoreductase (Rnf) from the acetogen Acetobacterium woodii requires Na+ and is reversibly coupled to the membrane potential. J Biol Chem 288: 31496–31502 10.1074/jbc.M113.510255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schlegel K, Welte C, Deppenmeier U, Müller V (2012) Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J 279: 4444–4452 10.1111/febs.12031 [DOI] [PubMed] [Google Scholar]
- 29. Imkamp F, Biegel E, Jayamani E, Buckel W, Müller V (2007) Dissection of the caffeate respiratory chain in the acetogen Acetobacterium woodii: identification of an Rnf-type NADH dehydrogenase as a potential coupling site. J Bacteriol 189: 8145–8153 10.1128/JB.01017-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Müller V, Imkamp F, Biegel E, Schmidt S, Dilling S (2008) Discovery of a ferredoxin:NAD+-oxidoreductase (Rnf) in Acetobacterium woodii: a novel potential coupling site in acetogens. Ann N Y Acad Sci 1125: 137–146 10.1196/annals.1419.011 [DOI] [PubMed] [Google Scholar]
- 31. Juárez O, Nilges MJ, Gillespie P, Cotton J, Barquera B (2008) Riboflavin is an active redox cofactor in the Na+-pumping NADH: quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J Biol Chem 283: 33162–33167 10.1074/jbc.M806913200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barquera B, Morgan JE, Lukoyanov D, Scholes CP, Gennis RB, et al. (2003) X- and W-band EPR and Q-band ENDOR studies of the flavin radical in the Na+ -translocating NADH:quinone oxidoreductase from Vibrio cholerae. J Am Chem Soc 125: 265–275 10.1021/ja0207201 [DOI] [PubMed] [Google Scholar]
- 33. Hayashi M, Nakayama Y, Yasui M, Maeda M, Furuishi K, et al. (2001) FMN is covalently attached to a threonine residue in the NqrB and NqrC subunits of Na(+)-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett 488: 5–8. [DOI] [PubMed] [Google Scholar]
- 34. Backiel J, Juárez O, Zagorevski DV, Wang Z, Nilges MJ, et al. (2008) Covalent binding of flavins to RnfG and RnfD in the Rnf complex from Vibrio cholerae. Biochemistry (Mosc) 47: 11273–11284 10.1021/bi800920j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wunsch P, Zumft WG (2005) Functional domains of NosR, a novel transmembrane iron-sulfur flavoprotein necessary for nitrous oxide respiration. J Bacteriol 187: 1992–2001 10.1128/JB.187.6.19922001.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cramer WA, Hasan SS, Yamashita E (2011) The Q cycle of cytochrome bc complexes: a structure perspective. Biochim Biophys Acta 1807: 788–802 10.1016/j.bbabio.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu S-S (2010) Mitochondrial Q cycle-derived superoxide and chemiosmotic bioenergetics. Ann N Y Acad Sci 1201: 84–95 10.1111/j.1749-6632.2010.05632.x [DOI] [PubMed] [Google Scholar]
- 38. Kim YC, Wikström M, Hummer G (2007) Kinetic models of redox-coupled proton pumping. Proc Natl Acad Sci U S A 104: 2169–2174 10.1073/pnas.0611114104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wikström M, Verkhovsky MI (2006) Towards the mechanism of proton pumping by the haem-copper oxidases. Biochim Biophys Acta 1757: 1047–1051 10.1016/j.bbabio.2006.01.010 [DOI] [PubMed] [Google Scholar]
- 40. Juárez O, Morgan JE, Nilges MJ, Barquera B (2010) Energy transducing redox steps of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Proc Natl Acad Sci U S A 107: 12505–12510 10.1073/pnas.1002866107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Katoh K, Standley DM (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol 30: 772–780 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Castresana J (2000) Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol Biol Evol 17: 540–552. [DOI] [PubMed] [Google Scholar]
- 43. Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247: 536–540 10.1006/jmbi.1995.0159 [DOI] [PubMed] [Google Scholar]
- 44. Rost B (1999) Twilight zone of protein sequence alignments. Protein Eng 12: 85–94. [DOI] [PubMed] [Google Scholar]
- 45. Koehl P, Levitt M (2002) Sequence variations within protein families are linearly related to structural variations. J Mol Biol 323: 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doolittle RF (1986) Of Urfs and Orfs: A Primer on how to Analyze Derived Amino Acid Sequences. University Science Books. 118 p. [Google Scholar]
- 47. Pessione E, Divari S, Griva E, Cavaletto M, Rossi GL, et al. (1999) Phenol hydroxylase from Acinetobacter radioresistens is a multicomponent enzyme. Purification and characterization of the reductase moiety. Eur J Biochem FEBS 265: 549–555. [DOI] [PubMed] [Google Scholar]
- 48. Le SQ, Gascuel O (2008) An Improved General Amino Acid Replacement Matrix. Mol Biol Evol 25: 1307–1320 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- 49. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinforma Oxf Engl 22: 2688–2690 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 50. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- 51. Han MV, Zmasek CM (2009) phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics 10: 356 10.1186/1471-2105-10-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Juárez O, Morgan JE, Barquera B (2009) The Electron Transfer Pathway of the Na+-pumping NADH:Quinone Oxidoreductase from Vibrio cholerae. J Biol Chem 284: 8963–8972 10.1074/jbc.M809395200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Quinn MJ, Resch CT, Sun J, Lind EJ, Dibrov P, et al. (2012) NhaP1 is a K+(Na+)/H+ antiporter required for growth and internal pH homeostasis of Vibrio cholerae at low extracellular pH. Microbiol Read Engl 158: 1094–1105 10.1099/mic.0.056119-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nakajo K, Komori R, Ishikawa S, Ueno T, Suzuki Y, et al. (2006) Resistance to acidic and alkaline environments in the endodontic pathogen Enterococcus faecalis. Oral Microbiol Immunol 21: 283–288 10.1111/j.1399-302X.2006.00289.x [DOI] [PubMed] [Google Scholar]
- 55. Skulachev VP (1989) The sodium cycle: a novel type of bacterial energetics. J Bioenerg Biomembr 21: 635–647. [DOI] [PubMed] [Google Scholar]
- 56. Xu Q, Dziejman M, Mekalanos JJ (2003) Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci U S A 100: 1286–1291 10.1073/pnas.0337479100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Häse CC, Mekalanos JJ (1999) Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A 96: 3183–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sorokin DY, Tourova TP, Mussmann M, Muyzer G (2008) Dethiobacter alkaliphilus gen. nov. sp. nov., and Desulfurivibrio alkaliphilus gen. nov. sp. nov.: two novel representatives of reductive sulfur cycle from soda lakes. Extrem Life Extreme Cond 12: 431–439 10.1007/s00792-008-0148-8 [DOI] [PubMed] [Google Scholar]
- 59. Crossman LC, Chen H, Cerdeño-Tárraga A-M, Brooks K, Quail MA, et al. (2013) A small predatory core genome in the divergent marine Bacteriovorax marinus SJ and the terrestrial Bdellovibrio bacteriovorus. ISME J 7: 148–160 10.1038/ismej.2012.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pereira IAC, Ramos AR, Grein F, Marques MC, da Silva SM, et al. (2011) A Comparative Genomic Analysis of Energy Metabolism in Sulfate Reducing Bacteria and Archaea. Front Microbiol 2. Available: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3119410/. Accessed 5 July 2013. [DOI] [PMC free article] [PubMed]
- 61. Strittmatter AW, Liesegang H, Rabus R, Decker I, Amann J, et al. (2009) Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ Microbiol 11: 1038–1055 10.1111/j.1462-2920.2008.01825.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Biegel E, Müller V (2010) Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc Natl Acad Sci U S A 107: 18138–18142 10.1073/pnas.1010318107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Koo M-S, Lee J-H, Rah S-Y, Yeo W-S, Lee J-W, et al. (2003) A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J 22: 2614–2622 10.1093/emboj/cdg252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Köpke M, Held C, Hujer S, Liesegang H, Wiezer A, et al. (2010) Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc Natl Acad Sci U S A 107: 13087–13092 10.1073/pnas.1004716107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, et al. (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282: 754–759. [DOI] [PubMed] [Google Scholar]
- 66. Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, et al. (1999) Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet 21: 385–389 10.1038/7716 [DOI] [PubMed] [Google Scholar]
- 67. Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, et al. (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287 10.1126/science.1123061 [DOI] [PubMed] [Google Scholar]
- 68. Williams KP, Gillespie JJ, Sobral BWS, Nordberg EK, Snyder EE, et al. (2010) Phylogeny of Gammaproteobacteria. J Bacteriol 192: 2305–2314 10.1128/JB.01480-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schauder R, Widdel F, Fuchs G (1987) Carbon assimilation pathways in sulfate-reducing bacteria II. Enzymes of a reductive citric acid cycle in the autotrophic Desulfobacter hydrogenophilus. Arch Microbiol 148: 218–225 10.1007/BF00414815 [DOI] [Google Scholar]
- 70. Fuchs G, Stupperich E, Eden G (1980) Autotrophic CO2 fixation in Chlorobium limicola. Evidence for the operation of a reductive tricarboxylic acid cycle in growing cells. Arch Microbiol 128: 64–71 10.1007/BF00422307 [DOI] [Google Scholar]
- 71. Weisburg WG, Hatch TP, Woese CR (1986) Eubacterial origin of chlamydiae. J Bacteriol 167: 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Moulder JW (1993) Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect Agents Dis 2: 87–99. [PubMed] [Google Scholar]
- 73. Fuerst JA, Sagulenko E (2011) Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9: 403–413 10.1038/nrmicro2578 [DOI] [PubMed] [Google Scholar]
- 74. Kerger BD, Mancuso CA, Nichols PD, White DC, Langworthy T, et al. (1988) The budding bacteria, Pirellula and Planctomyces, with atypical 16S rRNA and absence of peptidoglycan, show eubacterial phospholipids and uniquely high proportions of long chain beta-hydroxy fatty acids in the lipopolysaccharide lipid A. Arch Microbiol. 149: 255–260 10.1007/BF00422014 [DOI] [Google Scholar]
- 75.McClarty G (1999) Chlamydial metabolism as inferred from the complete genome sequence. Chlamydia Intracell Biol Pathog Immun Am Soc Microbiol Wash DC: 69–100.
- 76. Lambden PR, Pickett MA, Clarke IN (2006) The effect of penicillin on Chlamydia trachomatis DNA replication. Microbiology 152: 2573–2578 10.1099/mic.0.29032-0 [DOI] [PubMed] [Google Scholar]
- 77. Yagi T (1991) Bacterial NADH-quinone oxidoreductases. J Bioenerg Biomembr 23: 211–225. [DOI] [PubMed] [Google Scholar]
- 78. Hsiao C, Chou I-C, Okafor CD, Bowman JC, O’Neill EB, et al. (2013) RNA with iron(II) as a cofactor catalyses electron transfer. Nat Chem 5: 525–528 10.1038/nchem.1649 [DOI] [PubMed] [Google Scholar]
- 79. Beinert H, Holm RH, Münck E (1997) Iron-Sulfur Clusters: Nature’s Modular, Multipurpose Structures. Science 277: 653–659 10.1126/science.277.5326.653 [DOI] [PubMed] [Google Scholar]
- 80. Anbar AD (2008) Elements and Evolution. Science 322: 1481–1483 10.1126/science.1163100 [DOI] [PubMed] [Google Scholar]
- 81. Andrews SC, Robinson AK, Rodríguez-Quiñones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27: 215–237 10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- 82. Schaible UE, Kaufmann SHE (2004) Iron and microbial infection. Nat Rev Microbiol 2: 946–953 10.1038/nrmicro1046 [DOI] [PubMed] [Google Scholar]
- 83. Jouanneau Y, Jeong HS, Hugo N, Meyer C, Willison JC (1998) Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus-characterization of two membrane-bound iron-sulfur proteins. Eur J Biochem FEBS 251: 54–64. [DOI] [PubMed] [Google Scholar]
- 84. Bruggemann H, Baumer S, Fricke WF, Wiezer A, Liesegang H, et al. (2003) The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc Natl Acad Sci U S A 100: 1316–1321 10.1073/pnas.0335853100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Seedorf H, Fricke WF, Veith B, Brüggemann H, Liesegang H, et al. (2008) The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci U S A 105: 2128–2133 10.1073/pnas.0711093105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Rohlin L, Gunsalus RP (2010) Carbon-dependent control of electron transfer and central carbon pathway genes for methane biosynthesis in the Archaean, Methanosarcina acetivorans strain C2A. BMC Microbiol 10: 62 10.1186/1471-2180-10-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gifford CM, Wallace SS (2000) The genes encoding endonuclease VIII and endonuclease III in Escherichia coli are transcribed as the terminal genes in operons. Nucleic Acids Res 28: 762–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Frigaard N-U, Takaichi S, Hirota M, Shimada K, Matsuura K (1997) Quinones in chlorosomes of green sulfur bacteria and their role in the redox-dependent fluorescence studied in chlorosome-like bacteriochlorophyll c aggregates. Arch Microbiol 167: 343–349 10.1007/s002030050453 [DOI] [Google Scholar]
- 89. Kjaer B, Frigaard NU, Yang F, Zybailov B, Miller M, et al. (1998) Menaquinone-7 in the reaction center complex of the green sulfur bacterium Chlorobium vibrioforme functions as the electron acceptor A1. Biochemistry (Mosc) 37: 3237–3242 10.1021/bi973121t [DOI] [PubMed] [Google Scholar]
- 90. Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 45: 316–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Macy JM, Probst I (1979) The Biology of Gastrointestinal Bacteroides. Annu Rev Microbiol 33: 561–594 10.1146/annurev.mi.33.100179.003021 [DOI] [PubMed] [Google Scholar]
- 92.Jayamani E (2008) A unique way of energy conservat ion in glutamate fermenting clostridia Marburg: Philipps-Universität Marburg. Available: http://d-nb.info/1002526671/34.
- 93. Neehaul Y, Juárez O, Barquera B, Hellwig P (2012) Thermodynamic contribution to the regulation of electron transfer in the Na(+)-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry (Mosc) 51: 4072–4077 10.1021/bi300343u [DOI] [PubMed] [Google Scholar]
- 94. Bogachev AV, Bloch DA, Bertsova YV, Verkhovsky MI (2009) Redox properties of the prosthetic groups of Na(+)-translocating NADH:quinone oxidoreductase. 2. Study of the enzyme by optical spectroscopy. Biochemistry (Mosc) 48: 6299–6304 10.1021/bi900525v [DOI] [PubMed] [Google Scholar]
- 95. Varadarajan R, Zewert TE, Gray HB, Boxer SG (1989) Effects of buried ionizable amino acids on the reduction potential of recombinant myoglobin. Science 243: 69–72. [DOI] [PubMed] [Google Scholar]
- 96. Zhou Z, Swenson RP (1996) Evaluation of the electrostatic effect of the 5′-phosphate of the flavin mononucleotide cofactor on the oxidation-reduction potentials of the flavodoxin from desulfovibrio vulgaris (Hildenborough). Biochemistry (Mosc) 35: 12443–12454 10.1021/bi9610865 [DOI] [PubMed] [Google Scholar]
- 97. Breinlinger EC, Rotello VM (1997) Model Systems for Flavoenzyme Activity. Modulation of Flavin Redox Potentials through π-Stacking Interactions. J Am Chem Soc 119: 1165–1166 10.1021/ja9612110 [DOI] [Google Scholar]
- 98. Zhou Z, Swenson RP (1996) The cumulative electrostatic effect of aromatic stacking interactions and the negative electrostatic environment of the flavin mononucleotide binding site is a major determinant of the reduction potential for the flavodoxin from Desulfovibrio vulgaris [Hildenborough]. Biochemistry (Mosc) 35: 15980–15988 10.1021/bi962124n [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of NqrA and RnfC subunits. Secondary structure prediction of Anaerophaga thermohalophila (Bacteroidetes) RnfC and NqrA. Figure shows the secondary structure prediction consensus of eight algorithms, using the Network Protein Sequence Analysis software. The conserved residues of RnfC involved in NADH and FAD binding sites, as well as the cysteine residues involved in the 4Fe-4S centers attachement are highlighted in black.
(PDF)
Rooted phylogenetic tree of NqrB subunit. Maximum Likelihood phylogenetic analysis of the NqrB subunit of the Na+-NQR complex and the homolog RnfD subunit from the RNF complex. The tree was rooted using the RnfD clade as outgroup. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site. NCBI GI numbers precede the taxa identification.
(PDF)
Rooted phylogenetic tree of NqrF subunit. Maximum Likelihood phylogenetic analysis of the NqrF subunit of the Na+-NQR complex and sequences of the reductase subunit of aromatic monooxigenases (AMOr), which are the closes know homologues of NqrF. The tree was rooted using the AMOr (benC/ToL) proteins as outgroup. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site. NCBI GI numbers precede the taxa identification.
(PDF)
Rooted phylogenetic tree of NqrD subunit. Maximum Likelihood phylogenetic analysis of the NqrD subunit of the Na+-NQR complex and the homolog RnfE subunit from the RNF complex. The RnfE branch was defined as outgroup. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site. NCBI GI numbers precede the taxa identification.
(PDF)
Rooted phylogenetic tree of NqrE subunit. Maximum Likelihood phylogenetic analysis of the NqrE subunit of the Na+-NQR complex and the homolog RnfA subunit from the RNF complex. The tree was rooted using the RnfA branch (100% BS) as outgroup. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site. NCBI GI numbers precede the taxa identification.
(PDF)
Unrooted phylogenetic tree of NqrD subunit. Maximum Likelihood tree of the subunit D (NqrD) of the Na+-NQR complex. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site.
(PDF)
Unrooted phylogenetic tree of NqrE subunit. Maximum Likelihood tree of the subunit NqrE of the Na+-NQR complex. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site.
(PDF)
Unrooted phylogenetic tree of NqrA subunit. Maximum Likelihood tree of the subunit A (NqrA) of the Na+-NQR complex. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site.
(PDF)
Unrooted phylogenetic tree of NqrC subunit. Maximum Likelihood tree of the subunit NqrC of the Na+-NQR complex. Numbers near nodes indicate RaxML bootstrap branch support values (when ≥50%). Branch lengths are proportional to the number of substitutions per site.
(PDF)
List of the analyzed bacterial taxa harboring Na+-NQR complex.
(PDF)
Similarity profiles of the Na+-NQR subunits with the closest homologues in RNF and AMOr.
(PDF)