Abstract
Triplex structures generated by sequence-specific triplex-forming oligonucleotides (TFOs) have proven to be promising tools for gene targeting strategies. In addition, triplex technology has been highly utilized to study the molecular mechanisms of DNA repair, recombination and mutagenesis. However, triplex formation utilizing guanine-rich oligonucleotides as third strands can be inhibited by potassium-induced self-association resulting in G-quadruplex formation. We report here that guanine-rich TFOs partially substituted with 8-aza-7-deaza-guanine (PPG) have improved target site binding in potassium compared with TFOs containing the natural guanine base. We designed PPG-substituted TFOs to bind to a polypurine sequence in the supFG1 reporter gene. The binding efficiency of PPG-substituted TFOs to the target sequence was analyzed using electrophoresis mobility gel shift assays. We have determined that in the presence of potassium, the non-substituted TFO, AG30 did not bind to its target sequence, however binding was observed with the PPG-substituted AG30 under conditions with up to 140 mM KCl. The PPG-TFOs were able to maintain their ability to induce genomic modifications as measured by an assay for gene-targeted mutagenesis. In addition, these compounds were capable of triplex-induced DNA double strand breaks, which resulted in activation of apoptosis.
Keywords: triplex-forming oligonucleotides, 8-aza-7-deaza-guanine
Introduction
Triplex-forming oligonucleotides (TFOs) bind as third strands in a sequence-specific manner within the major groove of duplex DNA at polypurine/polypyrimidine stretches.1-3 The specificity of TFO binding is derived from the base triplets formed by either Hoogsteen or reverse Hoogsteen hydrogen bonds formed between the third strand and the purine strand of the duplex.4 Triplex formation follows a specific binding code imposed by several structural constraints. In the purine motif, a GA or GT-rich TFO binds antiparallel to the purine strand of the duplex through reverse Hoogsteen bonds with the canonical triplets being G.G:C, A.A:T or T.A:T. In the pyrimidine motif, a TC-rich TFO binds in a parallel orientation to the purine strand of the duplex through Hoogsteen bonds, with the canonical triplets being T.A:T and C+.G:C.
Due to their sequence specificity, TFOs have been used for targeted genome modifications. Studies have shown that TFOs can induce site-specific damage by delivering a mutagen, such as psoralen or nitrogen mustards, to chromosomal target sites.5-8 In addition, these compounds have been used to inhibit transcription and to stimulate site-directed recombination of a co-transfected donor DNA, resulting in correction of a mutation in the target gene.9-12 TFOs have also been used to study mechanisms of DNA damage and repair, recombination and structurally induced genomic instability. Previous studies have demonstrated that triplex formation itself induces mutagenesis and stimulates DNA repair.7,13 Furthermore, we have recently demonstrated that TFOs are capable of inducing DNA double strand breaks (DSBs) resulting in the activation of apoptotic pathways.14
Despite their versatility, some obstacles exist in the utilization of TFOs as DNA binding molecules, particularly the influence of the cellular environment on triplex formation. In the pyrimidine motif, triplex formation requires protonation of cytosines at the N3 position for proper Hoogsteen binding with N7 guanine. However, cytosine has a relatively high pKa that is dependent upon structural influences, including the position of the base within the TFO. Therefore pyrimidine oligonucleotides usually bind with low affinity to duplex DNA at physiological pH and form unstable triplexes.15,16 These limitations have been addressed by the incorporation of base and sugar modifications into pyrimidine TFOs.17-20 On the contrary, TFOs form stable triplexes in the purine motif in a pH-independent manner. However, several groups have shown that binding of these guanine-rich (G-rich) TFOs to duplex DNA is significantly reduced at physiologic concentrations of monovalent cations, such as potassium (K+).21-23 This observation can be attributed in part to the tendency of G-rich oligonucleotides to self-associate and form alternative structures such as G-quartets.24-26 Formation of these G-rich TFO aggregates most likely results in the sequestering of the TFOs thereby reducing their in vivo bioactivity.
Previous studies aimed at eliminating potassium-mediated self-association of G-rich TFOs have utilized TFOs substituted with modified guanine bases with minimal success. In one study, the substitution of guanine with 7-deaza-guanine reduced self-association of the G-rich TFO but did not improve triplex formation in the presence of K+.27 In a subsequent study examining the bioactivity of short G-rich TFOs, it was demonstrated that TFOs containing 7-deaza-xanthine substitutions displayed improved intracellular targeting compared with an unsubstituted G-rich TFO.28 Several groups have also tested the ability of the base analog 6-thioguanine to overcome K+ mediated oligonucleotide self-association.22,29,30 These studies demonstrated that incorporation of 6-thioguanine did reduce TFO self-association. Although, similar binding affinities were observed in the absence of K+ with buffer conditions that favor triplex formation, an increase in TFO binding was observed in the presence of physiological concentrations of K+.
Self-association of G-rich TFOs is mediated by the N7 position of the guanine ring.31 Hence, another base analog, 8-aza-7-deaza-guanine (PPG) has been synthesized with the N7 and C8 positions interchanged within the guanine ring, thus maintaining the electron density of the ring while preventing Hoogsteen-bonding at the N7 position. Studies have shown that short G-rich TFOs entirely substituted with PPG displayed increased targeting efficiencies.32,33 Further experimentation determined that G-rich olignucleotides partially substituted with PPG resulted in a reduction of self-association and improved performance in DNA hybridization assays.34 Significantly, the results from these studies revealed that partially PPG-substituted oligonucleotides did not exhibit a reduction in duplex stability when compared with natural G. Consequently, we hypothesized that in situtations where it may not be feasible to modify each G, such as within a TFO comprised of long runs of guanines, partial substitution with PPG may be sufficient to reduce self-association of TFOs.
Previous work in our lab has involved the use of the highly G-rich 30mer TFO, AG30, which binds with high affinity to its target sequence located in the supFG1 reporter gene.14 In order to investigate the potential benefits of partial PPG-substitution on TFO efficacy, we designed two AG30 TFOs with either every fourth guanine substituted with PPG (A6G30) or every third guanine substituted with PPG (A8G30). We tested the ability of the modified TFOs to bind to a duplex target compared with the non-substituted G-rich 30-mer, using conditions that mimic physiologic levels of potassium. Here, we report that in the absence of K+, the PPG-substituted TFOs displayed binding affinities comparable to the non-substituted TFO. Notably, in the presence of K+, the PPG-substituted TFOs displayed a 1000-fold higher affinity for the target site than the non-substituted TFO. Our gene targeting data correlates with our in vitro binding studies; the PPG-substituted TFOs have improved bioactivity as evaluated by an assay for targeted mutagenesis. In addition, we have determined that the PPG-substituted TFOs are capable of inducing DNA DSBs, which ultimately resulted in the activation of apoptosis. Thus revealing that PPG-substituted molecules are also useful tools for studying the molecular mechanisms involved in the response to triplex-induced DNA damage. These results suggest that partial modification of G-rich TFOs with the PPG guanine analog may provide an alternative strategy to overcome the potassium-induced self-aggregation phenomena that severely limits triplex formation under physiological conditions.
Results
Design of the PPG substituted G-rich triplex-forming oligonucleotides
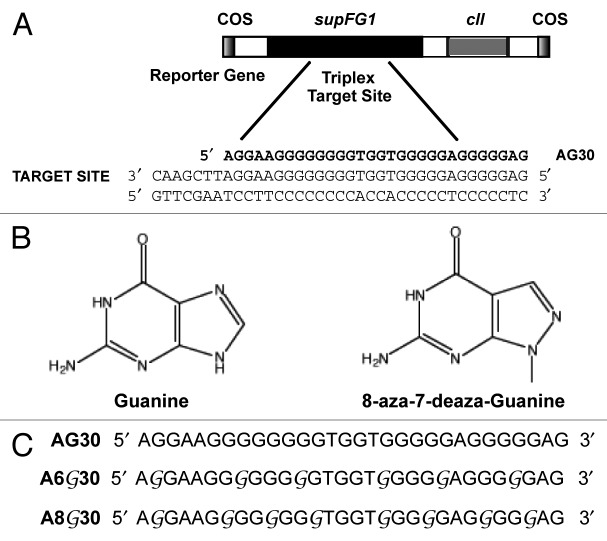
AG30, a G-rich TFO, binds as a third strand in the anti-parallel motif to form a triple helix at the polypurine/polypyrimidine duplex target located at the 3′ end of the supFG1 reporter gene at position 167–196 (Fig. 1A).5 Studies have demonstrated that this TFO is capable of inducing site-directed recombination when co-transfected with a DNA donor molecule.35,36 Additionally, triplex structures induced by AG30 have the ability to induce mutations at a chromosomal locus both in cell culture and an animal model.7,37 Furthermore, we have demonstrated that AG30-induced DSBs can activate apoptosis.14 As a result, AG30 represents an excellent model to study the potential limitations imposed upon triplex technology by the cellular environment. Given that 23 of the 30 bases in AG30 are guanine, we hypothesized that the guanines within AG30 could self-associate and thereby reduce its bioactivity. Previous studies determined that AG30 binds to the duplex DNA target site with high affinity, under buffer conditions that promote triplex formation.38 In order to investigate the potential effects of self-association on AG30, we utilized the guanine analog 8-aza-7-deaza-guanine (PPG, or G) in our studies (Fig. 1B). Two TFOs were designed with the same sequence as AG30 but with every fourth (A6G30) or third (A8G30) guanine substituted with the modified guanine base, PPG (Fig. 1C). The sequences of the PPG-substituted TFOs are depicted in Figure 1C.
Figure 1. Triplex formation by the PPG substituted TFOs. (A) The G-rich TFO, AG30 was designed to bind to a polypurine sequence at position 167–196 of the supFG1 reporter gene. The sequence of AG30 and its target duplex site is included. (B) Structure of the natural guanine base compared with the modified base 8-aza-7-deaza-guanine (PPG) base. (C) Design of the 30-mer G-rich TFOs. The G represents PPG.
Evaluation of triplex formation
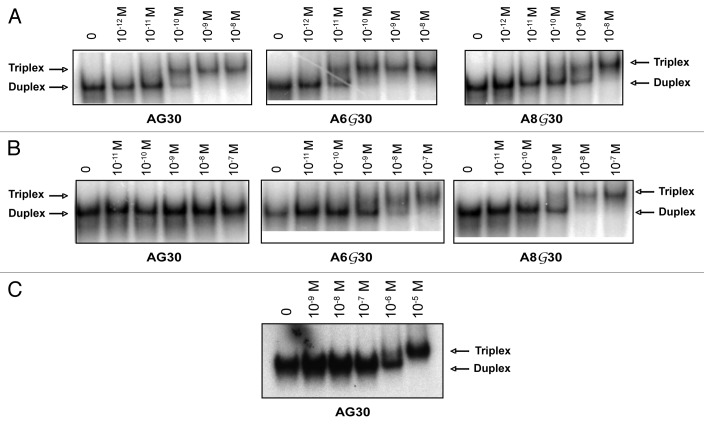
To determine the relative binding affinities of the PPG-substituted TFOs for the target duplex, a gel mobility shift assay was performed. Target duplex (10-11 M) was incubated overnight at 37 °C with increasing concentrations of TFOs in triplex binding buffer. The mixtures were separated by electrophoresis using a native acrylamide gel. The apparent Kd for each TFO was calculated as the concentration of TFO at which binding was one-half maximal. As shown in Figure 2A using buffer conditions that promote triplex formation, AG30 bound to the target site with high affinity (Kd apparent ~1 × 10-10 M). The PPG substituted TFOs, A6G30 (Kd apparent ~1 × 10-11 M) and A8G30 (Kd apparent ~1 × 10-10 M), also bound to the target site with relatively high affinities (Fig. 2A). In contrast, when the gel shifts were repeated in the presence of 140 mM KCl (Fig. 2B), AG30 displayed a much lower affinity for the target site (Kd apparent ~1 × 10-6 M) (Fig. 2C). Interestingly, the PPG-substituted TFOs, A6G30 (Kd apparent ~1 × 10-9 M) and the A8G30 (Kd apparent ~1 × 10-9 M) were able to maintain a high binding affinity for the target site even in the presence of K+ (Fig. 2B). These results suggest that partial incorporation of PPG into G-rich TFOs may provide a small contribution to improving TFO binding affinity in buffer conditions, which favor triplex formation, with a 10-fold increase observed with A6G30. However in buffer conditions that mimic physiologic conditions, the PPG-containing TFOs have a 103-fold increase in binding affinity compared with the non-substituted TFO, AG30. These in vitro binding studies suggest that substitution of even a few guanines with PPG bases in G-rich TFOs may provide enhanced bioactivity.
Figure 2. Triplex formation of the modified TFOs in the presence of potassium. (A) Gel mobility shift analysis of triplex formation in standard triplex buffer (10mM MgCl2/ no K+). The target duplex was end-labeled and incubated with increasing concentrations of the indicated TFO followed by native PAGE. The lane marked 0 represents duplex DNA alone with no added TFO. (B) Gel mobility shift analysis of triplex formation in the presence of 140 mM KCl. (C) Gel mobility shift assay of AG30 in the presence of 140 mM KCl.
Evaluation of self-association
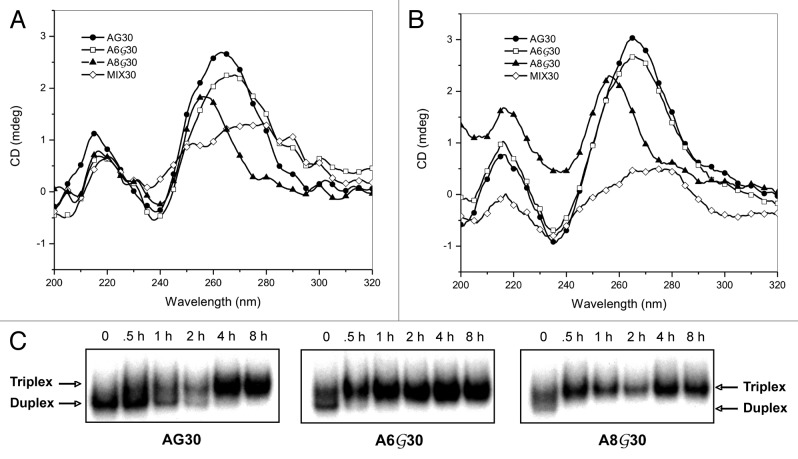
Studies have determined that G-rich oligonucleotides can aggregate and self-associate to form secondary G-quadruplex structures in the presence of certain concentrations of cations. CD experiments were performed to investigate the effect of the PPG-modification on the potential for AG30, A6G30, and A8G30 to form secondary structures. The conformational characteristics of AG30 were analyzed by CD spectra using buffer conditions that favor triplex formation (10 mM MgCl2) (Fig. 3A). The CD spectrum showed a negative peak near 240 nm and a positive peak near 260 nm, a typical spectrum for a parallel G-quadruplex structure. The depth of the minimum observed was larger for AG30 and A6G30, compared with A8G30 (Fig. 3A). A decrease in peak intensity at 260 nm was observed with an increase in PPG substitution, with A8G30 exhibiting a peak with the lowest intensity. In addition, there was a small wavelength shift in the maxima of A8G30. Furthermore, the mixed sequence oligonucleotide MIX30 did not form a secondary structure under the same buffer conditions. It is well established that monovalent cations, like potassium stabilize G-quadruplexes.39 Thus we investigated the effect of K+ on the structural stability of the secondary structures created by the G-rich TFOs in buffer conditions with physiological concentrations of K+ (10 mM MgCl2, 140 mM KCl). Inclusion of K+ resulted in an increase in the CD intensity at 260 nm for AG30. As shown in Figure 3B, strong CD peaks were observed in AG30 and A6G30, suggesting the formation of stable G-quadruplex structures. However, substitution with PPG led to a decrease in the peak intensity of A8G30 at 260 nm, with no observed increase in the CD spectra upon addition of K+. Additionally, the control mixed sequence oligonucleotide MIX30 did not show the two-banded spectroscopic pattern characteristic of a parallel G-quadruplex.
Figure 3. Potential effect of G-rich oligonucleotide self-aggregation on triplex formation. (A) CD spectra of AG30, A6G30, and A8G30 in standard triplex binding buffer (10 mM MgCl2/no K+). All spectra were collected at room temperature in a species concentration of 2μM. (B) CD spectra collected in the presence of 140mM KCl. (C) Triplex formation using PPG-substituted TFOs at various time points. AG30, A6G30 and A8G30 (10-8 M) were incubated with end-labeled target duplex (10-10 M) for the indicated times in standard triplex binding buffer (10 mM MgCl2/no K+).
Guanine substitution of our TFOs was designed to reduce G-quartet formation through self-association. A decrease in TFO self-association would be reflected in increased availability of the TFO for duplex targeting and binding. Binding assays were performed utilizing both the PPG-substituted and non-substituted AG30s at various incubation times with 10-10 M target duplex as indicated in Figure 3C. A TFO concentration of 10-8 M was used in these studies since we previously determined that it was an effective concentration to observe binding of duplex DNA with all three TFOs in the absence of K+. The PPG-substituted TFOs, A6G30 and A8G30, almost completely bound available duplex within 30 min as indicated by the complete shift of the target duplex DNA (Fig. 3C). The gel shifts also indicate that the duplex remained stably associated with the PPG-TFOs for as long as 8 h. Contrarily, the non-substituted AG30 began associating with the target duplex after 1 h incubation, with complete association of available duplex only at 4 h. These gel shift assays demonstrate that PPG-TFOs are capable of rapidly binding to their target site and is able to do so more efficiently than G-rich TFOs comprised of natural guanine bases. This may be attributed in part to a reduction in oligonucleotide self-association resulting in an increase in bioavailability.
Chromosomal mutagenesis induced by substituted TFOs
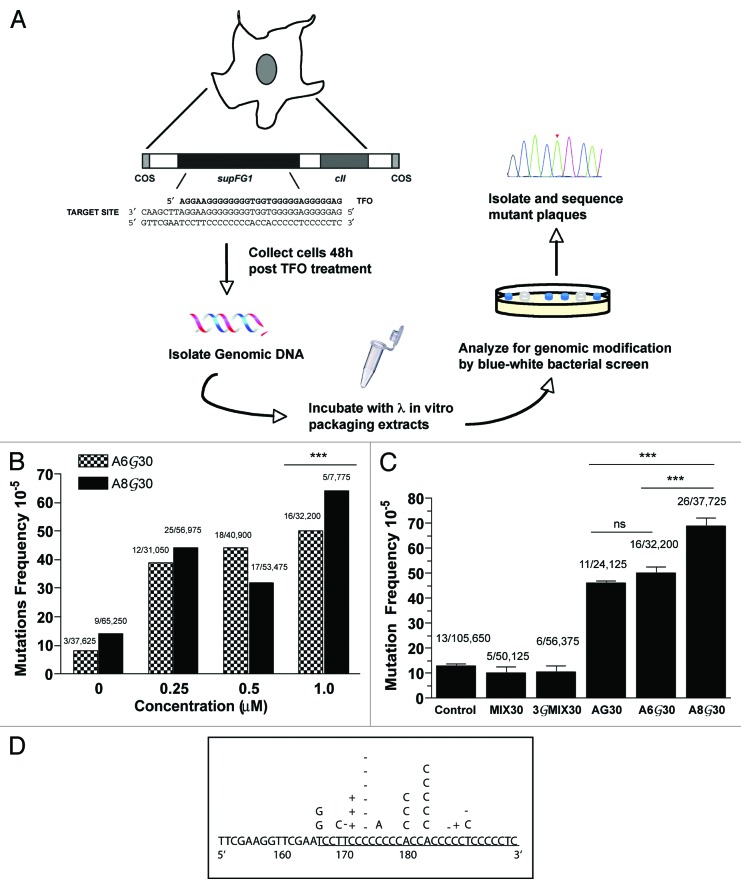
To investigate the ability of PPG base substitution to improve intracellular triplex formation of G-rich TFOs leading to improved bioactivity, an assay for targeted mutagenesis in mammalian cells was used (Fig. 4A). We utilized a mouse cell line (AV16), containing multiple copies of randomly integrated chromosomal λsupFG1 shuttle vector DNA. The supFG1 reporter gene contains the target site for third strand binding by either AG30, A6G30, or A8G0. Using packaging extracts, the vector DNA can be isolated from mouse genomic DNA into phage particles and subsequently analyzed for induced mutations.40 SupFG1 encodes an amber suppressor tRNA whose function can be scored in indicator bacteria. A targeted mutation in the supFG1 gene will result in production of a white plaque upon shuttle vector rescue, in contrast to the blue plaques produced by functional supFG1 genes. This is a useful system for evaluating improved triplex formation due to PPG substitution, because site-directed mutagenesis induced by non-substituted AG30 at the supFG1 gene has been established both in vitro and in vivo.7,37
Figure 4. Targeted chromosomal mutagenesis by PPG substituted G-rich TFOs. (A) Schematic describing gene-targeted mutagenesis assay. AV16 cells were transfected with TFOs using cationic lipids. Cells were collected for DNA isolation 48h post transfection to allot time for induction of mutations, before analysis of genomic DNA. (B) Dose dependence of PPG-substituted TFOs on targeted mutagenesis of the chromosomal supFG1 gene in AV16 cells. (C) The frequency of mutations in the supFG1 gene induced by AG30 or the PPG-substituted TFOs was calculated by dividing the number of colorless mutant plaques by the total number of plaques counted. Each experiment was performed three times and the standard errors were calculated for the mutation frequency values, as indicated by the error bars. *** P < 0.001 (D) Sequence analysis of the supFG1 gene mutations induced by the treatment of AV16 cells with PPG-TFOs, A6G30 and A8G30.
In order to evaluate whether PPG-substituted TFOs could generate triplex-induced mutations in AV16 cells and to establish the dose required for detecting targeted mutagenesis using these molecules, we performed a dose response assay. AV16 cells were treated with increasing concentrations of A6G30 and A8G30 and analyzed for induction of mutations. The results determined that both PPG-substituted TFOs were capable of generating triplex-induced mutations (Fig. 4B). As expected, mutation frequencies increased with increasing concentration of PPG-TFO, with targeted mutations being detected at concentrations as low as 0.25 μM. These studies determined that A8G30 has improved bioactivity compared with A6G30. Administration of A8G30 at 1 μM induced a mutation frequency of 64 × 10-5, while A6G30 treatment yielded a mutation frequency of 50 × 10-5 (P < 0.001). These data confirm that partial substitution of G-rich TFOs with PPG can improve target site binding without compromising the bioactivity of the molecules.
AV16 cells were then treated with a control oligonucleotide, MIX30 or 3GMIX30, or the TFO, AG30, A6G30 or A8G30. In order to provide an opportunity for triplex formation and mutation induction via repair and replication, the cells were harvested and the vector DNA was isolated for genetic analysis 48h post-treatment. The results indicate that treatment of the cells with the PPG-substituted AG30 induced mutations at frequencies higher than with AG30 alone (Fig. 4C). A8G30 induced a 6-fold increase in mutation frequency when compared with cells alone (Fig. 4C). In fact, A8G30 induced a mutation frequency of 69 × 10−5, which is almost 2-fold higher than the frequency observed with AG30 (46 × 10-5, P < 0.001) and A6G30 (50 × 10-5, P < 0.001). Although, increased mutation frequencies were also observed with the use of A6G30 (Fig. 4C), the observed induction was not significantly higher than that of the non-substituted AG30. The inability of the control oligonucleotide, MIX30, to induce mutations above background provides evidence to support a mechanism of induced mutagenesis dependent upon the binding of the TFO to its specific target site. Treatment with the PPG control oligonucleotide, 3GMIX30 also provides support that the induction of mutations did not arise from the PPG guanine modification.
Sequence analysis of the A8G30 and A6G30 induced mutations revealed base substitutions and single base pair insertion and deletion mutations precisely within the TFO binding site, consistent with previous studies (Fig. 4D).7 Because the majority of mutants analyzed were located in the triplex binding site, the sequencing data provides further evidence that the mutations were TFO-induced and site-specific. Taken together, the analysis of the pattern of mutations and the inability of the control oligonucleotide to induce mutations, supports a mechanism that attributes the increase in mutation frequency in the supFG1 gene to binding of the PPG-substituted TFOs to the chromosomal target site. These studies demonstrate that substitution of natural G with the guanine analog, PPG, can improve not only binding, but also the bioactivity of G-rich TFOs.
Triplex-induced DSBs
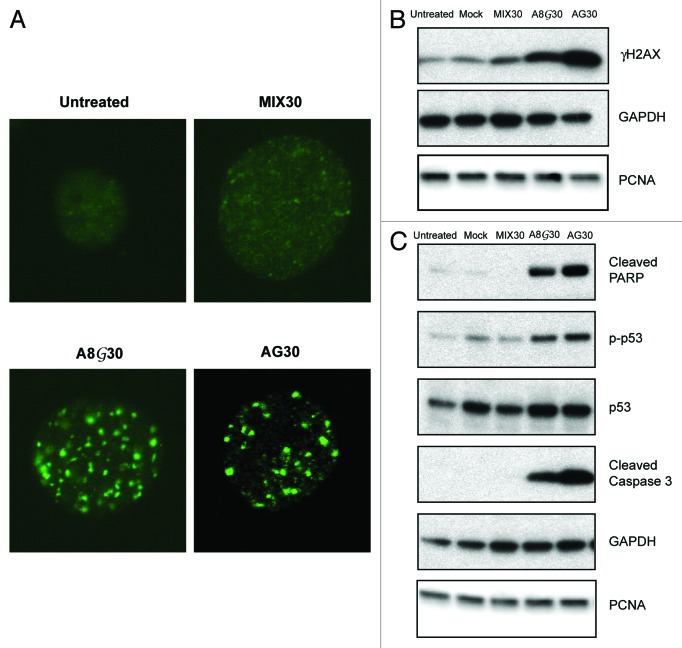
Triplex technology has been used extensively to study the molecular responses to structurally induced DNA damage. Our previous studies have determined that chromosomal triplex structures generated by TFOs are capable of inducing DSBs.14 In order to determine whether PPG-substitution may in some way alter this observation, we treated AV16 cells with MIX30, AG30, or A8G30 and evaluated for the induction of DSBs 24h following treatment. Phosphorylated histone variant H2AX (γH2AX) foci formation is commonly used as a quantitative marker for DSBs using immunofluorescence microscopy. The presence of triplex-induced DSBs following treatment with AG30 and A8G30 was determined by staining for γH2AX. AG30 treatment resulted in the formation of more γH2AX nuclear foci compared with untreated and MIX30 treated cells (Fig. 5A). This is consistent with previously reported results.14 Treatment with A8G30 also resulted in γH2AX foci formation. In fact, it appears that there is a slight increase in γH2AX foci formation after treatment with A8G30 compared with AG30-treatment. Western blot analysis of γH2AX also confirms the presence of H2AX (S139) phosphorylation in only the TFO treated cells in agreement with the immunofluorescence results (Fig. 5B). Combined these data imply that the induction of DSBs generated by TFO treatment can be attributed to triplex formation.
Figure 5. PPG-substituted TFOs induce DSBs. (A) Immunofluorescence of triplex-induced γH2AX foci 24h post-treatment with AG30 and A8G30. (B) Western blot analysis of γH2AX protein levels 24h after TFO treatment. (C) Detection of caspase-mediated cleavage of PARP as a measure of triplex-induced apoptosis. Treatment with the PPG-substituted TFO, A8G30 resulted in similar p53 phosphorylation levels at serine 15, as AG30.
Studies were then initiated to determine whether the TFO-induced DSB damage was extensive enough to induce pro-apoptotic pathways. AV16 cells were treated with a mock transfection, the control oligonucleotide, MIX30, or the TFOs, AG30 and A8G30 and then analyzed for the induction of apoptosis. Western blot analysis of caspase-mediated cleavage of Poly(ADP-ribose)polymerase (PARP) was used to confirm triplex activated apoptosis. PARP is a nuclear protein that recognizes DNA strand breaks and its cleavage by caspase 3 is an early event in apoptotic response. Caspase-mediated cleavage of PARP was only detected in the cell lysates isolated from AG30 and A8G30 treated AV16 cells (Fig. 5C). Similar levels of cleaved PARP were detected following treatment with either TFO, suggesting that the observed apoptosis can be attributed to triplex formation. Following DNA damage, phosphorylation of the tumor suppressor p53 at serine 15 is a central component in the activation of apoptosis. After TFO treatment, we determined by western blot analysis that an increase in p53 phosphorylation at serine 15 corresponded to an increase in caspase 3-mediated cleavage of PARP (Fig. 5C). These results confirm that triplex-induced DNA strand breaks via PPG-modified TFOs are capable of activating p53 dependent apoptosis. Furthermore, it confirms that PPG-substituted TFOs may serve as important tools in the study of structurally induced DNA damage, particularly under conditions where it is important to verify that the molecular response can be attributed to triplex formation and not the generation of secondary structures created by self-association.
Discussion
One current limitation in the use of purine-rich TFOs to target chromosomal DNA is the relatively poor duplex binding under physiologic conditions of monovalent cations. Physiologic potassium levels, for example, can stabilize the formation of intermolecular secondary structures, such as G-quartets, which in turn reduce the bioavailability of guanine-rich TFOs. In the present study, we have substituted a 30-mer purine-rich TFO with the guanine base analog 8-aza-7-deaza-guanine (PPG), in which the N7 and C8 positions of the guanine ring are interchanged. We show here that the partially PPG-substituted TFOs have increased binding affinity to a polypurine target site in vitro, even in the presence of physiological concentrations of potassium. Additionally, the PPG-substituted TFOs were able to associate with the target DNA suggesting that self-aggregation of the non-substituted TFO may reduce its bioavailability. Notably, the TFO containing PPG-substitutions at every third guanine A8G30 induced almost a 7-fold increase over background in mutation frequency in a chromosomal reporter gene in mammalian cells, almost twice the rate of a TFO containing natural guanine. Guanine modification, therefore, plays an important role in the on-going improvement of purine TFOs to enhance the efficiency of in vivo gene targeting and modification.
Triplex technology has been used extensively as a model to study the mechanisms involved in the recognition and repair of DNA damage induced by endogenous altered helical structures such as H-DNA. However, triplex formation via G-rich TFOs is not readily observed in vitro even at sub-physiological potassium levels. As a result, the question may often arise as to whether the observed molecular responses are actually mediated by triplex formation at the corresponding target site and not due to formation of secondary structures. Partial substitution of G-rich TFOs with PPG potentiates the capacity of TFOs to form stable triplexes in vitro under physiological potassium concentrations. In the work reported here, we demonstrate the utility of PPG-substituted TFOs as a model for endogenous triplex formation. We designed PPG-substituted TFOs that are capable of inducing DSBs as measured by γH2AX foci formation resulting in the activation of apoptosis. Similar levels of γH2AX foci when compared with the damage induced by the TFO comprised of natural guanine confirm that triplex formation can induce DSBs. In addition, it emphasizes that TFO-induced apoptosis is a result of triplex formation and cannot be attributed to non-specific toxicity generated by secondary structures.
Several strategies have been reported in the literature to circumvent self-association of G-rich oligonucleotides. For instance, it has been shown that incorporation of 6-thioguaine within purine oligonucleotides reduces G-quartet formation under physiological conditions. However, this base analog slightly destabilizes triplex formation.23 Other groups have demonstrated that replacement of the negatively charged phosphodiester backbone with the positively charged phosphoramidite linkage favors triplex formation in the presence of KCl.41 The present study demonstrates that even partial substitution of G-rich TFOs with the PPG base analog can reduce self-association and improve triplex formation under physiological potassium concentrations. These results suggest that PPG-substitution can be used to improve the bioactivity of gene-targeting TFOs. In addition, they may prove beneficial as tools to study the molecular mechanisms of DNA damage and repair of altered helical structures.
Materials and Methods
Oligonucleotides
Oligonucleotides were synthesized by the Midland Certified Reagent Company Inc. using cyanoethyl phosphoramidite chemistry and purified by RP-HPLC. The sequences of the 30mer G-rich oligonucleotides used in the studies are depicted in Figure 1C. The G-rich TFO, AG30, was synthesized with the following sequence 5′ AGGAAGGGGG GGGTGGTGG GGGAGGGGGA G 3′. The guanines in A6G30 and A8G30 were replaced with 8-aza-7-deaza-guanine (PPG, G, Glen Research) as indicated in Figure 1C. In order to prevent nuclease degradation all oligonucleotides were synthesized to contain a 3′ amine group, through the use of a 3′-amino-modifier C7 CPG (Glen Research).42 In addition, the mixed sequenced control oligonucleotides MIX30 and 3GMIX30 were synthesized with the following sequences, respectively: 5′ AGTCAGTCAG TCAGTCAGTC AGTCAGTCAG 3′ and 5′ AGTCAGTCAG TCAGTCAGTC AGTCAGTCAG 3′.
Triplex binding assays
Third strand binding to target duplexes was measured by gel mobility assays as previously described.43 Briefly, two complementary 57-mer oligonucleotides containing the sequence corresponding to bp 157–213 of the SupFG1 reporter gene were synthesized, mixed 1:1 in 25 mM NaCl, heated to 90 °C for 20 min and allowed to slowly cool to room temperature to form duplex DNA. Following end labeling of the duplex using T4 polynucleotide kinase and γ32P-ATP, the duplex was purified by gel electrophoresis. Binding reaction mixtures (20 μl) containing end-labeled duplex DNA (10−11 M) were incubated overnight at 37 °C with increasing concentrations of TFO in a triplex binding buffer (10 mM TRIS-HCl [pH 7.6], 10% glycerol, 10 mM MgCl2) and supplemented with 140 mM KCl, where indicated. The products were resolved by gel electrophoresis on a 12% native polyacrylamide gel at 70 V in 1× TB buffer, with 10 mM MgCl2. The apparent equilibrium dissociation constant (Kd apparent) was calculated as the concentration of TFO at which binding was one-half maximal.
Circular dichroism
DNA substrates were prepared using the TFOs described above. Oligonucleotides (2 μM) were prepared in triplex binding buffer (10 mM Tris, pH 7.6, 10 mM MgCl2, 10% glycerol) and supplemented with 140 mM KCl, when preparing K+ containing samples. The mixtures were boiled for 10 min and gradually cooled to room temperature. CD spectra were recorded by a Chirascan Circular Dichroism Spectrometer (Applied Photophysics), using a 1 mm path length cuvette in a volume of 500 μl at room temperature. For each sample, three scans were taken at wavelengths from 200 to 320 nm. An average value was then calculated from the scans and corrected for the spectrum of the buffer control.
Cells and mutagenesis assay
The established mouse epithelial cell line, AV16, containing ~100 copies of the λsupFG1 shuttle vector, was derived from C127 cells as previously described.44 AV16 cells were transfected with the selected TFOs at final concentrations of up to 1 μM using cationic lipid transfection. Following 48–72 h of TFO treatment, genomic DNA was isolated and incubated with λ in vitro packaging extracts for shuttle vector rescue and reporter gene analysis as previously described.44,45 Briefly, functional supFG1 gene suppress the nonsense mutations in the host β-galactosidase gene yielding blue plaques in the presence of IPTG and X-Gal. If however a mutation occurs in the supFG1 gene, the amber mutation will not be suppressed and the resulting plaque will be white. Mutation frequencies were determined by dividing the number of colorless plaques by the total number of plaques counted. Experiments were completed in triplicate and standard errors were calculated for the mutation frequencies as indicated by the error bars. Differences in the mutation frequencies were analyzed by one or two way Anova and tukey test as posthoc. All statistical analyses were performed using Graphpad Prism software. ***P < 0.001, **P < 0.01
Immunofluorescence
Cells, seeded onto UV-irradiated coverslips, were treated for 24hrs and samples were prepared under reduced light as previously described.46 Cells were incubated with the following antibodies: rabbit anti-γH2AX antibody (Cell Signaling) and FITC-conjugated F(ab′)2 fragment donkey anti-rabbit IgG (H+L) (Molecular Probes Inc.). Images were captured using an Axiovert 200 microscope (Carl Zeiss Micro Imaging, Inc.).
Western analysis
Floating and adherent cells were collected, cell pellets were lysed with RIPA buffer (150 mM NaCl, 0.1% SDS, and inhibitors), and 30–50 μg of total protein per sample was resolved by SDS-PAGE. Proteins were detected by a standard immunoblot protocol using the following primary antibodies: cleaved PARP; cleaved caspase3; phospho-p53 (ser15); phospho-H2AX (serine 139); (Cell Signaling Technology, Inc.) and PCNA (Santa Cruz). Each experiment was performed a minimum of three times, and representative western blots are shown.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank A. Gupta for assistance with the circular dichroism. This work was supported by grants from the National Institutes of Health [K22 CA120049, K22 CA120049-03S1 to F.A.R.]
Glossary
Abbreviations:
- TFO
triplex-forming oligonucleotides
- DSBs
double strand breaks
- PPG
8-aza-7-deaza-guanine
- PARP
Poly(ADP-ribose)polymerase
- G-rich
guanine-rich
- K+
potassium
Footnotes
Previously published online: www.landesbioscience.com/journals/artificialdna/article/27792
References
- 1.Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–50. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 2.Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;241:456–9. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 3.Le Doan T, Perrouault L, Praseuth D, Habhoub N, Decout JL, Thuong NT, Lhomme J, Hélène C. Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-[alpha]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987;15:7749–60. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.François JC, Saison-Behmoaras T, Hélène C. Sequence-specific recognition of the major groove of DNA by oligodeoxynucleotides via triple helix formation. Footprinting studies. Nucleic Acids Res. 1988;16:11431–40. doi: 10.1093/nar/16.24.11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang G, Levy DD, Seidman MM, Glazer PM. Targeted mutagenesis in mammalian cells mediated by intracellular triple helix formation. Mol Cell Biol. 1995;15:1759–68. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raha M, Wang G, Seidman MM, Glazer PM. Mutagenesis by third-strand-directed psoralen adducts in repair-deficient human cells: high frequency and altered spectrum in a xeroderma pigmentosum variant. Proc Natl Acad Sci U S A. 1996;93:2941–6. doi: 10.1073/pnas.93.7.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasquez KM, Wang G, Havre PA, Glazer PM. Chromosomal mutations induced by triplex-forming oligonucleotides in mammalian cells. Nucleic Acids Res. 1999;27:1176–81. doi: 10.1093/nar/27.4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagatsugi F, Sasaki S, Miller PS, Seidman MM. Site-specific mutagenesis by triple helix-forming oligonucleotides containing a reactive nucleoside analog. Nucleic Acids Res. 2003;31:e31. doi: 10.1093/nar/gng031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers FA, Vasquez KM, Egholm M, Glazer PM. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc Natl Acad Sci U S A. 2002;99:16695–700. doi: 10.1073/pnas.262556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lonkar P, Kim KH, Kuan JY, Chin JY, Rogers FA, Knauert MP, Kole R, Nielsen PE, Glazer PM. Targeted correction of a thalassemia-associated beta-globin mutation induced by pseudo-complementary peptide nucleic acids. Nucleic Acids Res. 2009;37:3635–44. doi: 10.1093/nar/gkp217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schleifman EB, Bindra R, Leif J, del Campo J, Rogers FA, Uchil P, Kutsch O, Shultz LD, Kumar P, Greiner DL, et al. Targeted disruption of the CCR5 gene in human hematopoietic stem cells stimulated by peptide nucleic acids. Chem Biol. 2011;18:1189–98. doi: 10.1016/j.chembiol.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faria M, Wood CD, Perrouault L, Nelson JS, Winter A, White MR, Helene C, Giovannangeli C. Targeted inhibition of transcription elongation in cells mediated by triplex-forming oligonucleotides. Proc Natl Acad Sci U S A. 2000;97:3862–7. doi: 10.1073/pnas.97.8.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–5. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 14.Kaushik Tiwari M, Rogers FA. XPD-dependent activation of apoptosis in response to triplex-induced DNA damage. Nucleic Acids Res. 2013;41:8979–94. doi: 10.1093/nar/gkt670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shindo H, Torigoe H, Sarai A. Thermodynamic and kinetic studies of DNA triplex formation of an oligohomopyrimidine and a matched duplex by filter binding assay. Biochemistry. 1993;32:8963–9. doi: 10.1021/bi00085a030. [DOI] [PubMed] [Google Scholar]
- 16.Singleton SF, Dervan PB. Influence of pH on the equilibrium association constants for oligodeoxyribonucleotide-directed triple helix formation at single DNA sites. Biochemistry. 1992;31:10995–1003. doi: 10.1021/bi00160a008. [DOI] [PubMed] [Google Scholar]
- 17.Lee JS, Woodsworth ML, Latimer LJ, Morgan AR. Poly(pyrimidine). poly(purine) synthetic DNAs containing 5-methylcytosine form stable triplexes at neutral pH. Nucleic Acids Res. 1984;12:6603–14. doi: 10.1093/nar/12.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escudé C, Sun JS, Rougée M, Garestier T, Hélène C. Stable triple helices are formed upon binding of RNA oligonucleotides and their 2′-O-methyl derivatives to double-helical DNA. C R Acad Sci III. 1992;315:521–5. [PubMed] [Google Scholar]
- 19.Torigoe H, Sato N, Nagasawa N. 2′-O,4′-C-ethylene bridged nucleic acid modification enhances pyrimidine motif triplex-forming ability under physiological condition. J Biochem. 2012;152:17–26. doi: 10.1093/jb/mvs049. [DOI] [PubMed] [Google Scholar]
- 20.Xodo LE, Manzini G, Quadrifoglio F, van der Marel GA, van Boom JH. Effect of 5-methylcytosine on the stability of triple-stranded DNA--a thermodynamic study. Nucleic Acids Res. 1991;19:5625–31. doi: 10.1093/nar/19.20.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng AJ, Van Dyke MW. Monovalent cation effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1993;21:5630–5. doi: 10.1093/nar/21.24.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivas WM, Maher LJ., 3rd Overcoming potassium-mediated triplex inhibition. Nucleic Acids Res. 1995;23:1936–41. doi: 10.1093/nar/23.11.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivas WM, Maher LJ., 3rd Competitive triplex/quadruplex equilibria involving guanine-rich oligonucleotides. Biochemistry. 1995;34:278–84. doi: 10.1021/bi00001a034. [DOI] [PubMed] [Google Scholar]
- 24.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–80. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 25.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–4. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 26.Cheng AJ, Van Dyke MW. Oligodeoxyribonucleotide length and sequence effects on intramolecular and intermolecular G-quartet formation. Gene. 1997;197:253–60. doi: 10.1016/S0378-1119(97)00269-2. [DOI] [PubMed] [Google Scholar]
- 27.Milligan JF, Krawczyk SH, Wadwani S, Matteucci MD. An anti-parallel triple helix motif with oligodeoxynucleotides containing 2′-deoxyguanosine and 7-deaza-2′-deoxyxanthosine. Nucleic Acids Res. 1993;21:327–33. doi: 10.1093/nar/21.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faruqi AF, Krawczyk SH, Matteucci MD, Glazer PM. Potassium-resistant triple helix formation and improved intracellular gene targeting by oligodeoxyribonucleotides containing 7-deazaxanthine. Nucleic Acids Res. 1997;25:633–40. doi: 10.1093/nar/25.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao TS, Durland RH, Seth DM, Myrick MA, Bodepudi V, Revankar GR. Incorporation of 2′-deoxy-6-thioguanosine into G-rich oligodeoxyribonucleotides inhibits G-tetrad formation and facilitates triplex formation. Biochemistry. 1995;34:765–72. doi: 10.1021/bi00003a009. [DOI] [PubMed] [Google Scholar]
- 30.Gee JE, Revankar GR, Rao TS, Hogan ME. Triplex formation at the rat neu gene utilizing imidazole and 2′-deoxy-6-thioguanosine base substitutions. Biochemistry. 1995;34:2042–8. doi: 10.1021/bi00006a026. [DOI] [PubMed] [Google Scholar]
- 31.Murchie AI, Lilley DM. Tetraplex folding of telomere sequences and the inclusion of adenine bases. EMBO J. 1994;13:993–1001. doi: 10.1002/j.1460-2075.1994.tb06344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belousov ES, Afonina IA, Kutyavin IV, Gall AA, Reed MW, Gamper HB, Wydro RM, Meyer RB. Triplex targeting of a native gene in permeabilized intact cells: covalent modification of the gene for the chemokine receptor CCR5. Nucleic Acids Res. 1998;26:1324–8. doi: 10.1093/nar/26.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebbinghaus SW, Fortinberry H, Gamper HB., Jr. Inhibition of transcription elongation in the HER-2/neu coding sequence by triplex-directed covalent modification of the template strand. Biochemistry. 1999;38:619–28. doi: 10.1021/bi980981g. [DOI] [PubMed] [Google Scholar]
- 34.Kutyavin IV, Lokhov SG, Afonina IA, Dempcy R, Gall AA, Gorn VV, Lukhtanov E, Metcalf M, Mills A, Reed MW, et al. Reduced aggregation and improved specificity of G-rich oligodeoxyribonucleotides containing pyrazolo[3,4-d]pyrimidine guanine bases. Nucleic Acids Res. 2002;30:4952–9. doi: 10.1093/nar/gkf631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datta HJ, Chan PP, Vasquez KM, Gupta RC, Glazer PM. Triplex-induced recombination in human cell-free extracts. Dependence on XPA and HsRad51. J Biol Chem. 2001;276:18018–23. doi: 10.1074/jbc.M011646200. [DOI] [PubMed] [Google Scholar]
- 36.Chan PP, Lin M, Faruqi AF, Powell J, Seidman MM, Glazer PM. Targeted correction of an episomal gene in mammalian cells by a short DNA fragment tethered to a triplex-forming oligonucleotide. J Biol Chem. 1999;274:11541–8. doi: 10.1074/jbc.274.17.11541. [DOI] [PubMed] [Google Scholar]
- 37.Vasquez KM, Narayanan L, Glazer PM. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–3. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 38.Wang G, Glazer PM. Altered repair of targeted psoralen photoadducts in the context of an oligonucleotide-mediated triple helix. J Biol Chem. 1995;270:22595–601. doi: 10.1074/jbc.270.38.22595. [DOI] [PubMed] [Google Scholar]
- 39.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–80. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 40.Glazer PM, Sarkar SN, Summers WC. Detection and analysis of UV-induced mutations in mammalian cell DNA using a lambda phage shuttle vector. Proc Natl Acad Sci U S A. 1986;83:1041–4. doi: 10.1073/pnas.83.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dagle JM, Weeks DL. Positively charged oligonucleotides overcome potassium-mediated inhibition of triplex DNA formation. Nucleic Acids Res. 1996;24:2143–9. doi: 10.1093/nar/24.11.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zendegui JG, Vasquez KM, Tinsley JH, Kessler DJ, Hogan ME. In vivo stability and kinetics of absorption and disposition of 3′ phosphopropyl amine oligonucleotides. Nucleic Acids Res. 1992;20:307–14. doi: 10.1093/nar/20.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasquez KM, Dagle JM, Weeks DL, Glazer PM. Chromosome targeting at short polypurine sites by cationic triplex-forming oligonucleotides. J Biol Chem. 2001;276:38536–41. doi: 10.1074/jbc.M101797200. [DOI] [PubMed] [Google Scholar]
- 44.Rogers FA, Manoharan M, Rabinovitch P, Ward DC, Glazer PM. Peptide conjugates for chromosomal gene targeting by triplex-forming oligonucleotides. Nucleic Acids Res. 2004;32:6595–604. doi: 10.1093/nar/gkh998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gunther EJ, Yeasky TM, Gasparro FP, Glazer PM. Mutagenesis by 8-methoxypsoralen and 5-methylangelicin photoadducts in mouse fibroblasts: mutations at cross-linkable sites induced by offoadducts as well as cross-links. Cancer Res. 1995;55:1283–8. [PubMed] [Google Scholar]
- 46.Stachelek GC, Dalal S, Donigan KA, Campisi Hegan D, Sweasy JB, Glazer PM. Potentiation of temozolomide cytotoxicity by inhibition of DNA polymerase beta is accentuated by BRCA2 mutation. Cancer Res. 2010;70:409–17. doi: 10.1158/0008-5472.CAN-09-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]