Abstract
The affinity-mediated selection of large libraries of DNA-encoded small molecules is increasingly being used to initiate drug discovery programs. We present universal methods for the encoding of such libraries using the chemical ligation of oligonucleotides. These methods may be used to record the chemical history of individual library members during combinatorial synthesis processes. We demonstrate three different chemical ligation methods as examples of information recording processes (writing) for such libraries and two different cDNA-generation methods as examples of information retrieval processes (reading) from such libraries. The example writing methods include uncatalyzed and Cu(I)-catalyzed alkyne-azide cycloadditions and a novel photochemical thymidine-psoralen cycloaddition. The first reading method “relay primer-dependent bypass” utilizes a relay primer that hybridizes across a chemical ligation junction embedded in a fixed-sequence and is extended at its 3′-terminus prior to ligation to adjacent oligonucleotides. The second reading method “repeat-dependent bypass” utilizes chemical ligation junctions that are flanked by repeated sequences. The upstream repeat is copied prior to a rearrangement event during which the 3′-terminus of the cDNA hybridizes to the downstream repeat and polymerization continues. In principle these reading methods may be used with any ligation chemistry and offer universal strategies for the encoding (writing) and interpretation (reading) of DNA-encoded chemical libraries.
Keywords: chemical ligation, click chemistry, combinatorial chemistry, drug discovery, modified nucleotide, photochemistry, polymerase, template-dependent polymerization, unimolecular information recording and retrieval, in vitro selection
Introduction
The use of DNA-encoded chemical libraries permits the simultaneous interrogation of large numbers of small-molecules using affinity-mediated selection methodologies.1-11 Such libraries have recently been reported to have yielded potent small-molecule inhibitors for a range of therapeutic target proteins10,12-18 including for challenging targets such as the inhibition of protein-protein interactions.19-22 During the chemical synthesis of DNA-encoded libraries of small-molecules the chemical history of individual library members is either recorded using the sequences of oligonucleotide tags that are ligated to the library member within each of multiple chemistry-specific compartments or directed using a pre-existing oligonucleotide template. During DNA-recorded chemical library generation successive cycles of split, encoding by tagging, chemistry and pool generate large combinatorial mixtures of different small-molecules, the identities of each of which may be inferred by determining the sequence of the attached concatenated oligonucleotide tag set. A range of oligonucleotide tagging strategies has been reported to be suitable for the generation of DNA-recorded libraries including solid-phase coupled peptide and oligonucleotide synthesis,1,23 the enzymatic ligation of single-stranded oligonucleotides,2 the enzymatic extension of hybridized single-stranded oligonucleotides,24 the enzymatic ligation of double-stranded oligonucleotides10 and the chemical ligation of single-stranded oligonucleotides.25 Related strategies have been reported for the generation of DNA-directed libraries using template-directed chemical synthesis on a pre-existing template3,5,11 or hybridization-dependent routing of pre-existing templates to chemistry-specific compartments.6-8
Many of these oligonucleotide tagging schemes suffer from significant limitations. Hybridization-mediated strategies are limited to small split sizes because large differences need to exist between the sequences of all members of the encoding oligonucleotide set in order to reduce mis-hybridization rates. And some of these strategies require the covalent conjugation of each chemical building-block to a different oligonucleotide tag which will become laborious for large sets of chemical building blocks. Also, the use of enzymes to effect ligation or extension requires the alternate establishment of chemistry-compatible and enzyme-compatible conditions often within large numbers of low-volume compartments and can suffer from the inhibition of enzyme activity by residual chemistry components.
The chemical ligation of oligonucleotides is an attractive alternative encoding strategy. Chemical reactions are generally more versatile than enzymatic reactions and in many instances may be performed under a range of solution conditions which could obviate the need for buffer exchange within thousands of low-volume compartments - as has been reported for some enzymatic tagging strategies.10 One of the more significant limitations of chemical ligation-mediated encoding schemes is the apparent need to generate a chemical linkage between two oligonucleotides that may be traversed by a template-dependent polymerase in order that tag identities and associations may be determined by sequencing. In this paper we demonstrate two different cDNA-generation (reading) strategies for chemically ligated oligonucleotides—neither of which require that a polymerase be able to translocate through the chemical ligation junction. We also demonstrate three different example chemical ligation strategies that may be read by these processes. In principle both of these cDNA-generation strategies permit the use of any chemical bond-forming reaction to support an oligonucleotide tagging strategy for DNA-encoded chemical library generation.
Strategies for generating chemically ligated oligonucleotides that polymerases may translocate through have also been reported and also offer alternative approaches for the synthesis of DNA-encoded chemical libraries. Examples include the use of cyanogen bromide or water-soluble carbodiimides to generate native phosphodiester linkages from 5′-monophospho and 3′-hydroxyl oligonucleotides,26,27 the use of 5′-Iodo and 3′-Phosphorothio oligonucleotides to generate phosphorothiodiester linkages,28 the use of Cu(I)-catalyzed triazole synthesis with 5′-propargyl and 3′-azido oligonucleotides29 and the use of Cu(I)-catalyzed triazole synthesis with 5′-azido and 3′-propargyl oligonucleotides.25,30 The utility of such approaches for the generation of DNA-encoded libraries of small-molecules is dependent upon the extent to which tag polymerization can be avoided by for example by making chemical ligations template-dependent, or the development of a suitable protection strategy, or the use of pairs of orthogonal chemistries. Challenges to these approaches are likely to be experienced with regard to yield, removal of unreacted tags and interference with the reaction scheme used to generate the encoded chemical entity, but we believe that these can be overcome and we are also exploring such approaches.25 In this paper we report on strategies for the generation of soluble unimolecular oligonucleotide-encoded information (writing) and the determination of this information (reading) using chemical linkages between oligonucleotides that are not traversable by polymerases. We present three chemical ligation methods and two cDNA-generation methods by which this may be achieved and support each with experimental demonstration of both the writing and the reading processes.
Results
Generation of cDNA from a chemically ligated conjugate using relay primer-dependent bypass
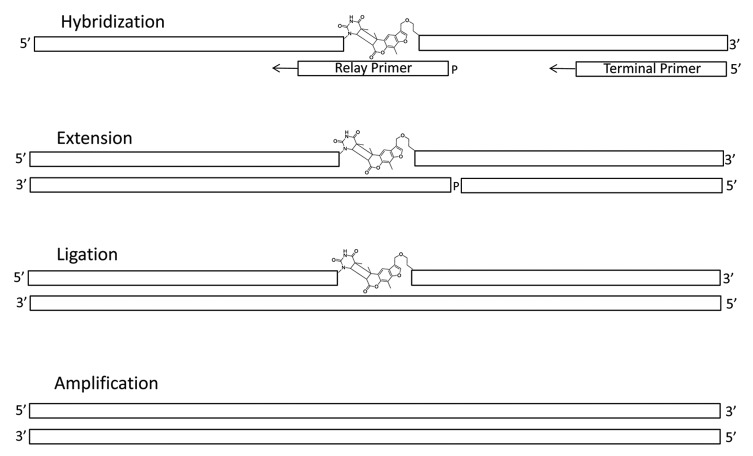
Relay-primer dependent bypass utilizes a “relay primer” and coupled template-dependent polymerization and ligation as a means to generate contiguous cDNA sequences that can in turn be amplified, cloned and sequenced. In this approach both termini of the concatenated tag set contain sufficient fixed sequence for the purposes of primer-binding and each chemical ligation junction is flanked on either side by sufficient fixed sequence for the purposes of relay primer-binding. A relay primer is our term for an oligonucleotide that hybridizes to a pair of chemically ligated tags in a manner that bridges their ligation junction. In the presence of a non-strand displacing template-dependent polymerase and annealed 5′-terminal and 5′-phosphorylated relay primers each primer is extended up to the 5′-terminus of the next. The addition of a ligase then seals the nick between these and generates a contiguous cDNA that may then be amplified, cloned and sequenced as part of the reading process. This process is shown in Figure 1. With multiple relay primers the sequences of multiple concatenated tags may be copied into a single cDNA. We refer to this method as relay primer-dependent bypass and note it’s similarity to the manner in which Okazaki fragments are generated on the lagging template strand and then ligated together during DNA replication.31
Figure 1. Mechanism of cDNA generation from chemically ligated tags using relay primer-mediated bypass on a photochemically ligated non-traversable template. A terminal primer is annealed to the 3′-terminus of the chemically ligated oligonucleotide and a second 5′-monophosphorylated primer (relay primer) is annealed across the chemical ligation junction. Extension of both primers and ligation generates full-length cDNA including potentially encoding sequences from both upstream and downstream of the photochemical ligation junction.
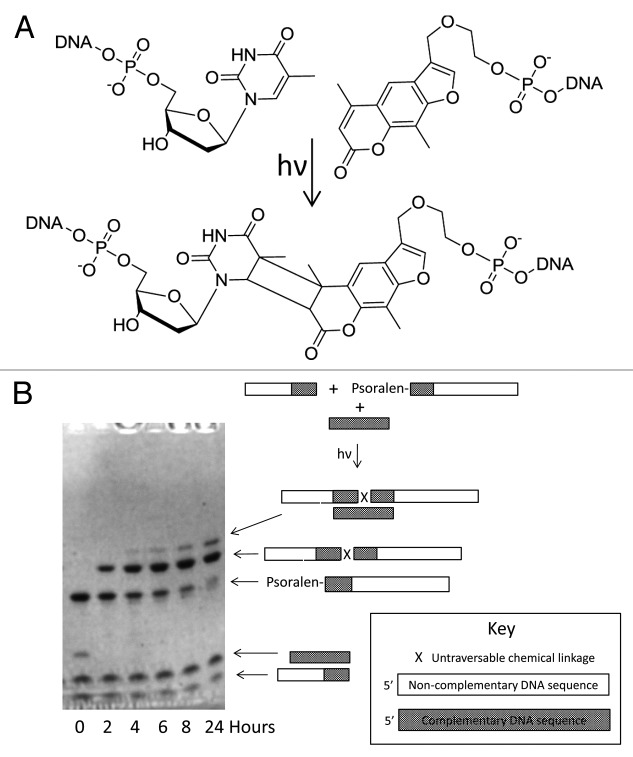
In order to demonstrate relay primer-dependent bypass we used a psoralen-mediated photochemical ligation strategy to generate a cyclobutane adduct between an adjacent 3′-thymidine and a 5′-psoralen in a template-dependent reaction using a splint oligonucleotide to co-locate the photo-reactive oligonucleotide termini as shown in Figure 2A and B. The stereochemistry of the thymidine-psoralen adduct may be either cis or trans.32 The linkage thus formed lacks a contiguous sugar-phosphate backbone and does not resemble a wild-type internucleotide linkage. This ligation is efficient as indicated in Figure 2B with conversions exceeding 90% and the conjugate of (1) and (4) was synthesized with a yield exceeding 80% and an observed deconvoluted MW of 15,819 Da (calculated MW 15,813 Da). Photochemistry of this kind is orthogonal to most other chemical reactions and may be performed under a range of solution conditions - we have determined that it occurs with high yield at pH values as low as 5.5 and as high as 9.4 as shown in Table 1.
Figure 2. Psoralen-mediated photochemical oligonucleotide ligation. (A) The photochemical ligation of a 3′-thymidine oligonucleotide to a 5′-psoralen oligonucleotide after co-annealing to a complementary splint oligonucleotide (not shown). The stereochemistry of the thymidine-psoralen adduct may be either cis or trans. (B) Denaturing PAGE analysis of the time-course of the photochemical reaction that results in the ligation of a 3′-thymidine oligonucleotide to a 5′-psoralen oligonucleotide with illumination at 365 nm, the gel is imaged by UV shadowing of a fluorescent plate. The appearance of the photochemical ligation product is indicated with a conversion of 50% occurring between two and four hours of illumination.
Table 1. Yields of psoralen-mediated oligonucleotide-templated photochemical oligonucleotide ligations under different solution conditions.
| Buffer | Psoralen-thymidine ligation conversion |
|---|---|
| pH 5.5 Phosphate | 65% |
| pH 7.0 Phosphate | 88% |
| pH 9.4 Borate | 80% |
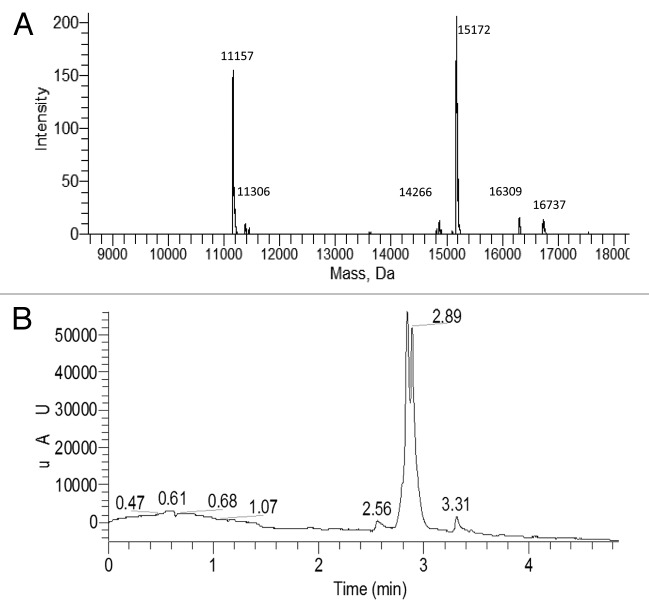
Oligonucleotides modified with psoralen are easily obtained commercially and photochemistry is readily applied to multi-well plates containing many low-volume samples. Accordingly, a photochemical ligation strategy such as this appears to be well-suited for the generation of DNA-recorded chemical libraries so long as tag identities and associations can be determined by sequencing. The first step in the process of determining the sequence of photoligated concatenated psoralen oligonucleotide tags is to generate a contiguous cDNA from which the sequences and associations of the photoligated tags can be deduced. To facilitate the subsequent separation of the cDNA from the photoligated template for analysis purposes the 5′-terminus of the template was biotinylated, and to facilitate detection of the cDNA the 5′-terminus of the terminal primer was fluoresceinylated. The photoligated conjugate was hybridized to the 5′-terminal primer and the 5′-monophosphorylated relay primer. This hybridized complex was then incubated with T4 DNA Polymerase to extend both the relay primer and the terminal primer. T4 DNA Polymerase was chosen because it does not exhibit strand-displacement activity27 and so was expected to extend the 3′-terminus of the terminal primer up to the 5′-terminus of the relay primer without displacing it. Because the 5′-terminus of the relay primer is 5′-monophosphorylated the junction between the extended relay primer and the extended terminal primer is a substrate for ligation with T4 DNA Ligase which was then added to the incubating mixture along with ATP. The steps in this process are outlined in Figure 1. Once the incubation was complete the hybridized complex was incubated with immobilized streptavidin to capture the chemically ligated conjugate and the cDNA was eluted with 0.1 M NaOH and neutralized. LCMS analysis is shown in Figure 3A and B with detection of the fluorescein at 495 nm indicating that approximately 50% of the terminal primer had now been extended to generate a product with a mass close to that of full-length cDNA (observed deconvoluted MW 15 172 Da, calculated MW 15 459 Da) with the observed mass suggesting the likely loss of one dT. The remaining approximately 50% corresponds to the extension of the terminal primer up to the psoralen ligation junction (observed deconvoluted MW 11 157 Da, calculated MW 11 154 Da). A control experiment in which the T4 DNA Ligase was omitted gave no full-length cDNA as expected. Sequencing indicated the cDNA had the expected sequence except for the absence of a 3′-terminal dT along with a shorter cDNA that was terminated at the psoralen ligation junction. Of 34 full-length cDNA, all were missing a single dT from the 3′-terminal region with 33 of these containing no more than one additional deletion or substitution.
Figure 3. cDNA generation from chemically ligated tags using relay primer-mediated bypass on a photochemically ligated non-traversable template. (A) LC analysis showing approximately equal amounts of two primer-extension products after purification with detection at 495 nm. (B) Deconvoluted MS analysis of mixed primer-extension products showing two principal components with masses of 15,172 Da and 11,157 Da which correspond to full-length cDNA minus one dT, and primer extension up to the photochemical ligation junction respectively.
Generation of cDNA from a chemically ligated conjugate using repeat-dependent bypass
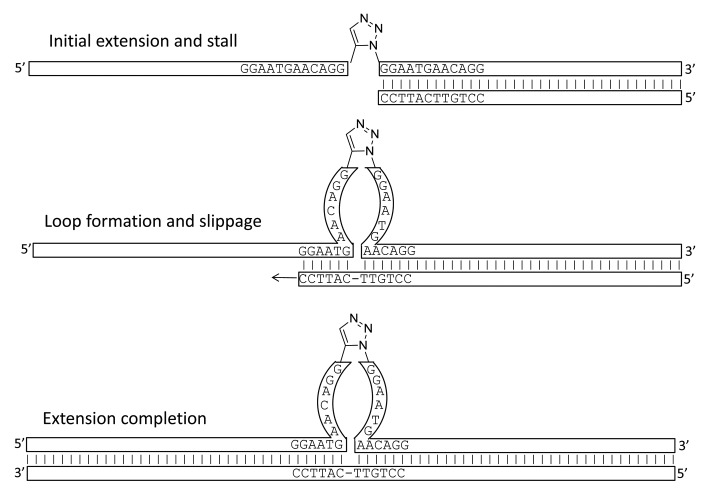
Repeat-dependent bypass utilizes a “repeat” architecture in which the chemical ligation junction is flanked upon either side by the same repeated sequence. In the presence of a hybridized upstream primer and a suitable polymerase the primer will be extended up to the chemical ligation junction and then stall having copied the first of the repeated sequences. If the rearrangement of this stalled complex results in hybridization of the 3′-terminus of the newly synthesized cDNA to the second of the repeated sequences then template-dependent polymerization may continue. This process is shown in Figure 4. With multiple different repeats flanking multiple chemical ligation junctions multiple concatenated tags may be copied into a single cDNA. We have shown by experiment that this process readily occurs, does not lead to the shuffling of tag combinations in free solution, and we propose a mechanism by which it may occur. We refer to this method as repeat-dependent bypass and note it’s similarity to the manner in which repeated sequences within genomic DNA can be contracted or expanded in processes that can lead to genomic instability33,34 and have been observed to occur in vitro to different extents with different polymerases.35
Figure 4. Proposed mechanism for the generation of cDNA from chemically ligated tags using repeat-dependent bypass. A terminal primer is annealed to the 3′-terminus of a chemically ligated oligonucleotide with a repeated sequence either side of the chemical ligation junction. Upon extension of this primer with a polymerase the newly generated cDNA stalls at the chemical ligation junction having copied the first of the repeated sequences. Rearrangement occurs with loop formation and the slippage of the terminus of the cDNA to become hybridized to the second instance of the repeated sequence, whereupon extension resumes and generates a full-length cDNA except for the deletion of one of the repeated sequences.
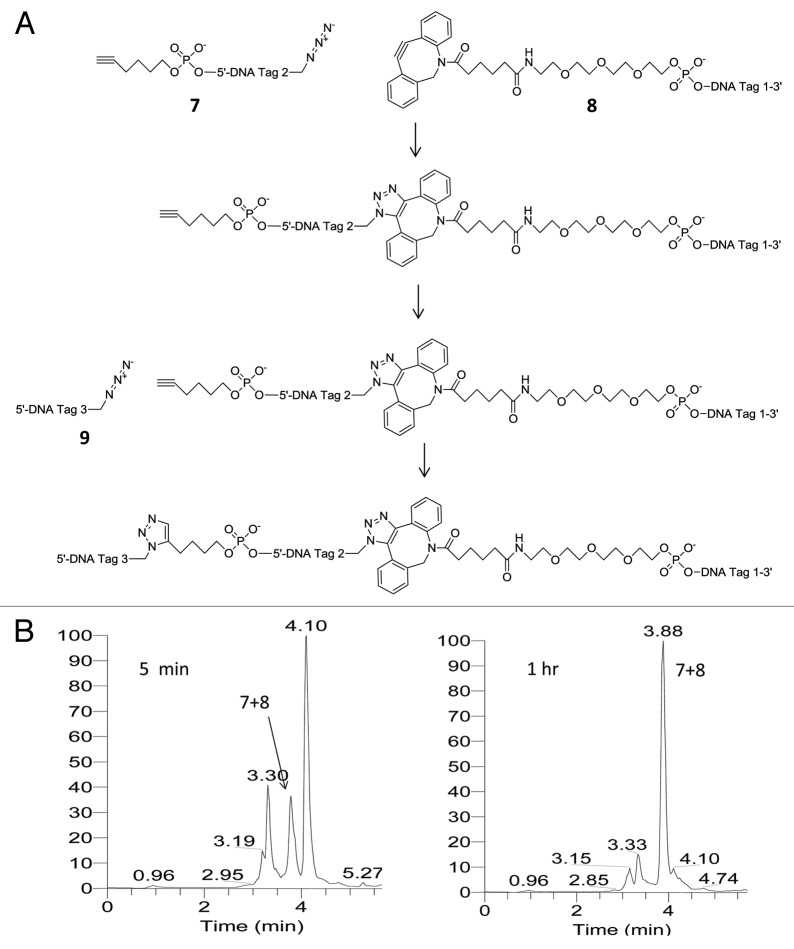
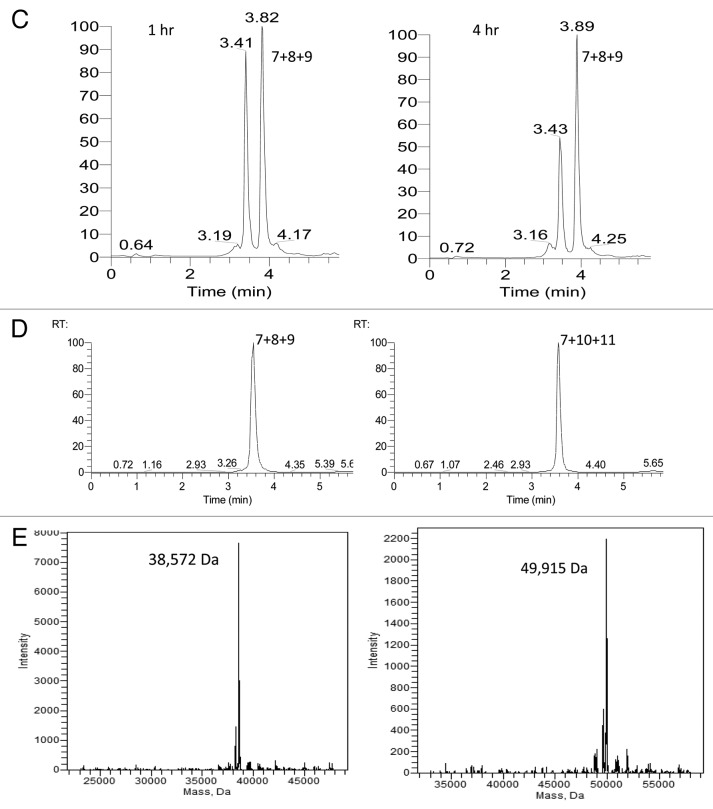
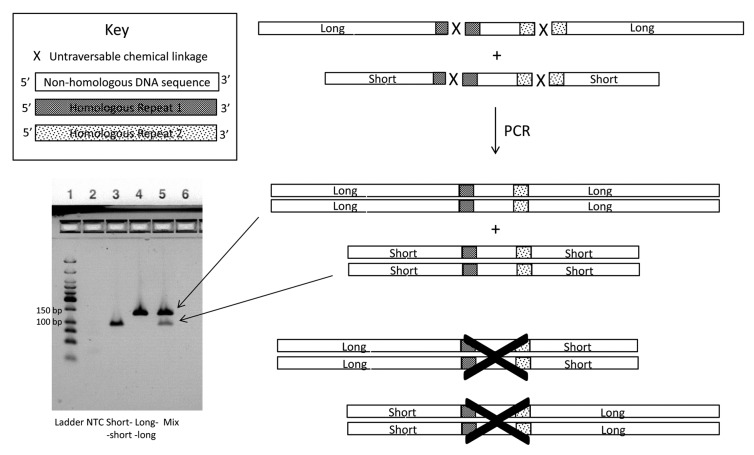
In order to demonstrate repeat-dependent bypass we successively used a Cu(I)-free triazole formation reaction followed by a Cu(I)-catalyzed triazole formation reaction thereby enabling the controlled sequential conjugation of a central oligonucleotide functionalized with both a 5′-alkynyl and a 3′-azide first by reacting it at its 3′-terminus and then at its 5′-terminus as shown in Figure 5A. The chemical structure of each linkage is different and neither was expected to be traversable by a polymerase in the absence of a bypass mechanism because of the relatively large distance between the ligated bases and the fact that similar length triazole linkages have been shown not to function as templates for PCR amplification.29 This was tested experimentally as described below. The Cu(I)-free ligation conversion was over 90% (Fig. 5B) and the Cu(I)-catalyzed ligation conversion was over 75% (Fig. 5C). After gel-purification and extraction we isolated 6 and 8 nmoles (40 and 53%) of each of two doubly conjugated oligonucleotides—one constructed by conjugating (7) to (8) and (9) (“short-short”) and the other by conjugating (7) to (10) and (11) (“long-long”). The final products were analyzed by LCMS with observed deconvoluted MW values of 38,572 and 49,915 Da (calculated MW 38 560 and 49 917 Da) as shown in Figure 5D and E. We screened a panel of DNA polymerases for their ability to read through the Cu(I)-catalyzed linkage formed by conjugating (7) and (9) including SuperScript III (Invitrogen), Taq (NEB), Thermoscript (Invitrogen), KOD (Novagen), Vent (NEB), Vent (exo-) (NEB), Deep Vent (NEB) and Deep Vent (exo-) (NEB). We observed that PCR products were only generated with Vent (exo-) and Deep Vent (exo-). Similar constructs without a repeated sequence flanking the chemical ligation junction gave no PCR products under the same conditions. PCR amplification was conducted for each conjugate separately and for an equimolar mixture of the two conjugates using Deep Vent (exo-) DNA Polymerase. Analysis of the PCR products by agarose gel electrophoresis showed that the amplification of each of the isolated conjugates generates double-stranded PCR products with electrophoretic mobilities that correspond in each case to the loss of one of each of the repeated sequences in each of the pairs of ligation junctions—as shown in lanes 3 and 4 of Figure 6. The electrophoretic mobility values observed are consistent with the operation of the repeat-dependent bypass mechanism shown in Figure 5. The short-short conjugate is 120 nucleotides in length with an expected PCR amplification product length of 96 base-pairs and the long-long conjugate is 156 nucleotides in length with an expected PCR amplification product length of 132 base-pairs. The mixed conjugate sample was also analyzed by cloning and sequencing. Of 35 sequence reads, 25 were observed to be of the long-long combination and 10 of the short-short combination with no chimeric sequences observed with all sequence reads showing only one of each of the repeated sequences at each junction.
Figure 5A and B. (A) Controlled concatenation of three oligonucleotides using successive Cu(I)-free and Cu(I)-catalyzed chemical ligation by triazole formation. First the central oligonucleotide (7) is chemically ligated to (8) using the copper-free reaction of the azide group in (7) with the strained alkyne in (8), thereby preserving the terminal alkyne in (7) which is subsequently reacted in a Cu(I)-catalyzed reaction with the azide in the subsequently added third oligonucleotide (9). (B) LC time-course of Cu(I)-free conjugation of (7) with (8) using absorbance at 260 nm.
Figure 5C–E. (C) LC time-course of Cu(I)-catalyzed conjugation of (9) with the conjugate of (7) and (8) using absorbance at 260 nm. (D) LC analysis of gel-purified doubly-conjugated products (7)+(8)+(9) and (7)+(10)+(11) using absorbance at 260 nm. (E) Deconvoluted MS of gel-purified doubly-conjugated products (7)+(8)+(9) “short-short” and (7)+(10)+(11) “long-long.”
Figure 6. Demonstration of the fidelity of cDNA generation from chemically ligated tags using repeat-dependent bypass. A pair of doubly-conjugated oligonucleotides was prepared - one by the chemical ligation of two short oligonucleotides to a central oligonucleotide and the other by the chemical ligation of two long oligonucleotides to the same central oligonucleotide. The corresponding junction in each conjugate was flanked by the same repeated sequence. cDNA generation and subsequent PCR and electrophoresis using ethidium-stained agarose indicates that each of the conjugates may be amplified using relay primer-mediated bypass and that amplification occurs without the occurrence of homology-mediated shuffling, as indicated by the presence of bands with mobilities corresponding to amplification products containing “long-long” and “short-short” tag combinations, and the absence of the chimeric amplification products “long-short” and “short-long.”
Discussion
Relay primer-dependent bypass
In order to demonstrate cDNA generation using relay primer-dependent bypass and a template interrupted by a non-polymerase traversable chemical ligation junction we first developed a method to generate a psoralen-thymidine ligated oligonucleotide. In contrast to the more well-known use of psoralen-modified oligonucleotides to generate psoralen-thymidine cyclobutane interstrand cross-links between hybridized oligonucleotides (see for example refs. 36 and 37) this method generates psoralen-thymidine cyclobutane linkages between adjacent nucleotides on the same strand and appears to be novel. This linkage is formed by co-hybridizing a 3′-thymidine oligonucleotide adjacent to a 5′-psoralen oligonucleotide to a complementary splint oligonucleotide that had no thymidines in closer proximity to the psoralen than four nucleotides away in the 3′-direction or two nucleotides away in the 5′-direction (closer proximity was not tested), and then irradiating the complex with UV light. We were able to generate this conjugate with isolated yields of approximately 80%. Yields of this magnitude translate into overall yields of 50% for the successive ligation of two tags and of 40% for the successive ligation of three tags. For DNA-encoded chemical library applications these numbers of encoded cycles are of most interest to the medicinal chemistry community as they are more likely to lead to Lipinski-compliant compounds.38 For cDNA generation a 5′-biotinylated version of the acceptor oligonucleotide was used (4) and the product was then isolated using denaturing PAGE. Using this template we were able to demonstrate the formation of cDNA with a yield of 50% and with the expected sequence. These observations demonstrate our ability to photochemically ligate 5′-psoralen oligonucleotides to 3′-thymidine oligonucleotides using a complementary oligonucleotide splint (writing) and to then generate cDNA that permits the determination of the sequences and associations of ligated tags (reading).
Repeat-dependent bypass
In order to demonstrate the generation of cDNA using repeat-dependent bypass using a conjugate interrupted by a pair of non-polymerase-traversable chemical ligation junctions we first developed a method that allows for the controlled generation of two triazole chemical ligation junctions within a single conjugate using a copper-free triazole formation from an azide and a strained alkyne39 followed by a Cu(I)-mediated triazole formation similar to other reported reactions.40,41 This approach permits the specific conjugation of three oligonucleotides without the need to use any protecting groups, also in high yield. For cDNA generation an oligonucleotide conjugate was generated by the chemical ligation of (7), (8) and (9) (“short-short”) and used as a template for PCR along with a similar second conjugate generated by the chemical ligation of (7), (10) and (11) (“long-long”). Each of these conjugates has two chemical ligation junctions, within each conjugate each junction is flanked by a different repeated sequence, but these repeated sequences are the same for each of the long-long and short-short conjugates. Both conjugates were used to demonstrate that conjugates of this general design can be used to program template-dependent polymerase-dependent amplifications such as PCR. Because the conjugates are of different lengths we were also able to use them to demonstrate whether information encoded in ligated tag set combinations would be preserved during repeat-dependent bypass amplification, or alternatively whether it would be lost via recombination-mediated processes similar to those seen during DNA shuffling.42 We were able to show that amplification using each of these templates resulted in amplification products containing only one of each repeated sequence at each junction, and that mixed amplification products were not generated when the templates were mixed prior to amplification. These data are consistent with our proposed loop-formation/slippage repeat-dependent bypass model (Fig. 4) and suggest that if recombination processes are occurring they are relatively infrequent. Accordingly sequence data derived from the amplification of chemically ligated conjugates using this cDNA-generation process can be used to infer the identities and co-association statistics of ligated oligonucleotides present in the input conjugates—as is required for the successful application of this strategy to the encoding of the chemical history of individual library members within DNA-encoded libraries.
Material and Methods
Synthesis of an oligonucleotide conjugate with one psoralen-thymidine photochemical ligation junction
Oligonucleotides (1), (2) and (3) (shown in Scheme 1) were acquired from Integrated DNA Technologies. All three were mixed together in aqueous 500 mM sodium phosphate at pH 7.0 with the psoralen oligonucleotide (1) at 1 mM and the other two oligonucleotides at 1.1 mM. The mixture was then briefly heated to 95 °C followed by slow cooling to room temperature to allow hybridization to occur. Ten microliters aliquots were then cooled to 4 °C and irradiated in 1.5 ml polypropylene microcentrifuge tubes (Fisher Scientific, 02-682-550) with UV light at 365 nm using a UVL-21 compact UV lamp (UVP). Aliquots of 1 uL were taken over a time-course and were analyzed by denaturing PAGE. The gel was visualized and photographed using the UV shadowing of a fluorescent TLC plate.
Scheme 1. Sequences and chemical modifications of oligodeoxynucleotides used in this study.
Generation of cDNA from an oligonucleotide conjugate with one psoralen-thymidine photochemical ligation junction
A gel-purified conjugate of (1) and (4) was then incubated in a 100 ul volume with a 5′-monophosphorylated relay primer (5) and a 5′-FAM-labeled terminal primer (6) each at 10 uM in T4 DNA Ligase Buffer (NEB) with 1 mM of each dNTP and 30 units of T4 DNA Polymerase (NEB). Following incubation for 1 h at 37 °C the mixture was supplemented with an additional 0.5 mM ATP and 10 units of T4 DNA Ligase (NEB) were added and it was then incubated for an additional 1 h at 37 °C. The reaction product was then purified by incubation with 200 μl of streptavidin-coated Dynabeads M280 (Invitrogen), washed, eluted with 35 μl of 0.1 M NaOH followed by neutralization with 1M pH 7.0 tris(hydroxymethyl)aminomethane. The product was then analyzed by LCMS on a Thermo Scientific LCQ Fleet using an ACE 3 C18–300 (50 × 2.1 mm) column and a 5 min gradient of 5–35% of buffer B using buffer A (1% hexafluoroisopropanol [HFIP], 0.1% di-isopropylethyl amine [DIEA], 10 μM EDTA in water) and buffer B (0.075% HFIP, 0.0375% DIEA, 10 μM EDTA, 65% acetonitrile/35% water). LC was monitored at 260 nm and 488 or 495 nm. MS was detected in the negative mode, and mass peak deconvolution was performed using ProMass software. In order to determine the sequence of both cDNA regions that were generated as a result of primer extensions, the 3′-terminus of the cDNA was first ligated to a new primer-binding oligonucleotide 5′-monophospho-GCTGTGCAGG TAGAGTGC-FAM-3′ using single-stranded enzymatic ligation with 1 unit/ul of T4 RNA Ligase 1 (NEB,) in 1× T4 RNA Ligase buffer (NEB), supplemented with 200 μM ATP (NEB), 1 mM hexamine cobalt chloride (Sigma-Aldrich) and 25% PEG8000 (NEB) for 16 h at room temperature. This newly ligated cDNA was then purified using denaturing PAGE with fluorescence to visualize the band of interest followed by excision, extraction and PCR amplification was then conducted using the ligated cDNA at 200 pM, with the forward primer 5′-GCACTCTACC TGCACAGC-3′ and the reverse primer 5′-GGCAGTACGC AAGCTCG-3′ each at 0.5 μM and Platinum Supermix PCR mix (Invitrogen). Sixteen thermal cycles were performed with denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s and extension at 72 °C for 60 s. Amplification products were visualized on ethidium bromide-stained 4% agarose E-Gels (Invitrogen) with illumination at 305 nm. Subsequently 0.2 units/μl of Klenow DNA Polymerase (NEB) were added to the PCR reaction followed by incubation for 15 min at 37 °C. PCR products were then cloned using a Zero Blunt TOPO cloning kit (Invitrogen) and these clones were Sanger sequenced (Genewiz).
Synthesis of an oligonucleotide conjugate with two different triazole chemical ligation junctions
Oligonucleotides (7), (8) and (9) were acquired from Integrated DNA Technologies (IA, USA). Oligonucleotides (7) and (8) were mixed together at 0.5 mM concentration in aqueous solution with 0.2 M sodium phosphate buffer at pH 7.0. The reaction progress was monitored by LCMS. To this mixture was then added oligonucleotide (9) and a Cu(I)-catalyzed triazole formation reaction was conducted by further incubation with two molar equivalents of Cu(II) acetate, four molar equivalents of sodium ascorbate and one molar equivalent of Tris-(BenzylTriazolylmethyl) Amine (TBTA) in a volume of 50 ul with oligonucleotides at a concentration of 0.5 mM. This mixture was incubated overnight at room temperature followed by LCMS and purification using denaturing PAGE.
Generation of cDNA from an oligonucleotide conjugate with two triazole chemical ligation junctions
PCR amplifications were conducted with conjugates at 40 pM, with the forward primer 5′-TGCGGTCTAA CTGTCTA-3′ and the reverse primer 5′- AAGCATAGCA CCCGATT-3′ each at 0.5 uM using 0.1 units/ul of Deep Vent (exo-) Polymerase (NEB) in 1× Thermopol Buffer (NEB) with dNTP at 0.5mM. Twenty-two thermal cycles were performed with denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s and extension at 72 °C for 60 s. Amplification products were visualized on ethidium bromide-stained 4% agarose E-Gels (Invitrogen) with illumination at 305 nm. PCR products were cloned using a Zero Blunt TOPO cloning kit (Invitrogen) and these clones were Sanger sequenced (Genewiz).
Conclusion
There is significant and increasing interest in the use of the affinity-mediated selection of DNA-encoded libraries of small molecules to discover inhibitors of proteins of therapeutic interest. In order to construct such libraries efficiently it is necessary to utilize robust methods for the ligation of oligonucleotides that are in turn able to support the determination of oligonucleotide tag sequences and associations while not interfering with the wide range of chemical reactions that will be used in the synthesis of the encoded library members. In this study we have demonstrated three different chemical ligation strategies—the uncatalyzed cycloaddition of a strained alkyne and an azide, the Cu(I)-catalyzed cycloaddition of a terminal alkyne and an azide and the photochemical cycloaddition of thymidine and psoralen. Each of these chemical ligations may be used as part of an encoding strategy for the chemical synthesis of encoded libraries. We have also developed two independent methods that may be used to generate cDNA that incorporates sequence information derived from oligonucleotide tags concatenated using these chemical ligations. cDNA generated by these methods may be amplified, cloned and sequenced and used to infer identity and statistical information about the output populations of molecules emerging from affinity-mediated selection experiments - and thereby used to identify small molecules that may inhibit therapeutic protein targets of interest. The presented methods for the generation of cDNA include two alternative strategies, neither of which depends upon the ability of a polymerase to translocate through the chemical ligation junction, and both of which may be considered universal. One method utilizes a 5′-phosphorylated relay primer that is hybridized across the ligation junction and extended by a polymerase downstream while an upstream primer is extended up to it to form a nick. Ligation then delivers a single contiguous cDNA that may then be amplified by PCR, cloned and sequenced. We call this method relay primer-dependent bypass and note it’s similarity to the manner in which Okazaki fragments are generated on the lagging template strand and then ligated together during DNA replication.31 A second method utilizes a conjugate architecture in which the same repeated sequence flanks the chemical ligation junction. We show that Deep Vent (exo-) Polymerase is able to extend a primer past this junction and we suggest a stem formation/slippage mechanism that is consistent with the observed loss of one of each of the repeated sequences in the derived cDNA. We call this second reading process repeat-dependent bypass and note it’s similarity to the manner in which repeated sequences within genomic DNA can be contracted.32,33 Both methods may be used to support oligonucleotide-tagging strategies to encode the chemical history of individual molecules that together comprise libraries of encoded variants synthesized using split-and-mix methodologies, similar for example to (10). Both methods also appear well-suited to support the use of libraries of such compounds in affinity-mediated selection techniques to discover small molecule inhibitors of proteins of therapeutic interest as has been reported for other related technologies, also similar for example to (10). Because both of these cDNA-generation methods are agnostic to the structural details of the chemical ligation junction they are in principle applicable to any chemical ligation oligonucleotide tagging method. Therefore the demonstrated instances may be considered examples of universal strategies. We expect that these methods will find utility in support of the generation and use of DNA-encoded chemical libraries to discover inhibitors of therapeutic protein targets within drug discovery programs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Christoph Dumelin for his careful reading and helpful comments during the preparation of this manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/artificialdna/article/27896
References
- 1.Brenner S, Lerner RA. Encoded combinatorial chemistry. Proc Natl Acad Sci U S A. 1992;89:5381–3. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinoshita Y, Nishigaki K. Enzymatic synthesis of code regions for encoded combinatorial chemistry (ECC) Nucleic Acids Symp Ser. 1995;34:201–2. [PubMed] [Google Scholar]
- 3.Gartner ZJ, Liu DR. The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules. J Am Chem Soc. 2001;123:6961–3. doi: 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gartner ZJ, Kanan MW, Liu DR. Expanding the reaction scope of DNA-templated synthesis. Angew Chem Int Ed Engl. 2002;41:1796–800. doi: 10.1002/1521-3773(20020517)41:10<1796::AID-ANIE1796>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 5.Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR. DNA-templated organic synthesis and selection of a library of macrocycles. Science. 2004;305:1601–5. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halpin DR, Harbury PB. DNA display I. Sequence-encoded routing of DNA populations. PLoS Biol. 2004;2:E173. doi: 10.1371/journal.pbio.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halpin DR, Harbury PB. DNA display II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution. PLoS Biol. 2004;2:E174. doi: 10.1371/journal.pbio.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halpin DR, Lee JA, Wrenn SJ, Harbury PB. DNA display III. Solid-phase organic synthesis on unprotected DNA. PLoS Biol. 2004;2:E175. doi: 10.1371/journal.pbio.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melkko S, Scheuermann J, Dumelin CE, Neri D. Encoded self-assembling chemical libraries. Nat Biotechnol. 2004;22:568–74. doi: 10.1038/nbt961. [DOI] [PubMed] [Google Scholar]
- 10.Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat Chem Biol. 2009;5:647–54. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 11.Hansen MH, Blakskjaer P, Petersen LK, Hansen TH, Højfeldt JW, Gothelf KV, Hansen NJV. A yoctoliter-scale DNA reactor for small-molecule evolution. J Am Chem Soc. 2009;131:1322–7. doi: 10.1021/ja808558a. [DOI] [PubMed] [Google Scholar]
- 12.Deng H, O’Keefe H, Davie CP, Lind KE, Acharya RA, Franklin GJ, Larkin J, Matico R, Neeb M, Thompson MM, et al. Discovery of highly potent and selective small molecule ADAMTS-5 inhibitors that inhibit human cartilage degradation via encoded library technology (ELT) J Med Chem. 2012;55:7061–79. doi: 10.1021/jm300449x. [DOI] [PubMed] [Google Scholar]
- 13.Gentile G, Merlo G, Pozzan A, Bernasconi G, Bax B, Bamborough P, Bridges A, Carter P, Neu M, Yao G, et al. 5-Aryl-4-carboxamide-1,3-oxazoles: potent and selective GSK-3 inhibitors. Bioorg Med Chem Lett. 2012;22:1989–94. doi: 10.1016/j.bmcl.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 14.Disch JS, Evindar G, Chiu CH, Blum CA, Dai H, Jin L, Schuman E, Lind KE, Belyanskaya SL, Deng J, et al. Discovery of thieno[3,2-d]pyrimidine-6-carboxamides as potent inhibitors of SIRT1, SIRT2, and SIRT3. J Med Chem. 2013;56:3666–79. doi: 10.1021/jm400204k. [DOI] [PubMed] [Google Scholar]
- 15.Podolin PL, Bolognese BJ, Foley JF, Long E, 3rd, Peck B, Umbrecht S, Zhang X, Zhu P, Schwartz B, Xie W, et al. In vitro and in vivo characterization of a novel soluble epoxide hydrolase inhibitor. Prostaglandins Other Lipid Mediat. 2013;104-105:25–31. doi: 10.1016/j.prostaglandins.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Scheuermann J, Dumelin CE, Melkko S, Zhang Y, Mannocci L, Jaggi M, Sobek J, Neri D. DNA-encoded chemical libraries for the discovery of MMP-3 inhibitors. Bioconjug Chem. 2008;19:778–85. doi: 10.1021/bc7004347. [DOI] [PubMed] [Google Scholar]
- 17.Kleiner RE, Dumelin CE, Tiu GC, Sakurai K, Liu DR. In vitro selection of a DNA-templated small-molecule library reveals a class of macrocyclic kinase inhibitors. J Am Chem Soc. 2010;132:11779–91. doi: 10.1021/ja104903x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buller F, Steiner M, Frey K, Mircsof D, Scheuermann J, Kalisch M, Buhlmann P, Supuran CT, Neri D. Selection of carbonic anhydrase IX inhibitors from one million DNA-encoded compounds. ACS Chem Biol. 2011;6:336–44. doi: 10.1021/cb1003477. [DOI] [PubMed] [Google Scholar]
- 19.Buller F, Zhang Y, Scheuermann J, Schäfer J, Bühlmann P, Neri D. Discovery of TNF inhibitors from a DNA-encoded chemical library based on diels-alder cycloaddition. Chem Biol. 2009;16:1075–86. doi: 10.1016/j.chembiol.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Melkko S, Mannocci L, Dumelin CE, Villa A, Sommavilla R, Zhang Y, Grütter MG, Keller N, Jermutus L, Jackson RH, et al. Isolation of a small-molecule inhibitor of the antiapoptotic protein Bcl-xL from a DNA-encoded chemical library. ChemMedChem. 2010;5:584–90. doi: 10.1002/cmdc.200900520. [DOI] [PubMed] [Google Scholar]
- 21.Leimbacher M, Zhang Y, Mannocci L, Stravs M, Geppert T, Scheuermann J, Schneider G, Neri D. Discovery of small-molecule interleukin-2 inhibitors from a DNA-encoded chemical library. Chemistry. 2012;18:7729–37. doi: 10.1002/chem.201200952. [DOI] [PubMed] [Google Scholar]
- 22.Kollmann CS, Bai X, Tsai CH, Yang H, Lind KE, Skinner SR, Zhu Z, Israel DI, Cuozzo JW, Morgan BA, et al. Application of Encoded Library Technology (ELT) to a Protein-Protein Interaction target: Discovery of a Potent Class of Integrin Lymphocyte Function-associated Antigen 1 (LFA-1) J Med Chem. 2013 doi: 10.1016/j.bmc.2014.01.050. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen J, Brenner S, Janda KD. Synthetic methods for the implementation of encoded combinatorial chemistry. J Am Chem Soc. 1993;115:9812–3. doi: 10.1021/ja00074a063. [DOI] [Google Scholar]
- 24.Buller F, Mannocci L, Zhang Y, Dumelin CE, Scheuermann J, Neri D. Design and synthesis of a novel DNA-encoded chemical library using Diels-Alder cycloadditions. Bioorg Med Chem Lett. 2008;18:5926–31. doi: 10.1016/j.bmcl.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 25.Keefe AD, Wagner RW, Litovchick A, Clark MA, Cuozzo JW, Zhang Y, Centrella PA, Hupp CD. Methods for Tagging DNA-Encoded Libraries. Patent Cooperation Treaty Patent Application 2013; PCT/WO/2013/036810.
- 26.Naylor R, Gilham PT. Studies on some interactions and reactions of oligonucleotides in aqueous solution. Biochemistry. 1966;5:2722–8. doi: 10.1021/bi00872a032. [DOI] [PubMed] [Google Scholar]
- 27.Shabarova ZA, Merenkova IN, Oretskaya TS, Sokolova NI, Skripkin EA, Alexeyeva EV, Balakin AG, Bogdanov AA. Chemical ligation of DNA: the first non-enzymatic assembly of a biologically active gene. Nucleic Acids Res. 1991;19:4247–51. doi: 10.1093/nar/19.15.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Kool ET. Chemical and enzymatic properties of bridging 5′-S-phosphorothioester linkages in DNA. Nucleic Acids Res. 1998;26:3159–64. doi: 10.1093/nar/26.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Sagheer AH, Brown T. Synthesis and polymerase chain reaction amplification of DNA strands containing an unnatural triazole linkage. J Am Chem Soc. 2009;131:3958–64. doi: 10.1021/ja8065896. [DOI] [PubMed] [Google Scholar]
- 30.El-Sagheer AH, Sanzone AP, Gao R, Tavassoli A, Brown T. Biocompatible artificial DNA linker that is read through by DNA polymerases and is functional in Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:11338–43. doi: 10.1073/pnas.1101519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugimoto K, Okazaki T, Okazaki R. Mechanism of DNA chain growth, II. Accumulation of newly synthesized short chains in E. coli infected with ligase-defective T4 phages. Proc Natl Acad Sci U S A. 1968;60:1356–62. doi: 10.1073/pnas.60.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanne D, Straub K, Hearst JE, Rapoport H. Isolation and Characterization of Pyrimidine-Psoralen-Pyrimidine Photodiadducts from DNA. J Am Chem Soc. 1982;104:6764–9. doi: 10.1021/ja00388a046. [DOI] [Google Scholar]
- 33.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/SQB.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Bzymek M, Lovett ST. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A. 2001;98:8319–25. doi: 10.1073/pnas.111008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canceill D, Viguera E, Ehrlich SD. Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J Biol Chem. 1999;274:27481–90. doi: 10.1074/jbc.274.39.27481. [DOI] [PubMed] [Google Scholar]
- 36.Takasugi M, Guendouz A, Chassignol M, Decout JL, Lhomme J, Thuong NT, Hélène C. Sequence-specific photo-induced cross-linking of the two strands of double-helical DNA by a psoralen covalently linked to a triple helix-forming oligonucleotide. Proc Natl Acad Sci U S A. 1991;88:5602–6. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurz M, Gu K, Lohse PA. Psoralen photo-crosslinked mRNA-puromycin conjugates: a novel template for the rapid and facile preparation of mRNA-protein fusions. Nucleic Acids Res. 2000;28:E83. doi: 10.1093/nar/28.18.e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 39.Shelbourne M, Chen X, Brown T, El-Sagheer AH. Fast copper-free click DNA ligation by the ring-strain promoted alkyne-azide cycloaddition reaction. Chem Commun (Camb) 2011;47:6257–9. doi: 10.1039/c1cc10743g. [DOI] [PubMed] [Google Scholar]
- 40.Gierlich J, Burley GA, Gramlich PM, Hammond DM, Carell T. Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA. Org Lett. 2006;8:3639–42. doi: 10.1021/ol0610946. [DOI] [PubMed] [Google Scholar]
- 41.Kumar R, El-Sagheer A, Tumpane J, Lincoln P, Wilhelmsson LM, Brown T. Template-directed oligonucleotide strand ligation, covalent intramolecular DNA circularization and catenation using click chemistry. J Am Chem Soc. 2007;129:6859–64. doi: 10.1021/ja070273v. [DOI] [PubMed] [Google Scholar]
- 42.Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–91. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]