Abstract
Thrombin-binding aptamer (TBA) is a 15-nt DNA oligomer that efficiently inhibits thrombin. It has been shown that TBA folds into an anti-parallel unimolecular G-quadruplex. Its three-dimensional chair-like structure consists of two G-tetrads connected by TT and TGT loops. TBA undergoes fast degradation by nucleases in vivo. To improve the nuclease resistance of TBA, a number of modified analogs have been proposed. Here, we describe anomeric modifications of TBA. Non-natural α anomers were used to replace selected nucleotides in the loops and core. Significant stabilization of the quadruplex was observed for the anomeric modification of TT loops at T4 and T13. Replacement of the core guanines either prevents quadruplex assembly or induces rearrangement in G-tetrads. It was found that the anticoagulant properties of chimeric aptamers could be retained only with intact TT loops. On the contrary, modification of the TGT loop was shown to substantially increase nuclease resistance of the chimeric aptamer without a notable disturbance of its anticoagulant activity.
Keywords: thombin binding aptamer, anomer, quadruplex, anticoagulant properties, nuclease resistance
Introduction
DNA G-quadruplexes (GQ) have attracted much attention in recent years primarily due to the discovery of their protective role in telomeres and regulatory functions in the promoter regions of some biologically relevant genes.1 Another reason for the increased interest in GQ relates to their roles in the formation of three-dimensional structures of many DNA aptamers. Aptamers are most often referred to as relatively short single-stranded DNA or RNA oligonucleotides that are prepared by systematic evolution of ligands by exponential enrichment (SELEX) and show an ability to bind to certain cellular proteins with high affinity and specificity.2,3 It has been shown that thrombin binding aptamer (TBA) and aptamers binding to a number of HIV-1 proteins fold into GQ structures.4
TBA is probably the most widely known DNA aptamer, as evidenced by a large number of publications related to its biochemical and structural properties. It forms intramolecular GQ with two G-quartets in the core and three loops, one TGT and two TT loops.5,6 It has been established that aptamer loops are involved in recognition of the fibrinogen-binding site of the protein (exosite I) in a TBA-thrombin complex.7 The most recent model for the aptamer binding motif suggests that the TT loops act as a pincer-like system that embraces the protruding region of thrombin exosite I while the TGT loop is being fully exposed to the solvent.8 Thrombin plays an important role in thrombosis and hemostasis, functioning as the key enzyme in the conversion of fibrinogen to fibrin. Therefore, thrombin is considered a primary target in the development of aptamer therapeutics for certain cardiovascular disorders. TBA specifically inhibits thrombin in vivo at nanomolar concentrations.9 The drawback of using an unmodified TBA oligonucleotide is its sensitivity to serum nucleases and fast clearance in the kidneys.
Modification of an aptamer with non-natural nucleotides is a reasonable way to increase the in vivo stability of TBA. Because chemically modified nucleotides are not natural substrates of the polymerases used in SELEX, only a limited number of modified aptamer analogs could be developed using this procedure. Non-canonical NTPs that are well tolerated by polymerases are mostly 2′-substituted RNA derivatives, including 2′-fluoro, 2′-O-methyl and others.10 The requirements for the type of polymerase show wide variations depending on the nature of the modified NTPs.
An alternative route to modified aptamer analogs consists in the selective modification of a natural prototype. A number of TBA modifications have been developed to address thrombin activity, nuclease resistance and lifetime issues. In particular, studies on TBA analogs containing 3′-3′ and 5′-5′ junctions, LNA residues, internucleotide phosphorothioates, 2′-fluoroarabinose, isodeoxyguanosine and several other modified nucleotides have been reported.11-18 Enhanced binding affinity to thrombin and improved anticoagulant and antithrombotic properties have been observed for certain TBA modifications.11-13
In this study, we report the modification of TBA with anomeric α-nucleotides (Fig. 1). Synthetic α-oligonucleotides have been shown to be highly resistant to nuclease hydrolysis.19 Anomeric oligonucleotides may form hybrid parallel-stranded duplexes with their unmodified counterparts. Various types of DNA triplexes with anomeric and chimeric α-β strands have also been reported.19 However, there is only one previously reported study on the formation of anomeric GQ. Anomeric phosphorothioate oligonucleotide d(T2G4T2) was claimed to form parallel GQ and to inhibit HIV with an IC50 of 0.5 μM.20 Here, we studied a set of chimeric α-β TBA analogs. Modified aptamers with α-nucleotides at different positions were evaluated for their ability to form GQ structures and to inhibit thrombin. We demonstrate that modification of loop sites in TBA with α nucleotides induces changes in both the thermal stability of GQ and the inhibitory activity of chimeric aptamers. Nonetheless, loop modifications still fit within the same type of unimolecular GQ. In contrast, modification of the core guanines either prevents GQ assembly or induces formation of an intramolecular GQ structure with a different organization of G-quartets.
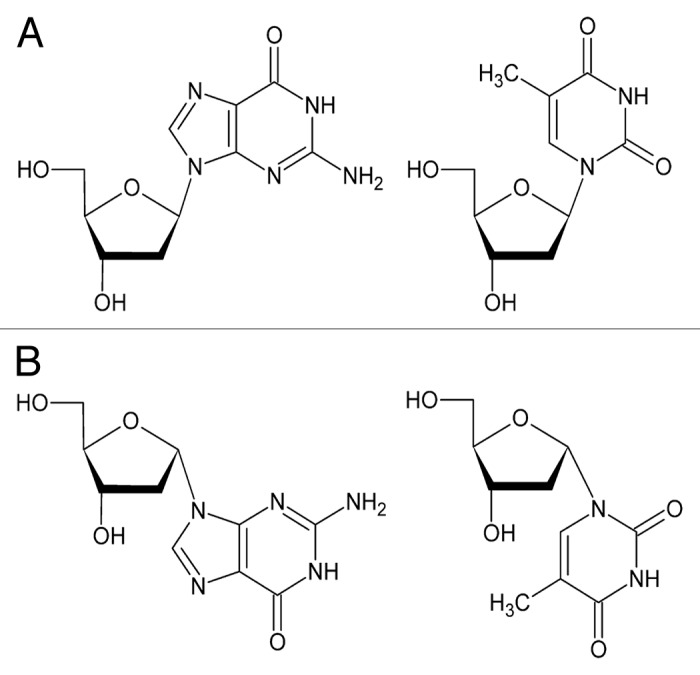
Figure 1. Natural β deoxyguanosine and thymidine (A), and their α anomeric analogs (B).
Results
Thermal stability of the chimeric TBAs
We differentiated loop regions from the quadruplex core for selection of particular nucleotides to be replaced in the natural prototype. Replacements were made separately in the core of the GQ, in the central TGT loop, and in the TT loops. Two oligonucleotides were designed to have modifications in both loop regions or to contain only α nucleotides. Thus, 13 modified aptamers were synthesized and studied in UV melting experiments in 100 mM KCl at 295 nm as summarized in Table 1.
Table 1. Sequences, biophysical and biochemical properties of modified aptamers.
| ID | Sequencea |
Tmb(UV), °C |
Tmb(CD), °C |
CD (positive bands), nm | Clotting timec, s |
|---|---|---|---|---|---|
| TBA | GGTTGGTGTGGTTGG | 51.0 | 51.3 | 245/295 | > 200 |
| A1 | GGTTGGTGTGGTTGG | < 30 | nd | nd | 13.9 |
| A2 | GGTTGGTGTGGTTGG | 51.9 | 52.3 | 245/295 | 14.0 |
| A3 | GGTTGGTGTGGTTGG | 60.3 | 60.8 | 245/295 | 14.4 |
| A4 | GGTTGGTGTGGTTGG | 40.7 | 39.7 | 245/295 | 15.1 |
| A5 | GGTTGGTGTGGTTGG | 49.6 | 48.9 | 245/295 | 14.4 |
| A6 | GGTTGGTGTGGTTGG | 41.7 | 42.5 | 245/295 | > 200 |
| A7 | GGTTGGTGTGGTTGG | 42.8 | 45.4 | 245/295 | 48 |
| A8 | GGTTGGTGTGGTTGG | nd | nd | nd | 14.6 |
| A9 | GGTTGGTGTGGTTGG | nd | nd | nd | 14.3 |
| A10 | GGTTGGTGTGGTTGG | nd | nd | nd | 14.3 |
| A11 | GGTTGGTGTGGTTGG | 54.0 | 55.9 | 245/295 | 14.0 |
| A12 | GGTTGGTGTGGTTGG | 38.9 | 39.8d | 265 | 14.8 |
| A13 | GGTTGGTGTGGTTGG | nd | nd | nd | 15.3 |
Alpha nucleotides are underlined. bAt 5 μM oligonucleotide concentration. cAt a final aptamer concenration of 1.5 μM. dAt 265 nm.
Transformation of nucleotides in either loop did not affect the ability of chimeric TBAs to fold into GQ structures. Thermal stabilities of modified aptamers fell within a wide 20 °C range and showed dependence on the location of the nucleotide replacement. Two modified aptamers (A3 and A11) were characterized by increased Tm values compared with the natural prototype. Modified oligonucleotides A2 and A5 were nearly as stable as TBA. A hallmark of the three stable variants (A2, A3, and A11) was the replacement with α thymidine at positions 4 and 13 in the TT loop regions, while aptamer A5 had one modification at position 13. The anomeric transformation in the other positions of the loop regions did not benefit the stability of the modified TBAs. The most notable stabilization was observed for aptamer A3 (ΔTm = 9.3 °C), with modifications at positions 4 and 13.
Modification of the core guanines displayed an entirely different effect. Only aptamer A12 was able to form a stable GQ. The arrangement of α deoxyguanosines in this aptamer gave us the first indication of the different type of GQ it may form. Indeed, alternating anomeric GG pairs in the sequence of aptamer A12, αα, and ββ most likely argue in favor of similar conformations adopted by each type of deoxyguanosine in both G-quartets. This is not true for the unmodified TBA with alternating syn and anti deoxyguanosines in every GG pair.5
Similar to the natural prototype, GQs modified in the loop region did not show any signs of hysteresis in heating-cooling cycles at the temperature ramp rate of 0.2 °C/min. Slow or fast annealing of the samples before melting also did not affect the observed denaturation profiles.
Aptamer A12 with modified guanines demonstrated more complicated melting behavior (Fig. S1). In 100 mM KCl, melting and annealing curves were not superimposable and appeared to be biphasic. Melting curves showed dependence on the sample preparation technique. The slow annealing rate during sample preparation favored notable hysteresis and a minor transition at approximately 60 °C. Instead, quick annealing of the sample in ice decreased both hysteresis and high-temperature transition. This high-temperature transition becomes larger at higher concentrations of aptamer A12. The opposite effect was observed at a lower K+ concentration. When 30 mM KCl was used in UV melting experiments, the denaturation curve showed only one major transition at 38 °C. These results most likely support the formation of an intramolecular GQ A12 with an admixture of intermolecular species at relatively high concentrations of KCl or the aptamer.
To estimate the contribution of intermolecular admixture in the folding of aptamer A12, we compared concentration dependence of the Tm value for modified oligonucleotides A2 and A12 in 100 mM KCl. Melting temperature of aptamer A12 was estimated at the minimum of first derivative for the major transition. Similar to aptamer A2, oligonucleotide A12 did not show any significant Tm dependence on the aptamer concentration (Fig. S2), thus suggesting the formation of unimolecular structure as a major folding mode.
Thermal difference spectra of the anomeric quadruplexes
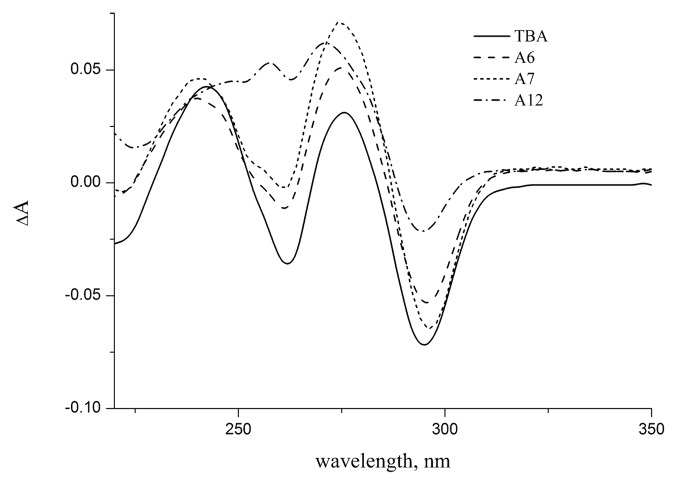
All TBA analogs with detected secondary structure were further characterized by their thermal difference spectra. A thermal difference spectrum (TDS) is easily generated by subtracting the UV spectra recorded above and below the melting temperature. The TDS has a specific pattern that is unique for each type of nucleic acid structure and reflects the conditions of base stacking.21 Unmodified TBA and all loop modified GQs showed very similar TDS signatures (Fig. 2; Fig. S3). Typical spectrum had two major positive peaks around 240 and 275 nm and a negative peak around 295 nm. Aptamer A12 with modified core showed different TDS spectrum implying other character of stacking interactions. TDS pattern of the aptamer A12 matches, nevertheless, specific signature of GQ family.
Figure 2. Thermal difference spectra (80 °C vs. 20 °C) of TBA and modified aptamers A6, A7, A12 in 10 mM sodium cacodylate (pH 7.2) and 100 mM KCl.
CD spectra of stable chimeric GQs
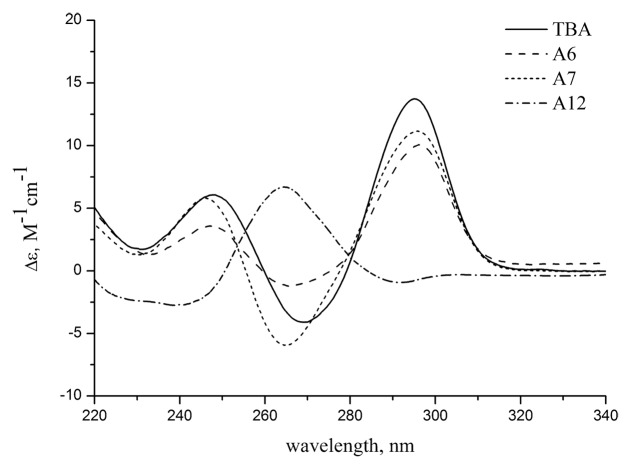
CD spectra are known to be highly sensitive to the type of GQ structure.22 Two basic types of GQs, parallel and anti-parallel, display very different CD patterns depending on the conformation of the guanines with respect to the deoxyribose. CD spectra of unmodified TBA and loop-modified GQs gave identical curve patterns at 20 °C in 10 mM sodium cacodylate (pH 7.2) and 100 mM KCl (Fig. 3; Fig. S4; Table 1). Two positive bands at 245 nm and 295 nm and a negative band at 265 nm refer to the anti-parallel folding of aptamers with an unmodified G-core. This conclusion is consistent with the data obtained from the TDS measurements.
Figure 3. CD spectra of unmodified TBA and aptamers A6, A7, A12 at 20 °C in 10 mM sodium cacodylate (pH 7.2) and 100 mM KCl.
The CD spectrum of oligonucleotide A12 with a modified G-core was typical for the parallel family of GQs, showing a major positive band at 265 nm. Based on the commonly used assignment model, the shape of the CD profile as observed in Figure 3 indicates an anti conformation for all bases in the G-quartets. However, more cautious conclusion suggests only that both G-quartets in the aptamer A12 have the same polarity.23 Relative strand directions and base conformations (syn or anti) cannot be resolved from CD data in this case.
CD spectra provide a sensitive measure of nucleic acid secondary structural changes. We used the temperature dependence of the CD bands to characterize the melting transitions of modified GQs. No significant differences in melting temperatures were found between the two different methods, CD and UV (Table 1).
Folding mode of modified GQs
Spectroscopic studies of the oligonucleotide A12 gave few indications on the formation of intermolecular species as an admixture. To address the question of molecularity of modified GQs using a different experimental technique, we analyzed fluorescence polarization of ethidium bromide (EtBr) bound to TBA aptamers and rotational relaxation times of GQ-EtBr complexes. The latter parameter was used in our earlier studies to characterize various types of DNA complexes with respect to their relative molecular hydrodynamic volume.24,25 Upon excitation with plane-polarized light a fluorophore bound to a larger molecule emits light, which is depolarized to the extent the molecule is free to rotate during the period between excitation and emission. The fluorescence rotational relaxation time is proportional to hydrodynamic volume of a molecule or a complex.26 We examined fluorescence polarization for unmodified TBA and those chimeric aptamers with detected UV melting transitions at 5 μM concentration of oligonucleotides. Calculation of rotational relaxation times gave us a value of 10.5 ± 1.5 ns for TBA and chimeric aptamers regardless of the type of modification. These findings support an intramolecular folding mode for both loop- and core-modified aptamers.
NMR studies of the aptamer A12
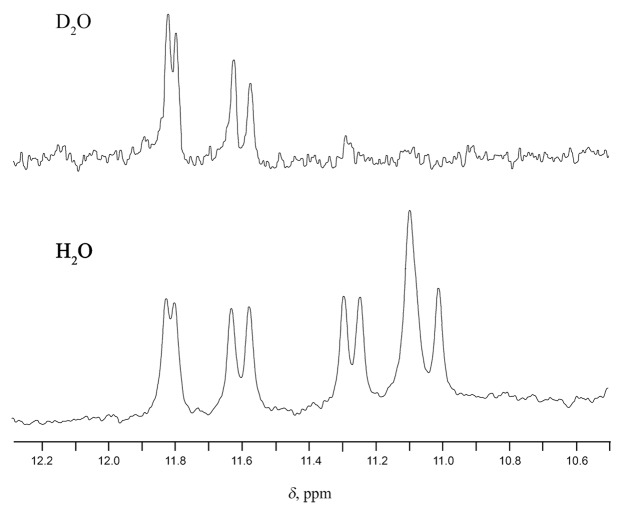
Aptamer A12 was found to form a folding structure, consisting of two G quartets with the same polarity. We further characterized this aptamer in NMR experiments. The imino proton region in the 1H NMR spectrum of aptamer A12 contains 10 resonances, including 7 well-resolved and 3 overlapping peaks. This observation is consistent with a single GQ structure containing two G tetrads and one TT base pair. Upon heating the sample from 10 to 30 °C, the intensity of the signals from imino protons dramatically decreased as a result of GQ unfolding. Two unresolved signals at 11.1 ppm showed a much faster decrease compared with the rest of the imino protons, leaving only eight imino resonances at 20 °C. The complete disappearance of imino resonances was observed at 30 °C and above. Unequal rates of imino signal fading certainly relate to specific pattern of GQ unfolding and may be presumably associated with the collapse of the TT base pair. Four low-field imino protons in the 11.5–11.9 ppm area were characterized by notably reduced exchange rates with D2O as shown in Figure 4. Selective deuterium exchange was observed within one hour after D2O addition. This effect is highly characteristic for shielded imino protons located in central G-tetrads.27 Like TBA, A12 has only two tetrads, but the G2-G5-G11-G14 one may be regarded as “central” since it is sandwiched between the G1-G6-G10-G15 one and TT base pair.
Figure 4. Imino proton NMR spectra of aptamer A12 at 10 °C in 10 mM KH2PO4 and 30 mM KCl, pH 7.0. Unequal exchange rate for imino protons.
In the last years diffusion ordered NMR spectroscopy (DOSY) is becoming very popular as a tool for the indirect determination of molecular weights of compounds in solution. Typically, DOSY experiment results in a 2D plot with a chemical shift in one direction and a diffusion coefficient (D) in the other. Comparing the diffusion coefficients of target molecules to that of references, the molecular weight can be calculated.28 Recently, DOSY has been used to detect dimerization of GQ structures.29 We applied this new technique to confirm the molecularity of the folding of aptamer A12. PEG 400, 6000, and 15 000 were used as external molecular weight markers. The diffusion coefficient measured for aptamer A12 was ~8.2 × 10–11 m2s–1. This value best matches unimolecular folding of the aptamer as it fell between D values for PEG 400 and PEG 6000 (Fig. S5). It appeared to be somewhat higher than the value derived from the PEG calibration graph for the mass 4726 (~5.6 × 10–11 m2s–1). This difference certainly relates to the different nature of PEG calibrants and A12. The aptamer is characterized by the compact folding and therefore has relatively small hydrodynamic volume of the molecule. As a result an increased value for the diffusion coefficient was observed.
Anticoagulant effect of chimeric aptamers
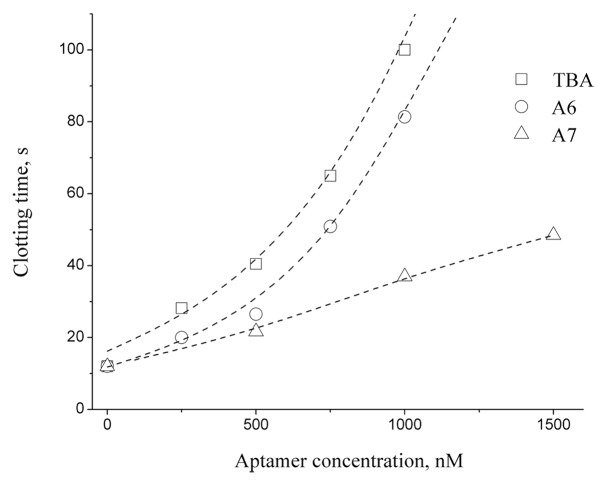
Clotting time measurements of modified aptamers taken at 37 °C are summarized in Table 1. These data suggest that oligonucleotide A6, with a modified central loop, retained the anticoagulant properties of TBA. Additionally, weak anticoagulant activity was observed for aptamer A7. The plot of clotting time vs. aptamer concentration showed similar activities for both TBA and aptamer A6 up to a 1 μM concentration (Fig. 5). Estimation of IC50 for aptamers A6, A7 and TBA yielded the values of 320, 480, and 170 nM, respectively (Fig. S6) Interestingly, modified aptamer A6 was characterized by a relatively low Tm value by UV melting, which is almost 10 °C lower than that of TBA. It has been discussed in the literature that GQ thermal stability is not associated directly with the anticoagulant activity of a modified aptamer.15 Our findings entirely agree with this conclusion. In this way, the most stable aptamer A3 (Tm 60.3 °C) was shown to be inactive in clotting time tests.
Figure 5. The plot of clotting time vs. aptamer concentration.
Resistance to nuclease cleavage
Formation of a GQ structure protects oligonucleotides from rapid nuclease cleavage.30 However, loop regions expose single-stranded fragments and make them accessible for digestion. The protective effect of anomeric modifications was estimated in comparative studies of selected aptamers by analyzing S1 nuclease cleavage. This endonuclease selectively cleaves single-stranded DNA and has been used for probing single-stranded regions in different DNA structures, for example, in the studies of cytidine tetraplexes.31 For the S1 nuclease resistance experiment, we selected aptamers A6 and A7, which showed anticoagulant activity, and aptamer A11, bearing α residues in all three loops. The results of nuclease cleavage at 37 °C are shown in Figure 6. The stability of unmodified TBA in selected conditions was extremely low. It took less than one minute to completely degrade TBA. Oligonucleotides A6 and A11 showed increased resistance compared with modified aptamer A7 after 1 h of exposure to S1 nuclease. The arrangement of modified nucleotides in aptamers A6, A7, and A11 does not provide any rationale for the observed pattern of resistance. Nevertheless, it is evident that the nuclease resistance of modified aptamers depends on not only the nature of the nucleotides in the loop but also the extent of exposure of looped regions. To further support this conclusion, we added completely modified oligonucleotide A10 to the cleavage study. The nuclease stability of the non-natural α random coil was lower than that of structurally organized aptamers A6 and A11. For 1 h at 37 °C, we observed a virtually complete digest of α oligomer A10 by S1 nuclease.
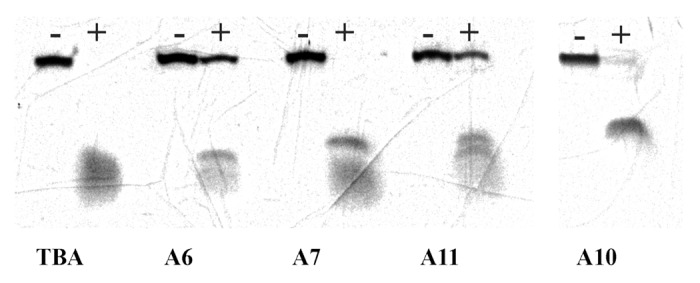
Figure 6. Digest of TBA and modified aptamers by S1 nuclease for 1 h at 37 °C in the reaction buffer containing 20 mM KCl. Complete cleavage of unmodified TBA proceeds in less than 1 min.
Discussion
Alpha anomeric modification of DNA has two important implications for artificial DNA structure and properties. First, anomeric DNA is unique in its ability to mimic strand reversal. Anomeric oligodeoxynucleotides are known to form parallel heteroduplexes with natural DNA strands.19 We have shown recently that this mimetic ability may be exploited for the recognition of base pair inversions in triplex DNA.32 In antiparallel GQs, syn guanines play a role of reversed strand mimetics. In this way, α guanosines can presumably replace selected core nucleotides. Therefore, core modifications represent substantial interest from the structural point of view even regardless of their impact on TBA activity. Particularly, the inability of α nucleotides to adopt syn conformation33 makes the design of modified GQs more predictable in terms of folding topology. Thus, artificial GQ structures may emerge useful tools in the design of new therapeutic GQ scaffolds.
The second implication of anomeric modification relates to an exceptional nuclease resistance of modified DNA strands. This feature makes α nucleotides very promising as modifying units for addressing lifetime issues of therapeutic GQ in vivo. We demonstrated that, in respect to TBA, the nuclease resistance may be greatly increased by selective modification in loop regions.
Considering specific structural characteristics of α nucleotides, we could expect the formation of GQ structures by oligonucleotides A1, A8, A12, and A13 with the anomeric core. However, anomeric modification of the TBA quadruplex core was mostly incompatible with GQ assembly. Aptamer A12 was the only oligonucleotide that formed GQ with mixed anomeric core. The later circumstance raises particular interest to the structural organization of this GQ. Formation of intramolecular GQ was found to be the major folding mode of this oligonucleotide, as confirmed by three different experimental methods. CD studies suggest that two G-quartets of this GQ have the same base polarity. NMR data support the formation of two G quartets and one TT base pair, thus implying the presence of two lateral loops.34 Additionally, it should be taken into account that unlike natural β anomers, α nucleosides cannot adopt a syn conformation.33 Therefore, two α-deoxyguanosine strands must be arranged in parallel. In this case one of two unimolecular topologies can be proposed for GQ structure formed by aptamer A12, i.e., TBA-like anti-parallel chair, or anti-parallel basket (Fig. 7) with all anti guanines in either model. Unambiguous structure determination of this GQ requires further NMR or X-ray studies.
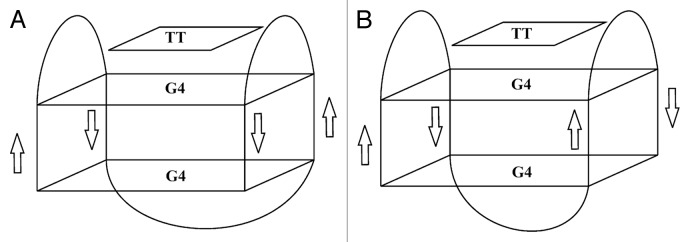
Figure 7. Presumable folding topologies of aptamer A12. (A) Anti-parallel chair. (B) anti-parallel basket.
In the case of loop modifications, our primary goal was to improve nuclease resistance, and we did not expect profound changes in aptamer folding. Indeed, all loop-modified aptamers formed TBA-like intramolecular GQs. The most stable chimeric structures had anomeric thymines at positions 4 and 13. Based on an NMR model of TBA, these bases are stacked with the neighboring G-quartet in an unmodified aptamer and hydrogen bonded to each other.5 Other NMR and crystallographic studies of TBA highlighted an important role of T4-T13 base pair stacking on the stabilization of GQ structure.8,35 Modifications of the TT loops that favor the optimal pairing of stacked thymines may considerably increase the thermal stability of non-natural TBA. The stabilization effect, similar in origin to that observed with α anomers in TT loops, has been described previously for unlocked nucleotide analogs.36 Substitution of unpaired T3 or T12 with a flexible unlocked residue induced the optimization of stacking between the adjacent T4-T13 base pair and the neighboring G quartet.
The importance of TT loop interactions in TBA structure stability is accompanied by their exclusive role in specific thrombin recognition. T4 and T13 are mainly involved in polar interactions with thrombin residues, whereas T3 and T12 form essentially hydrophobic contacts.8 Thus, the specific thrombin activity of TBA should be highly sensitive to the modification of TT loops. Indeed, anomeric substitutions in TT loops did not provide a benefit for retaining aptamer activity. Conversely, anticoagulant activity was essentially unaffected by the anomeric modification of the central TGT loop in aptamer A6. It has been discussed recently that the TGT loop is not involved in thrombin binding.8 This circumstance would certainly account for the biochemical tolerance of the central loop to modifications. There have been few examples of modifications in TBA loops in the literature, which allow aptamer analogs to retain or to exceed natural prototype activity. Typically, the modifications affected either central loop only or in combination with selected thymines in TT loops as described for isodeoxyguanosine, 2′-fluoroarabinose, 4-thio, and unlocked TBA analogs.13,14,17,36
Because the central loop is the most exposed region in the TBA structure8,35 its modification may considerably improve nuclease resistance of chimeric aptamers. Anomeric modification in the TGT loop was shown to dramatically increase nuclease resistance. Degradation of chimeric oligonucleotides by S1 nuclease was approximately two orders of magnitude less efficient compared with unmodified TBA. We also observed certain differences in nuclease resistance between modified aptamers A6, A7, A10, and A11. Cleavage results suggest that the protective effect of modification is only partially due to the α nucleotide substitution. Rearrangement in the loop that provides more effective interaction of the loop nucleotides with each other and the core also seems to be significant.
Conclusion
We have demonstrated that the effect of anomeric modification of TBA is strongly dependent on the location of α nucleotides. Aptamers modified in the loop regions retain their ability to fold into unimolecular GQ structure. Modifications of the GQ core in most instances prevent GQ assembly. Thermal stability of the loop-modified aptamers varies within wide temperature range. Considerable stabilization effect was observed for anomeric substitutions at positions 4 and 13. Biochemical studies of loop-modified aptamers showed that replacements in the TGT loop substantially increases the nuclease resistance of a chimeric aptamer, providing the least interference with its anticoagulant properties. We have found that a single core-modified aptamer A12 was able to fold into unimolecular GQ structure. Results of biophysical studies of this oligonucleotide suggest that either TBA-like anti-parallel chair or anti-parallel basket topology can be proposed for this GQ.
Materials and Methods
Oligonucleotides
DNA oligomers were synthesized using an ABI 3400 DNA/RNA synthesizer. Modified phosphoramidites were purchased from ChemGenes. Purification of oligodeoxynucleotides was performed on a Hypersil ODS column (5 μm; 4.6 × 250 mm) using 0.05 M TEAA (pH 7.0) and a linear gradient of MeCN (10–50% and 30 min for DMTr protected oligodeoxynucleotides and 0–25% and 30 min for fully deblocked oligodeoxynucleotides). The flow rate was 1 mL/min. Removal of the 5′-O-DMTr group was achieved by treatment with 2% aqueous TFA for 1 min followed by Et3N neutralization and precipitation with 2% LiClO4 in acetone. Oligonucleotide composition was confirmed by MALDI mass spectrometry using a Bruker Reflex IV mass spectrometer with hydroxypicolinic acid or 2-amino-5-nitropyridine as a matrix.
UV thermal denaturation experiments and thermal difference spectra
Absorbance vs. temperature profiles were obtained with a Shimadzu UV160A spectrophotometer equipped with a water-jacketed cell holder. Dry nitrogen gas was flushed through the cuvette to prevent the condensation of water at low temperatures. Melting experiments were measured at 295 nm in 10 mM sodium cacodylate (pH 7.2) and 30 mM or 100 mM KCl. The solutions were heated at a rate of 0.2 °C/min. Melting points were determined from derivative plots of the melting curves. Thermal difference spectra were calculated from UV spectra of aptamers at 80 and 20 °C. The oligonucleotide concentration was 5 μM.
Fluorescence polarization and lifetime of ethidium bromide bound to GQs
Fluorescence polarization (P) of ethidium bromide (EtBr) bound to the aptamers was measured using a Cary Eclipse spectrofluorimeter at 20 °C. The excitation wavelength was 540 nm, and the fluorescence intensity was registered at 610 nm. Fluorescence polarization was calculated as described.37 The free dye contribution was taken into account as described elsewhere.38 The concentration of EtBr was 4 μM, and the oligonucleotide concentration was in the range of 4–6 μM. The fluorescence lifetime (τ) of EtBr:TBA complexes was evaluated using the Easy Life V fluorometer. Fluorescence decay was registered using an RG610 long pass filter and LED excitation at 525 nm. Rotational relaxation time was estimated using the Perrin–Weber equation.37
CD spectroscopy
Circular dichroism measurements were performed with a Jasco-715 CD spectrometer equipped with a Peltier temperature controller. The spectra were obtained at a bandwidth of 1 nm and a spectral resolution of 0.2 nm; three scans were performed. CD melting profiles were obtained at 295 nm (at 260 nm for aptamer A12) in 10 mM sodium cacodylate (pH 7.2) and 100 mM KCl. The solutions were heated at a rate of 1 °C/min. Melting points were determined from derivative plots of the melting curves.
1H NMR of the oligonucleotide A12
NMR samples were prepared at a concentration of ~0.3 mM in 0.6 ml H2O+D2O (10%) or D2O buffer solution having 10 mM KH2PO4, 30 mM KCl, pH 7.0. 1H NMR spectra were recorded with Bruker AVANCE II 300 MHz, Bruker AMX III 400 MHz and Bruker AVANCE II 600 MHz spectrometers (300.1, 400.1, and 600.1 MHz).1H chemical shifts were referenced relative to external sodium 2, 2-dimethyl-2-silapentane-5-sulfonate (DSS). Proton spectra of samples in H2O+D2O were recorded using pulsed-field gradient WATERGATE W5 pulse sequence (zggpw5 from Bruker library) for H2O suppression at different temperatures. Measurements of the diffusion coefficients were performed using 2D 1H DOSY NMR spectroscopy (Diffusion Ordered SpectroscopY39) in D2O and H2O+D2O solutions at 10 °C. The diffusion measurements were performed using a bipolar gradient pulse pair-stimulated echo sequence incorporating a longitudinal eddy current delay (BPP-LED40) with the diffusion time (Δ) 100 ms and a longitudinal eddy current delay (Te) 5 ms. For samples in a H2O+D2O solution, presaturation for H2O suppression was used. The DOSY spectra were processed by Monoexponential Fitting using Bruker TopSpin software. Polyethylene glycols (PEG) in the same buffer solutions with average molecular weights 400, 6000, and 15 000 were used as the external calibrants of the molecular weight in DOSY spectra in ~1:1 molar ratio to studied structure. The NMR data were processed using Bruker XWIN-NMR and Bruker TopSpin 1.4, 2.0, and 2.1 software.
Clotting time measurements
Clotting time was measured according to the published procedure41 and the protocol of the assay kit manufacturer (“Thrombin-TEST,” Renam). The citrate-stabilized serum (100 µL) was incubated for 120 s at 37 °C, and then thrombin and an aptamer were added to the final concentration of the latter, 0.25–1.5 μM, and the clotting time was measured using a coagulation analyzer (MiniLab-701, Unimed).
S1 nuclease cleavage
Oligonucleotides were dissolved at a concentration of 50 μM in 30 μL of reaction buffer containing 20 mM KCl. Samples were heated to 100 °C and cooled slowly before the digest. Ten units of S1 nuclease (Thermo Scientific) were added to the aptamer samples. After incubation for 1 h at 37 °C, reaction mixtures were precipitated with 2% LiClO4 in acetone and analyzed by electrophoresis in polyacrylamide gel. Electrophoresis was performed with a 20% polyacrylamide gel in 7 M urea and 0.1 M TBE at 25 °C.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Vladimir L Florentiev passed away on October 24, 2012. This work was supported by the Russian Fund for Basic Research (11-04-01544, 14-04-01035, 14-04-01244) and Molecular and Cell Biology program of the Russian Academy of Sciences.
Glossary
Abbreviations:
- TEAA
triethylammonium acetate
- DMTr
4,4′-dimethoxytrityl
- TFA
trifluoroacetic acid
- PEG
polyethylene glycol
- MeCN
acetonitrile
Footnotes
Previously published online: www.landesbioscience.com/journals/artificialdna/article/28422
References
- 1.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–71. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Tucker WO, Shum KT, Tanner JA. G-quadruplex DNA aptamers and their ligands: structure, function and application. Curr Pharm Des. 2012;18:2014–26. doi: 10.2174/138161212799958477. [DOI] [PubMed] [Google Scholar]
- 5.Kelly JA, Feigon J, Yeates TO. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J Mol Biol. 1996;256:417–22. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 6.Schultze P, Macaya RF, Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J Mol Biol. 1994;235:1532–47. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 7.Russo Krauss I, Merlino A, Giancola C, Randazzo A, Mazzarella L, Sica F. Thrombin-aptamer recognition: a revealed ambiguity. Nucleic Acids Res. 2011;39:7858–67. doi: 10.1093/nar/gkr522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo Krauss I, Merlino A, Randazzo A, Novellino E, Mazzarella L, Sica F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012;40:8119–28. doi: 10.1093/nar/gks512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avino A, Fabrega C, Tintore M, Eritja R. Thrombin binding aptamer, more than a simple aptamer: chemically modified derivatives and biomedical applications. Curr Pharm Des. 2012;18:2036–47. doi: 10.2174/138161212799958387. [DOI] [PubMed] [Google Scholar]
- 10.Lauridsen LH, Rothnagel JA, Veedu RN. Enzymatic recognition of 2′-modified ribonucleoside 5′-triphosphates: towards the evolution of versatile aptamers. Chembiochem. 2012;13:19–25. doi: 10.1002/cbic.201100648. [DOI] [PubMed] [Google Scholar]
- 11.Pagano B, Martino L, Randazzo A, Giancola C. Stability and binding properties of a modified thrombin binding aptamer. Biophys J. 2008;94:562–9. doi: 10.1529/biophysj.107.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esposito V, Galeone A, Mayol L, Randazzo A, Virgilio A, Virno A. A mini-library of TBA analogues containing 3′-3′ and 5′-5′ inversion of polarity sites. Nucleosides Nucleotides Nucleic Acids. 2007;26:1145–9. doi: 10.1080/15257770701526978. [DOI] [PubMed] [Google Scholar]
- 13.Peng CG, Damha MJ. G-quadruplex induced stabilization by 2′-deoxy-2′-fluoro-D-arabinonucleic acids (2’F-ANA) Nucleic Acids Res. 2007;35:4977–88. doi: 10.1093/nar/gkm520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nallagatla SR, Heuberger B, Haque A, Switzer C. Combinatorial synthesis of thrombin-binding aptamers containing iso-guanine. J Comb Chem. 2009;11:364–9. doi: 10.1021/cc800178m. [DOI] [PubMed] [Google Scholar]
- 15.Bonifacio L, Church FC, Jarstfer MB. Effect of locked-nucleic acid on a biologically active g-quadruplex. A structure-activity relationship of the thrombin aptamer. Int J Mol Sci. 2008;9:422–33. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saneyoshi H, Mazzini S, Aviñó A, Portella G, González C, Orozco M, Marquez VE, Eritja R. Conformationally rigid nucleoside probes help understand the role of sugar pucker and nucleobase orientation in the thrombin-binding aptamer. Nucleic Acids Res. 2009;37:5589–601. doi: 10.1093/nar/gkp598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendelboum Raviv S, Horváth A, Aradi J, Bagoly Z, Fazakas F, Batta Z, Muszbek L, Hársfalvi J. 4-thio-deoxyuridylate-modified thrombin aptamer and its inhibitory effect on fibrin clot formation, platelet aggregation and thrombus growth on subendothelial matrix. J Thromb Haemost. 2008;6:1764–71. doi: 10.1111/j.1538-7836.2008.03106.x. [DOI] [PubMed] [Google Scholar]
- 18.Zaitseva M, Kaluzhny D, Shchyolkina A, Borisova O, Smirnov I, Pozmogova G. Conformation and thermostability of oligonucleotide d(GGTTGGTGTGGTTGG) containing thiophosphoryl internucleotide bonds at different positions. Biophys Chem. 2010;146:1–6. doi: 10.1016/j.bpc.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Morvan F, Debart F, Vasseur JJ. From anionic to cationic alpha-anomeric oligodeoxynucleotides. Chem Biodivers. 2010;7:494–535. doi: 10.1002/cbdv.200900220. [DOI] [PubMed] [Google Scholar]
- 20.Wyatt JR, Vickers TA, Roberson JL, Buckheit RW, Jr., Klimkait T, DeBaets E, Davis PW, Rayner B, Imbach JL, Ecker DJ. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc Natl Acad Sci U S A. 1994;91:1356–60. doi: 10.1073/pnas.91.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kypr J, Kejnovská I, Renciuk D, Vorlícková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–25. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray DM, Wen JD, Gray CW, Repges R, Repges C, Raabe G, Fleischhauer J. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008;20:431–40. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 24.Shchyolkina AK, Timofeev EN, Borisova OF, Il’icheva IA, Minyat EE, Khomyakova EB, Florentiev VL. The R-form of DNA does exist. FEBS Lett. 1994;339:113–8. doi: 10.1016/0014-5793(94)80396-X. [DOI] [PubMed] [Google Scholar]
- 25.Timofeev EN, Borisova OF, Shchyolkina AK. Structural polymorphism of oligo(dC) with mixed alpha,beta-anomeric backbone. J Biomol Struct Dyn. 2000;17:655–64. doi: 10.1080/07391102.2000.10506556. [DOI] [PubMed] [Google Scholar]
- 26.Jameson DM, Ross JA. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem Rev. 2010;110:2685–708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adrian M, Heddi B, Phan AT. NMR spectroscopy of G-quadruplexes. Methods. 2012;57:11–24. doi: 10.1016/j.ymeth.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Waldeck AR, Kuchel PW, Lennon AJ, Chapman BE. NMR diffusion measurements to characterise membrane transport and solute binding. Prog Nucl Magn Reson Spectrosc. 1997;30:39–68. doi: 10.1016/S0079-6565(96)01034-5. [DOI] [Google Scholar]
- 29.Ambrus A, Yang D. Diffusion-ordered nuclear magnetic resonance spectroscopy for analysis of DNA secondary structural elements. Anal Biochem. 2007;367:56–67. doi: 10.1016/j.ab.2007.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Z, Huang CC, Tan W. Nuclease resistance of telomere-like oligonucleotides monitored in live cells by fluorescence anisotropy imaging. Anal Chem. 2006;78:1478–84. doi: 10.1021/ac0517601. [DOI] [PubMed] [Google Scholar]
- 31.Kumar P, Verma A, Maiti S, Gargallo R, Chowdhury S. Tetraplex DNA transitions within the human c-myc promoter detected by multivariate curve resolution of fluorescence resonance energy transfer. Biochemistry. 2005;44:16426–34. doi: 10.1021/bi051452x. [DOI] [PubMed] [Google Scholar]
- 32.Kolganova NA, Shchyolkina AK, Chudinov AV, Zasedatelev AS, Florentiev VL, Timofeev EN. Targeting duplex DNA with chimeric α,β-triplex-forming oligonucleotides. Nucleic Acids Res. 2012;40:8175–85. doi: 10.1093/nar/gks410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latha YS, Yathindra N. Stereochemical studies on nucleic acid analogues. I. Conformations of alpha-nucleosides and alpha-nucleotides: interconversion of sugar puckers via O4′-exo. Biopolymers. 1992;32:249–69. doi: 10.1002/bip.360320306. [DOI] [PubMed] [Google Scholar]
- 34.Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chemistry. 2007;13:9738–45. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- 35.Mao XA, Gmeiner WH. NMR study of the folding-unfolding mechanism for the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) Biophys Chem. 2005;113:155–60. doi: 10.1016/j.bpc.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Pasternak A, Hernandez FJ, Rasmussen LM, Vester B, Wengel J. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011;39:1155–64. doi: 10.1093/nar/gkq823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber G, Anderson SR. The effects of energy transfer and rotational diffusion upon the fluorescence polarization of macromolecules. Biochemistry. 1969;8:361–71. doi: 10.1021/bi00829a050. [DOI] [PubMed] [Google Scholar]
- 38.Shchyolkina AK, Borisova OF, Livshits MA, Pozmogova GE, Chernov BK, Klement R, Jovin TM. Parallel-stranded DNA with mixed AT/GC composition: role of trans G.C base pairs in sequence dependent helical stability. Biochemistry. 2000;39:10034–44. doi: 10.1021/bi9913909. [DOI] [PubMed] [Google Scholar]
- 39.Johnson CS., Jr. Diffusion ordered nuclear magnetic resonance spectroscopy: principles and applications. Prog Nucl Magn Reson Spectrosc. 1999;34:203–56. doi: 10.1016/S0079-6565(99)00003-5. [DOI] [Google Scholar]
- 40.Wu D, Chen A, Johnson CS., Jr. An improved diffusion ordered spectroscopy experiment incorporating bipolar gradient pulses. J Magn Reson A. 1995;115:260–4. doi: 10.1006/jmra.1995.1176. [DOI] [Google Scholar]
- 41.Clauss A. [Rapid physiological coagulation method in determination of fibrinogen] Acta Haematol. 1957;17:237–46. doi: 10.1159/000205234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.