Abstract
The bird cherry-oat aphid (Rhopalosiphum padi), an important pest of cereal crops, not only directly sucks sap from plants, but also transmits a number of plant viruses, collectively the yellow dwarf viruses (YDVs). For quantifying changes in gene expression in vector aphids, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is a touchstone method, but the selection and validation of housekeeping genes (HKGs) as reference genes to normalize the expression level of endogenous genes of the vector and for exogenous genes of the virus in the aphids is critical to obtaining valid results. Such an assessment has not been done, however, for R. padi and YDVs. Here, we tested three algorithms (GeNorm, NormFinder and BestKeeper) to assess the suitability of candidate reference genes (EF-1α, ACT1, GAPDH, 18S rRNA) in 6 combinations of YDV and vector aphid morph. EF-1α and ACT1 together or in combination with GAPDH or with GAPDH and 18S rRNA could confidently be used to normalize virus titre and expression levels of endogenous genes in winged or wingless R. padi infected with Barley yellow dwarf virus isolates (BYDV)-PAV and BYDV-GAV. The use of only one reference gene, whether the most stably expressed (EF-1α) or the least stably expressed (18S rRNA), was not adequate for obtaining valid relative expression data from the RT-qPCR. Because of discrepancies among values for changes in relative expression obtained using 3 regions of the same gene, different regions of an endogenous aphid gene, including each terminus and the middle, should be analyzed at the same time with RT-qPCR. Our results highlight the necessity of choosing the best reference genes to obtain valid experimental data and provide several HKGs for relative quantification of virus titre in YDV-viruliferous aphids.
Introduction
Rhopalosiphum padi L. (Hemiptera: Aphididae) is an important pest of wheat, oat, barley, rye and other gramineous crops in the world [1], [2]. It can sometimes directly affect grain yield by sucking nutrients from these plants, but causes the most serious damage when it transmits a group of viruses that are collectively called yellow dwarf viruses (YDVs) to cereal crops and grasses [3], [4]. R. padi is considered a competent vector of the YDVs, which comprise Barley yellow dwarf virus (BYDV)-PAV and BYDV-PAS (genus Luteovirus) and Cereal yellow dwarf virus (CYDV)-RPV, CYDV-RPS (genus Polerovirus) and ungrouped BYDV-GPV [renamed Wheat yellow dwarf virus (WYDV)-GPV] [4], [5], [6], [7], all of which belong to the family Luteoviridae [8].
R. padi transmits YDVs in a persistently circulative and nonpropagative mode [9], [10] after piercing a YDV-infected plant and ingesting virions while sucking up the phloem sap [11]. Receptor-mediated endocytosis and release must then occur in the midgut and/or the hindgut and in the accessory salivary glands (ASG) [12], [13]. Otherwise, the virions will be excreted along with the feces or be retained in the aphid and exposed to a destructive environment or immune elements of the aphid or ill-defined metabolic processes so that it cannot complete the circulative route in the aphid [14]. The propagation of YDVs in their plant hosts has been investigated and elaborated using the prevailing absolute and relative quantification methods [15], [16], but rarely has research focused on fluctuation in the virus titre and accumulation in the vector aphid during the acquisition access period (AAP), latent period or inoculation access period (IAP), all pivotal issues that affect rates of transmission, duration of transmissibility and epidemiology of the plant virus in the field. With the entry of the virus into the vector aphid, many proteins are incorporated via a transcytosis mechanism associated with virus transmission [17], [18], [19], [20], followed by up- or down-regulation of the expression of numerous genes [21]. Using reverse transcription-quantitative polymerase chain reaction (RT-qPCR), we can rapidly assess the influence of virus entry on molecular functions and biological processes and on the real-time concentration of virus [22], [23].
A relative quantification method for many aspects of molecular research has come into favor to eliminate systemic errors among experimental treatments, developmental stages, RNA extraction efficiency, and RNA integrity [24], [25]. With this method, changes in the expression level of genes of interest (GOIs) can be determined by comparing the expression of the GOIs with that of housekeeping genes (HKGs) [24]. HKGs are constitutive in all cells under both normal and disturbed conditions and expressed constantly in natural situations [26]. If the expression of HKGs is validated as remaining constant under a defined set of conditions, relative quantification of GOIs can be realized with convincing results [24]. The selection of HKGs for normalization of the expression of the GOIs has become routine in studies on human disease diagnosis [27], [28]. With the publication of the complete genomes of different insects, from Drosophila melanogaster [29] to Plutella xylostella L. [30], these holometabolous insects have been the focus of searches for HKGs to use as reference genes [31], [32], [33]. Estimating the level of expression of HKGs to assay differences in expression in hemimetabolous insects, such as aphids, which transmit many plant viruses, has started recently with Aphis glycines [34]. For detecting YDV virions in the vector R. padi and determining the relative level of expression of vector genes using relative RT-qPCR, no HKG has been validated as a suitable reference gene to normalize expression. Although ACT1 was used to determine two YDVs in R. padi, it was actually from a coordinate of Bombyx mori, but the ACT1 gene sequence of R. padi was unknown [35], [36].
Selecting appropriate and reliable HKGs as reference genes is required for RT-qPCR experiments. Software tools to estimate and validate reference genes include GeNorm [25], Normfinder [37], and BestKeeper [38]. HKGs such as 18S rRNA, ACT1, EF-1α, GAPDH, UBI, RPSs, RPLs and TUB are commonly used reference genes for standardization when analyzing the expression of insect genes using RT-qPCR [34], [39], [40], [41], [42], [43], [44].
The announcement of the complete genome of the pea aphid, Acyrthosiphon pisum (Harris) [45], has opened a new avenue for finding homologous genes in other aphid species. The HKGs of R. padi can be chosen and cloned based on sequences from A. pisum, then used as reference genes to quantify the relative titre of viruses ingested by R. padi and the relative expression values of endogenous genes in the YDV-viruliferous aphid, replacing methods for absolute quantification [46], which may lead to incorrect, misleading conclusions [15].
Previously, actin protein and GAPDH protein of Myzus persicae were found to possess interaction with Beet western yellows virus in vitro [18]. In a recent study, some ACT genes (ACT1 to ACT4) and one GAPDH gene, together with many other genes from A. pisum were considered to be potentially related to the transmission of Pea enation mosaic virus and Soybean dwarf virus [21]. Here, we selected these two doubtful HKG genes, ACT1 and GAPDH, as well as EF-1α and 18S rRNA which were always used as reference genes for normalization of target genes in other aphids [42], [47], as four candidate reference gene for relative RT-qPCR analysis in R. padi. Firstly, these four HKGs from R. padi were cloned, and then their nucleic acid sequences were aligned with coordinates from other insect species on NCBI. We evaluated the expression stability of these four candidate reference genes using 3 tools (GeNorm, NormFinder, and BestKeeper) in 6 experimental virus–aphid morph combinations after various durations of aphid feeding on oat plants infected with the respective virus. To further explore the applicability of the optimal reference genes selected by GeNorm, we monitored the relative titre of the three viruses in the two morphs of R. padi during AAP and compared virus titre among the feeding durations using a one-way ANOVA. Also, the expression of two endogenous aphid genes, ago-1a and dcr1, whose encoded proteins are involved in miRNA pathway in aphids and have similar functional domains with the RNAi pathway-related Ago-2 and Dcr2 proteins [48], [49], [50], were normalized using the selected reference genes to detect any change in expression.
Results and Discussion
Sequencing of Candidate Reference Genes from R. padi
The total RNAs extracted from plants and aphids were all of good integrity (Figure S1) and purity (A260/A280 values: 1.95 ∼ 2.10; A260/A230 values > 2.0). All genes were amplified by RT-PCR (Figure S2) with the primers described in Table 1. Nucleic acid sequences of 4 candidate reference genes yielded good matches in a BLAST search of the NCBI database with high similarity to corresponding genes from other aphids and insects: 18S rRNA, 90–100%; EF-1α, 83–94%; ACT1, 82–96%; GAPDH, 78–95%, respectively (Table S1).
Table 1. Primers for RT-PCR amplification used in this study.
| Gene | Forward (F) and reverse (R) primer (5′-3′) | EST size (bp) | Accession number |
| 18s rRNA | F: CTGGTTGATCCTGCCAGTAGTCATATG | 571 | KJ612093 |
| R: TTCCGATTACGGGGCCTCGGATG | |||
| ACT1 | F: ATGTGTGACGAAGAAGTAGC | 1131 | KJ612090 |
| R: TTAGAAGCACTTTCTGTGC | |||
| EF-1α | F: ATTGATATTGCTTTATGGAAATTCG | 837 | KJ612092 |
| R: ACCAGGGTGGTTCAATACAATAAC | |||
| GAPDH | F: ATGTCAAACATTGGTATCAATGGATTTGG | 999 | KJ612091 |
| R: TTTAATCCTTAGATTGCATGTACTTGAT | |||
| ago-1a-1 | F: AATCATTTCCAAATTTCAATGCCTCG | 727 | KJ612094 |
| R: TTGTAACATTACAAACTCTATATTTTC | |||
| ago-1a-2 | F: ATTAATTCTTTGGTTCGCCGAGCTG | 906 | KJ612095 |
| R: GTACAATATAATTCGATGAGGCTTATAAC | |||
| ago-1a-3 | F: ATAAAGTTGGAAGGTGATTACAAAC | 278 | KJ612096 |
| R: TACGTTAAGCATTGTAATTCATCTG | |||
| dcr1 | F: TTATGGACTTCCAGACATTAATATTTTATC | 957 | KJ612097 |
| R: TGAGTCATTGCTTGTAATAGATAACTACG | |||
| BYDV-GPV CP | F: ATGAGTACGGTCGCCCTTAGAAATG | 606 | |
| R: CTATTTCGGGTTTTGAAACAGGACC | |||
| BYDV-GPV RTD | F: GTAGACGCGGAACCCGGTCCTAG | 1215 | |
| R: TTAGGAACGGCCACCACCAAATG | |||
| BYDV-PAV CP a | F: ATGAATTCAGTAGGCCGTAGAGGAC | 600 | |
| R: GGTTCCGGTGTTGAGGAGTCTAC | |||
| BYDV-PAV RTD b | F: GCCAAATAGGTAGACTCCTCAACA | 1341 | |
| R1: TCACGAAAATTGTGCCTTGTACTC | |||
| R2: TCACTGAAACTGCGCCTTGTATTC | |||
| R3: TCACGAGAATTGAGCCTTGTACGC | |||
| BYDV-GAV CP | F: ATGAATTCAGTAGGCCGTAG | 600 | |
| R: CTATTTGGGAGTCATGTTG | |||
| BYDV-GAV RTD | F: GTAGACTCCTCAACACCAGAG | 1377 | |
| R: TCACCATGTGTCAGCTAAAC |
With this primer pair, RT-PCR product included less than 100 nt of the 5′-terminus of the RTD gene.
With this primer pair, RT-PCR product included less than 100 nt of the 3′-terminus of the CP gene.
Expression Profiles of Candidate Reference Genes
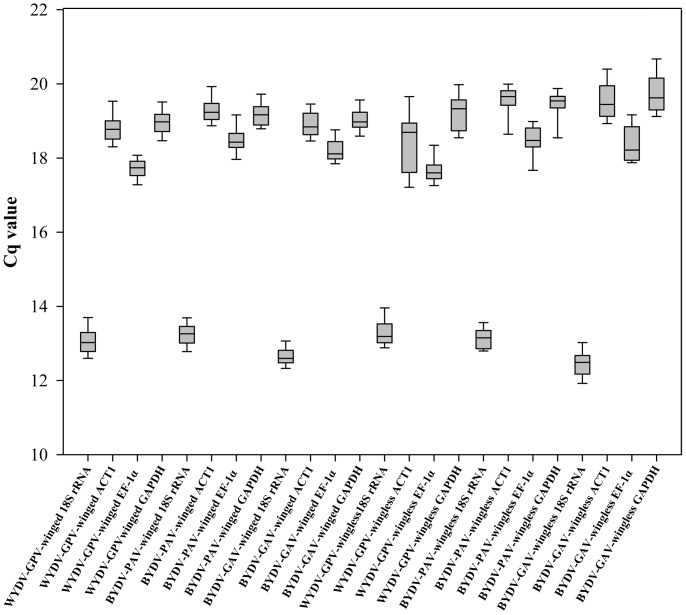
In the two-step RT-qPCR, the amplification efficiencies of the 4 candidate reference genes ranged from 91% to 97% (Table 2; Figure S3). Relative standard curves for determining the amplification efficiencies of these genes are shown in Figure S4, and all Cq values of the dilution series of the cDNA for each gene were less than 30, while those of the no-template controls were either higher than 36 or undetermined. Consequently, the raw Cq value of the individual gene reflected the actual mRNA level of this gene and could be compared directly among the 6 experimental groups. The distribution of the Cq values of the 4 candidate reference genes showed that the expression of 18S rRNA was maintained at extremely high levels with the lowest Cq values (Cq = 12∼14), higher levels with moderate Cq values (Cq 16∼22) were obtained for ACT1, EF-1α and GAPDH, but EF-1α was at the lower edge of this range in all experimental groups (Figure 1). The high expression level of 18S rRNA in R. padi was expected, in agreement with the lowest Cq value in other insects [44]. EF-1α, a well-known HKG in various organisms [51] and an evolutionarily conserved nuclear gene in aphids [52], [53], [54], can be used to classify the population of R. padi into various subgroups depending on EF-1α diversity in the population [52], [53]. Its expression level was consistent between winged aphids and wingless aphids, lower than that of 18S rRNA but slightly higher than that of ACT1 and GAPDH. The expression levels of ACT1 and GAPDH were similar in R. padi, but the mRNA level of ACT1 largely differs from GAPDH in planthoppers [44]. ACT1 and GAPDH encode a cytoskeletal structural protein and a catalytic enzyme, respectively [44]. Thus, it was difficult to interpret here why these two genes with diverse functions actually have similar expression levels in R. padi.
Table 2. Primers for RT-qPCR amplification used in this study.
| Gene | Forward and reverse primer sequencea | Amplicon | Efficiencyc |
| sizeb (bp) | (winged/wingless) | ||
| 18S rRNA | 5′-CGGGAGGAACGCTTTTATTAGA-3′ | 262 | 90.506/91.014 |
| 5′-TTGGATGTGGTAGCCGTTTCTC-3′ | |||
| ACT1 | 5′-AACGGAAGCACCTTTGAACC-3′ | 385 | 93.665/95.067 |
| 5′-GGAAGAAGCAGCAGTAGCCAT-3′ | |||
| EF-1α | 5′-TAGACGCTATCCTACCCCCCA-3′ | 328 | 95.902/97.157 |
| 5′-GTGAAATCAGCAGCACCCTTG-3′ | |||
| GAPDH | 5′-ACTACTGTTCACGCTACCACCG-3′ | 272 | 94.814/96.216 |
| 5′-GCTGCTTCCTTGACCTTACCTT-3′ | |||
| ago-1a-1 | 5′-ACCAGTCTGTTCGTCCATCTCAAT-3′ | 138 | 91.891/91.594 |
| 5′-GTTTTCTTTGTTCACCAATGTCCC-3′ | |||
| ago-1a-2 | 5′-ACCGAGCATCAGACCTAAAGTATT-3′ | 215 | 98.391/99.05 |
| 5′- TTAGAAGTTCCCTGACCATAGAGC-3′ | |||
| ago-1a-3 | 5′-TATTCTGTGCCGACAAAAAGG-3′ | 184 | 97.263/91.749 |
| 5′-GATTCAAAATGGTTGTCATCCC-3′ | |||
| dcr1 | 5′-CTACCACCCTGTTACTTTGTCCC-3′ | 168 | 93.875/101.698 |
| 5′-GTCGTCATTTGAGAAGCCCAC-3′ | |||
| BYDV-GPV CP | 5′-TTCGTTTTCGCAAAGGATTCA-3′ | 359 | 94.75/101.875 |
| 5′-GTGTTGGCGTCACCGTTACC-3′ | |||
| BYDV-GPV | 5′-GACCATCACCCACTCCACCT-3′ | 369 | 102.875/96.203 |
| RTD | 5′-CCTCGCTTGAATCATCATACG-3′ | ||
| BYDV-PAV CP | 5′-CGGGGCTGAGGTATTCGTAT-3′ | 281 | 93.65/98.105 |
| 5′-AGGACTTTGAGGCGGATTTG-3′ | |||
| BYDV-PAV | 5′-CATTGCCTACTCCAACTCATCG-3′ | 288 | 100.303/92.586 |
| RTD | 5′-ATTCTTTTGTTCGTGTACCCTCC-3′ | ||
| BYDV-GAV CP | 5′-CAGGCAGGACTGAGGTATTCGTA-3′ | 278 | 96.752/100.563 |
| 5′-GGTTGCTGATTTTGAGATGGTGA-3′ | |||
| BYDV-GAV | 5′-GTGCTGACCCCAAGTTTTACCTC-3′ | 191 | 98.585/100.127 |
| RTD | 5′-GTTTTCTGTTTTCCTAACCGTGCT-3′ |
Designed by an automated search in Primer Premier 5 software. Parameters for each primer pair: G+C content = 40–60%, annealing temperature between 58° and 62°C, and no false priming.
Amplicon size and specificity were checked using melt curve and 2% agarose gel electrophoresis (Figure S3).
Efficiency % = 10(−1/slope) −1. R 2 values of all relative standard curves were more than 0.989.
Figure 1. Box and whisker plots of Cq values for the 4 candidate reference genes in 6 experimental groups (one of three viruses in either winged or wingless adults of Rhopalosiphum padi).
Each box shows the lower 25th and upper 75th percentiles with median Cq values; the whiskers mark the lower 5th and upper 95th percentiles of the Cq values in each data set. Experimental groups from left to right: WYDV-GPV in winged adult, BYDV-PAV in winged adult, BYDV-GAV in winged adult, WYDV-GPV and wingless adult, BYDV-PAV and wingless adult, and BYDV-GAV and wingless adult.
The influence of virus and aphid morph on the Cq values of the 4 candidate reference genes was statistically analyzed with a two-way ANOVA (Table 3). When comparing Cq values for each gene (18S rRNA, ACT1 and EF-1α) among the different YDV-viruliferous aphids, Cq differed significantly (all p<0.001); values for GAPDH differed less among the three viruses (0.01<p<0.05). For the aphid morph factor, wing morph had no significant effect on the expression of 18S rRNA (p = 0.285) or EF-1α (p = 0.874), but did contribute to the significant difference in ACT1 (p = 0.04) and in GAPDH (p<0.001). For the interaction of YDV and wing morph, Cq values of 18S rRNA and EF-1α revealed no significant difference (both p>0.05), as opposed to the Cq values for ACT1 (p<0.001) and GAPDH (p = 0.02), probably due to the significant differences for both virus and wing morph.
Table 3. Influence of YDV entry and wing morph on raw Cq value for individual genes.
| Gene | Virus | Wing morph | Interaction |
| 18S rRNA | 0.000*** | 0.285ns | 0.276ns |
| ACT1 | 0.000*** | 0.040* | 0.000*** |
| EF-1α | 0.000*** | 0.874ns | 0.504ns |
| GAPDH | 0.010* | 0.000*** | 0.020* |
Two-way ANOVA analysis: ns, nonsignificant, p>0.05; * p<0.05, ** p<0.01; *** p<0.001.
Since the virus was considered the main effect and we had more than 2 viruses, we used Tukey’s honestly significant difference (HSD) post-hoc test to compare the influence of (1) BYDV-GAV vs. WYDV-GPV, (2) BYDV-GAV vs. BYDV-PAV, and (3) WYDV-GPV vs. BYDV-PAV on each candidate reference gene (Table 4). For the influence of infection by BYDV-GAV vs. WYDV-GPV on the expression of 18S rRNA, ACT1 or EF-1α of R. padi, the expression levels of these candidate reference genes differed significantly (all p<0.001), and less so for GAPDH (p = 0.009). Differences in 18S rRNA expression for BYDV-GAV vs. BYDV-PAV was highly significant (p<0.001), but not significant for the other 3 reference genes (ACT1, p = 0.310; EF-1α, p = 0.139; GAPDH, p = 0.590). WYDV-GPV and BYDV-PAV differed significantly in expression of ACT1 and of EF-1α (both p<0.001), but consistently lacked differences for 18S rRNA (p = 0.879) and for GAPDH (p = 0.109).
Table 4. Paired comparisons for reference genes between the WYDV-GPV (GPV), BYDV-PAV (PAV) and BYDV-GAV (GAV)-viruliferous aphids (Rhopalosiphum padi).
| GAV | GPV | PAV | ||||
| Gene | GPV | PAV | GAV | PAV | GAV | GPV |
| 18S rRNA | 0.000*** | 0.000*** | 0.000*** | 0.879 ns | 0.000*** | 0.879 ns |
| ACT1 | 0.000*** | 0.310 ns | 0.000*** | 0.000** | 0.310 ns | 0.000*** |
| EF-1α | 0.000*** | 0.139 ns | 0.000*** | 0.000** | 0.139 ns | 0.000*** |
| GAPDH | 0.009** | 0.590 ns | 0.009** | 0.109 ns | 0.590 ns | 0.109 ns |
Tukey’s HSD post hoc tests: ns, nonsignificant, p>0.05; * p<0.05; ** p<0.01; *** p<0.001.
In summary, the Cq values for expression of 18S rRNA in WYDV-GPV- and BYDV-PAV-viruliferous R. padi fell into one group (subset 2), the BYDV-GAV-viruliferous R. padi into another (subset 1). As for the Cq values of ACT1 and EF-1α, the WYDV-GPV-viruliferous R. padi was in one group (subset 1), the others in a second (subset 2). The Cq values for the GAPDH in BYDV-PAV- and BYDV-GAV-viruliferous R. padi are logically within one group, rather than with WYDV-GPV-viruliferous R. padi, because of the lack of significant differences between the infection with BYDV-PAV and BYDV-GAV (p = 0.590), with p-values lower than that for the comparison between WYDV-GPV and BYDV-PAV (p = 0.109). The lack of unanimous grouping of Cq values for these 4 candidate reference genes might be attributed to the differing transmission properties of the YDVs by R. padi because successful transmission depends on the compatibility between the virus and vector insects [9].
Expression Stability Analysis of Reference Genes in YDV-viruliferous R. padi
For analyzing the expression stability of the 4 candidate reference genes, a stability value for each individual gene was calculated using three Excel-based tools (GeNorm [25] and NormFinder [37] and BestKeeper [38]). The amplification efficiencies of 4 candidate genes in winged R. padi differed somewhat from that of the wingless (Table 2). Therefore, the stability ranking fell into two sets.
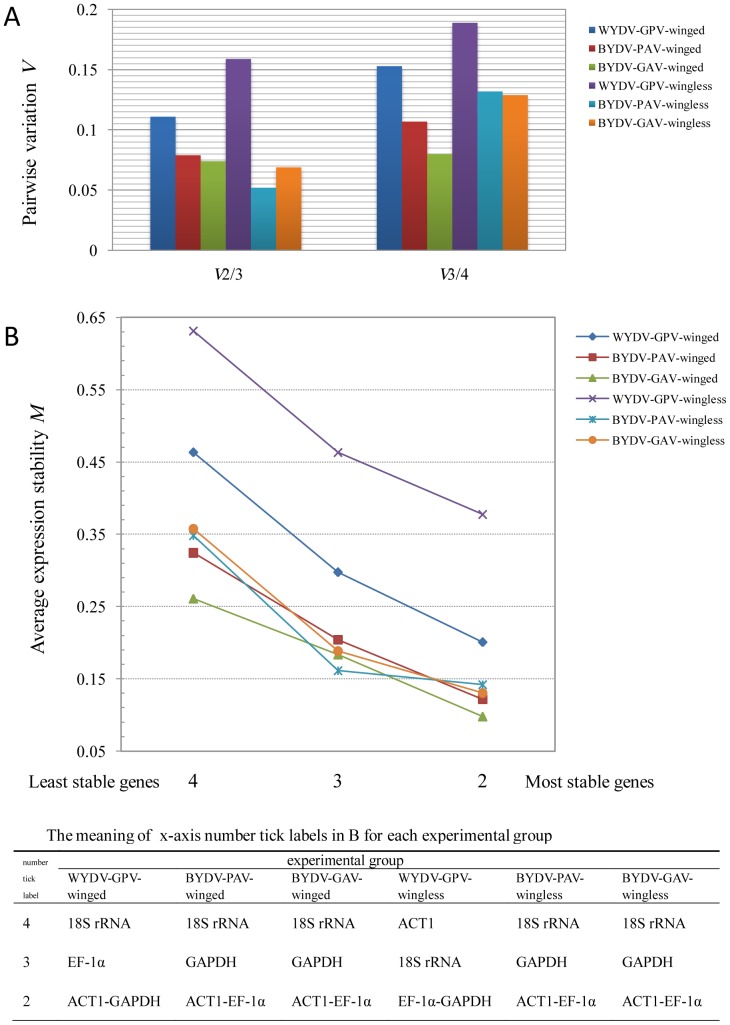
GeNorm only recognizes linear scale quantities, obtained by transforming raw Cq values using the delta-Cq method (described in the methods) or a standard curve; an expression stability value M for all genes contained in the InputData file is automatic calculated by an algorithm [25] and displayed. The smallest M value indicates the most stable gene expression; the largest M value indicates the most unstable expression. Importantly, expression of a gene with M >1.5 is considered to be inconsistent [25]. After the Cq values for the RT-qPCR of all 4 candidate reference genes under the 6 experimental conditions were transformed into a valid data set (see formula in the methods), the M values were calculated by a GeNorm applet. As shown in Tables 5 and 6, 4 virus–aphid combinations (WYDV-GPV-winged, BYDV-GAV-winged, BYDV-PAV-wingless and BYDV-GAV-wingless) had the same order of expression stability from the most stable to the least stable gene: EF-1α, ACT1, GAPDH and 18S rRNA. For the BYDV-PAV-winged combination, ACT1 was ranked as the best reference gene (lowest M), followed by EF-1α, GAPDH and 18S rRNA. However, ACT1 was the worst in the case of WYDV-GPV-wingless, and the stability sequence of the other 3 genes was the same for BYDV-PAV-winged. In brief, EF-1α (5 first places and 1 second place) seemed to be the best reference gene to normalize the GOIs in all YDV-viruliferous types of R. padi.
Table 5. Stability rankings according to three software tools for individual endogenous reference genes in winged adults of Rhopalosiphum padi carrying one of three YDVs.
| Virus and reference gene | GeNorm a | NormFinderb | BestKeeperc |
| M | stability value | Cq set SD | |
| GPV | |||
| EF-1α | 0.396 | 0.080 | 0.29 |
| ACT1 | 0.406 | 0.085 | 0.34 |
| GAPDH | 0.421 | 0.110 | 0.30 |
| 18S rRNA | 0.629 | 0.192 | 0.45 |
| PAV | |||
| ACT1 | 0.270 | 0.054 | 0.31 |
| EF-1α | 0.274 | 0.049 | 0.31 |
| GAPDH | 0.309 | 0.094 | 0.28 |
| 18S rRNA | 0.444 | 0.165 | 0.26 |
| GAV | |||
| EF-1α | 0.211 | 0.051 | 0.29 |
| ACT1 | 0.226 | 0.074 | 0.31 |
| GAPDH | 0.259 | 0.076 | 0.27 |
| 18S rRNA | 0.338 | 0.151 | 0.21 |
According to GeNorm, the smallest M value indicates the most stable gene expression, the largest M value the most unstable; expression of a gene with M >1.5 is considered inconsistent.
Like GeNorm, the most stable gene has the lowest stability value, and vice versa.
Genes with SD <1 can be ranked according to their Cq SD values. The gene most stably expressed has the lowest SD value; the highest SD value indicates the gene with the most unstable expression.
Table 6. Stability rankings according to three software tools for individual endogenous reference genes in wingless adults of Rhopalosiphum padi carrying one of three YDVs.
| Virus and reference gene | GeNorma | NormFinderb | BestKeeperc |
| M | stability value | Cq set SD | |
| GPV | |||
| EF-1α | 0.508 | 0.033 | 0.26 |
| GAPDH | 0.570 | 0.115 | 0.46 |
| 18S rRNA | 0.648 | 0.202 | 0.29 |
| ACT1 | 0.799 | 0.231 | 0.68 |
| PAV | |||
| EF-1α | 0.277 | 0.017 | 0.36 |
| ACT1 | 0.285 | 0.028 | 0.38 |
| GAPDH | 0.296 | 0.060 | 0.34 |
| 18S rRNA | 0.535 | 0.213 | 0.24 |
| GAV | |||
| EF-1α | 0.293 | 0.053 | 0.42 |
| ACT1 | 0.294 | 0.054 | 0.49 |
| GAPDH | 0.317 | 0.078 | 0.48 |
| 18S rRNA | 0.527 | 0.212 | 0.29 |
According to GeNorm, the smallest M value indicates the most stable gene expression, the largest M value the most unstable; expression of a gene with M >1.5 is considered inconsistent.
Like GeNorm, the most stable gene has the lowest stability value, and vice versa.
Genes with SD <1 can be ranked according to their Cq SD values. The gene most stably expressed has the lowest SD value; the highest SD value indicates the gene with the most unstable expression.
A different algorithm, NormFinder, only analyzes linear scale data [37]. The stability values of the reference genes can be calculated for one experimental group or for multiple groups in one data set. The most stable gene has the lowest stability value, and vice versa [37]. When the transformed linear data (the same Q values of the 4 candidate reference gene analyzed by GeNorm) was analyzed with NormFinder, sorting for expression stability of the candidate reference genes (Tables 5 and 6) for the WYDV-GPV-winged, BYDV-PAV-winged, BYDV-GAV-winged and BYDV-PAV-wingless groups was highly uniform: EF-1α was the most stable gene, but 18S rRNA was the most unstable. EF-1α was also the first choice of reference genes for the WYDV-GPV-wingless group, but sorted as the third most stable gene for the BYDV-GAV-wingless.
Finally, the raw Cq values of candidate reference genes in each group were assessed directly by BestKeeper; when the SD of the Cq is higher than 1, expression for that is considered as inconsistent, and only genes with a SD less than 1 can be ordered using their Cq SD values [38]. The gene most stably expressed thus has the lowest SD value; the gene most unstably expressed has the highest SD value. In this study, Cq SD values from the qPCRs of the 4 candidate reference genes in all groups were less than 1 (Tables 5 and 6) and can be compared with each other as individual groups. Unlike the results from GeNorm and NormFinder, 18S rRNA was evaluated as the first and second most stable gene in the following 5 groups: BYDV-PAV-winged, BYDV-GAV-winged, BYDV-PAV-wingless, BYDV-GAV-wingless and WYDV-GPV-wingless, but as the least stably expressed gene in the GPV-winged group. EF-1α, assessed as a nearly ideal reference gene by GeNorm and NormFinder tools, was ranked as the optimal reference gene only in the WYDV-GPV-winged and -wingless groups. For the other 4 groups, the performance of EF-1α was mediocre.
As discussed already, GeNorm and NormFinder yielded very similar results with respect to sorting expression stability in each group and in identifying the best reference gene among 4 candidate reference genes. We inferred that the two programs analyzed the same linear scale data set, which was transformed from raw Cq values and corrected by amplification efficiency of the respective gene (see formula in the methods). The results from the BestKeeper tool were nearly opposite to those obtained with GeNorm and NormFinder. Perhaps the number and variety of candidate reference genes that we used here are not very diversified.
Selection of Reference Genes for Normalization
There were 3 cases for calculating the best number of reference genes in our 6 experimental groups: (1) V2/3<0.15, V3/4<0.15; (2) V2/3<0.15, V3/4>0.15; and (3) V2/3>0.15, V3/4>0.15 (Figure 2A). The first case meant the best number of reference genes could be 2, 3 or 4; simply, the 2 most stable genes, ACT1 and EF-1α, (Figure 2B) were the ideal combination of reference genes for the normalization analysis of target genes in the respective groups, BYDV-PAV-winged, BYDV-GAV-winged, BYDV-PAV-wingless, and BYDV-GAV-wingless. The second case demonstrated that the optimal choice of reference genes was ACT1 and GAPDH for the WYDV-GPV-winged group (Figure 2B), while adding another one or two candidate reference genes increased the risk of misinterpreting the expression profile of the GOIs. The last case indicated that using just these 4 candidate reference genes would not form a dependable reference gene group for the relative quantification of GOIs in WYDV-GPV-viruliferous wingless R. padi.
Figure 2. Determination of optimal reference genes for normalization by GeNorm.
The optimal number of reference genes was determined by pairwise variation V (A) between two sequential normalization factors containing an increasing number of reference genes (n and n +1 reference genes). The pairwise variation begins with the first two and three (V2/3) most stable candidate reference genes (B), terminating with the addition of the least stable one, here V3/4. The cut-off V value is set at 0.15 by the program GeNorm, below which an additional reference gene does not need to be included for calculating the normalization factor.
Relative Quantification of Endogenous Genes
Since no up- or down-regulation of these candidate internal control genes was observed in the 6 combinations (SD <1 from BestKeeper and stability value M <1.5 from GeNorm), and the optimal number of reference genes had been selected by GeNorm, these genes can be used in the expression profile analysis of endogenous and low-abundance genes. Because the expression levels of ago-1a and dcr1a (dcr1) gene varied significantly among four reproductive morphs of A. pisum when measured with semi-quantitative RT-PCR [49], miRNA machinery and alternative transcription are thought to play a key role in morph transformation in the life cycle of A. pisum.
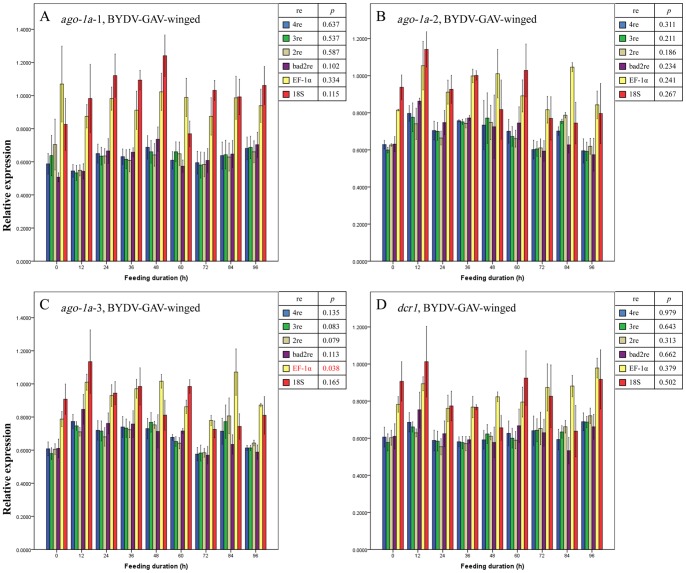
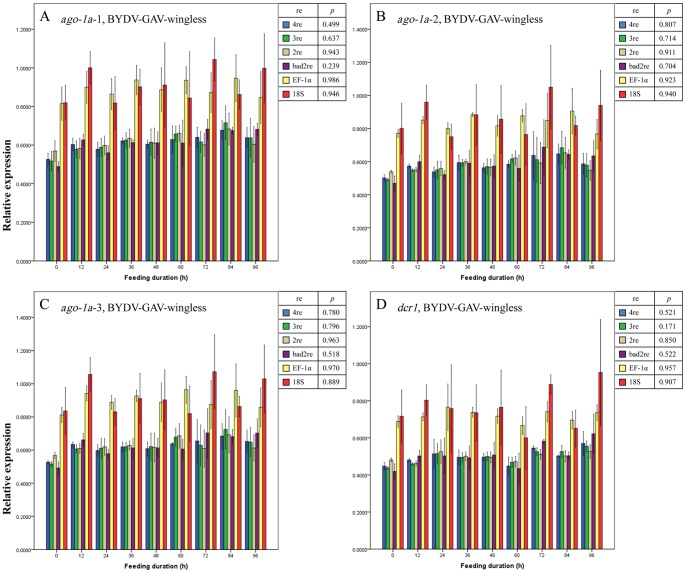
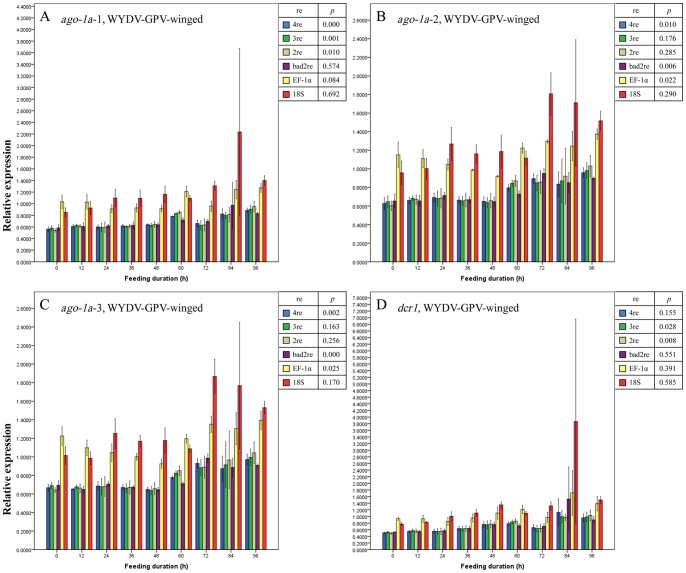
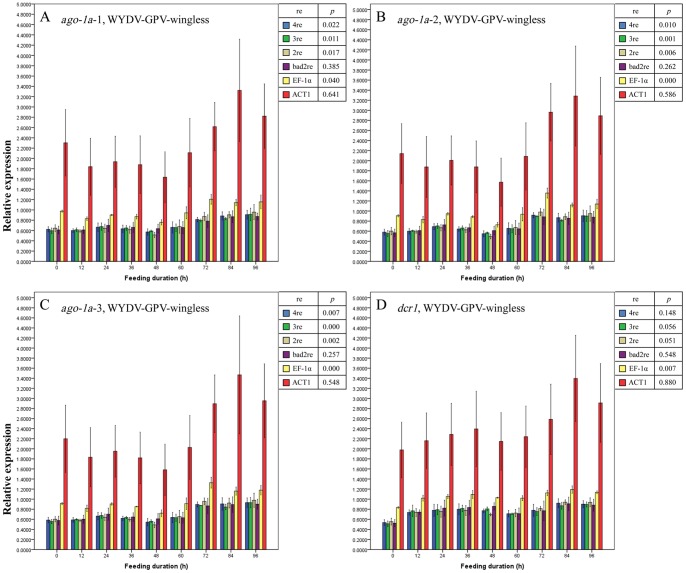
Regardless of the normalization method used (described in Materials and Methods), no significant differences were observed in the expression level for the 3 regions of the ago-1a and dcr1 genes before and after the entry of BYDV-GAV into winged (Figure 3 A–D) or wingless (Figure 4 A–D) adults of R. padi (all p>0.05, except for when EF-1α was used as the reference gene to normalize the expression change of ago-1a-3 region [p = 0.038] [Figure 3C]). We thus concluded that the expression of the ago-1a and dcr1 genes did not respond to the entry of BYDV-GAV into R. padi and remained unchanged. This finding conformed to the facts that BYDV-GAV could scarcely be attached by receptor proteins of R. padi, and has rare chance to access to hindgut apical plasmalemma [12].
Figure 3. Mean relative expression values (± SE, n = 3) for endogenous aphid genes ago-1a-1 (A), ago-1a-2 (B), ago-1a-3 (C) and dcr1 (D) in BYDV-GAV-viruliferous winged adults of Rhopalosiphum padi after different virus-feeding durations.
A–D, Expression values for each gene or each region of one gene at each duration were normalized with the reference gene(s) selected by GeNorm and then compared with a one-way ANOVA (p) among these durations with each normalization condition. 2re = 2 best reference genes; 3re = 3 best reference genes; 4re = all 4 reference genes; bad2re = 2 least stable reference genes; EF-1α = EF-1α as the reference gene; 18S = 18S rRNA as the reference gene.
Figure 4. Mean relative expression values (± SE, n = 3) for endogenous aphid genes ago-1a-1 (A), ago-1a-2 (B), ago-1a-3 (C) and dcr1 (D) in BYDV-GAV-viruliferous wingless adults of Rhopalosiphum padi after different virus-feeding durations.
A–D, Expression values for each gene or each region of one gene at each duration were normalized with the reference gene(s) selected by GeNorm and then compared with a one-way ANOVA (p) among these durations with each normalization condition. 2re = 2 best reference genes; 3re = 3 best reference genes; 4re = all 4 reference genes; bad2re = 2 least stable reference genes; EF-1α = EF-1α as the reference gene; 18S = 18S rRNA as the reference gene.
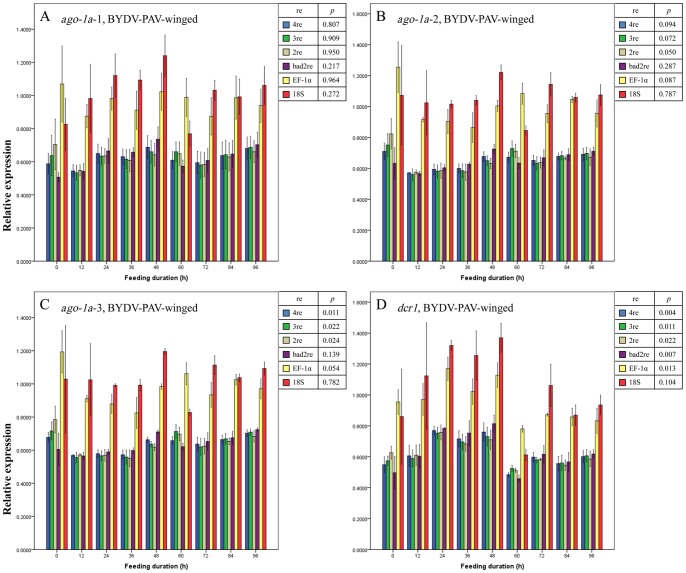
The entry of BYDV-PAV did not seem to affect the expression profile of ago-1a in either of the wing morphs of R. padi on the basis of the results in Figures 5A and 6A, for relative expression values of ago-1a-1 among these feeding durations approached the same (all p>0.05 under all normalizations). However, for expression levels of ago-1a-2 and ago-1a-3 (the middle region and 3′-terminus of ago-1a gene [49]) using all the same normalization methods, we drew different conclusions: after being normalized with a single reference gene (EF-1α or 18S rRNA) or with bad reference genes, the expression of ago-1a-2 and -3 in BYDV-PAV-viruliferous winged R. padi showed no significant changes from 0 h to 96 h feeding duration (purple, yellow and red bars in Figure 5B and C, all p>0.05); thus, ago-1a-2 and -3 appeared to be unaffected by BYDV-PAV acquisition; when the best 2 or 3 reference genes or all 4 reference genes were used to normalize the expression of these two regions in winged R. padi, a difference in the expression of ago-1a-2 among the different feeding durations emerged slightly above the significance level (blue, green, and brown bars in Figure 5B, 0.1>p≥0.05), but the expression values for ago-1a-3 already declined significantly before (0 h) and immediately after entry of BYDV-PAV (blue, green, and brown bars in Figure 5C, p<0.05) in spite of recover to initial level afterwards. In wingless R. padi, the changes in expression for ago-1a-3 higher than that for ago-1a-2 (Figure 6B and C, both p<0.1, the p value for ago-1a-3 was lower than for ago-1a-2) was observed when the best 2 or 3 reference genes or all 4 reference genes were used as normalization methods, with a similar trend in the expression change for ago-1a-2 and ago-1a-3 in the winged morph. On the basis of these findings, we concluded that although no change in expression was detected for ago-1a-1 in R. padi, the expression of the full-length ago-1a was disturbed by the entry of BYDV-PAV particles as seen by the obvious expression change in ago-1a-2 and ago-1a-3. For the expression change in dcr1 in BYDV-PAV-viruliferous winged and wingless R. padi (Figures 5D and 6D), the significance level (both p<0.1) was near that of ago-1a-2 and -3 (all p<0.1) with the normalization factors calculated by the best 2, or 3 reference genes or all 4 reference genes. The detected region for the dcr1 gene is also in the middle region and the 3′-terminus of the full-length dcr1 gene [49], so we speculated that the entry of BYDV-PAV also interfered with the expression of dcr1. The unexpected discrepancies among values for changes in relative expression obtained using 3 regions of the same gene in BYDV-PAV-viruliferous R. padi suggested that mechanisms on the splicing and reconstruction of these three regions in ago-1a were differences in aphids, probably because the pre-mRNA of region 2 and region 3 were recruited and then translate into the PAZ and PIWI domains of antivirus defense-associated AGO2 protein in aphids.
Figure 5. Mean relative expression values (± SE, n = 3) for endogenous aphid genes ago-1a-1 (A), ago-1a-2 (B), ago-1a-3 (C) and dcr1 (D) in BYDV-PAV-viruliferous winged adults of Rhopalosiphum padi after different virus-feeding durations.
A–D, Expression values for each gene or each region of one gene at each duration were normalized with the reference gene(s) selected by GeNorm and then compared with a one-way ANOVA (p) among these durations with each normalization condition. 2re = 2 best reference genes; 3re = 3 best reference genes; 4re = all 4 reference genes; bad2re = 2 least stable reference genes; EF-1α = EF-1α as the reference gene; 18S = 18S rRNA as the reference gene.
Figure 6. Mean relative expression values (± SE, n = 3) for endogenous aphid genes ago-1a-1 (A), ago-1a-2 (B), ago-1a-3 (C) and dcr1 (D) in BYDV-PAV-viruliferous wingless adults of Rhopalosiphum padi after different virus-feeding durations.
A–D, Expression values for each gene or each region of one gene at each duration were normalized with the reference gene(s) selected by GeNorm and then compared with a one-way ANOVA (p) among these durations with each normalization condition. 2re = 2 best reference genes; 3re = 3 best reference genes; 4re = all 4 reference genes; bad2re = 2 least stable reference genes; EF-1α = EF-1α as the reference gene; 18S = 18S rRNA as the reference gene.
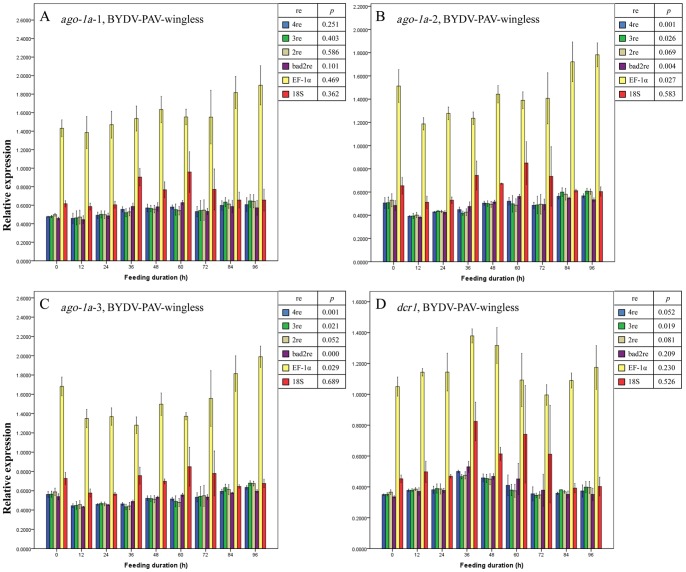
For WYDV-GPV-viruliferous R. padi, we could not give an explicit inference that the relative expression in the 3 regions of ago-1a and dcr1 gene changed or unchanged with the virus acquisition, since these normalization methods provided no concurrent and regular results (Figures 7 and 8). We inferred that the complex interactions between WYDV-GPV, oat plant and vector R. padi led to the unsuitability of the selected reference genes (Figure 2A, V 2/3 and V3/4 near or above the cut-off value of 0.15).
Figure 7. Mean relative expression values (± SE, n = 3) for endogenous aphid genes ago-1a-1 (A), ago-1a-2 (B), ago-1a-3 (C) and dcr1 (D) in BYDV-GPV-viruliferous winged adults of Rhopalosiphum padi after different virus-feeding durations.
A–D, Expression values for each gene or each region of one gene at each duration were normalized with the reference gene(s) selected by GeNorm and then compared with a one-way ANOVA (p) among these durations with each normalization condition. 2re = 2 best reference genes; 3re = 3 best reference genes; 4re = all 4 reference genes; bad2re = 2 least stable reference genes; EF-1α = EF-1α as the reference gene; 18S = 18S rRNA as the reference gene.
Figure 8. Mean relative expression values (± SE, n = 3) for endogenous aphid genes ago-1a-1 (A), ago-1a-2 (B), ago-1a-3 (C) and dcr1 (D) in BYDV-GPV-viruliferous wingless adults of Rhopalosiphum padi after different virus-feeding durations.
A–D, Expression values for each gene or each region of one gene at each duration were normalized with the reference gene(s) selected by GeNorm and then compared with a one-way ANOVA (p) among these durations with each normalization condition. 2re = 2 best reference genes; 3re = 3 best reference genes; 4re = all 4 reference genes; bad2re = 2 least stable reference genes; EF-1α = EF-1α as the reference gene; 18S = 18S rRNA as the reference gene.
The miRNA pathway has close relationship with RNAi pathway in eukaryotes, and the two pathways possess extremely parallel mechanism of RNA cleavage in cell cytoplasm [55]. The machinery proteins of miRNA pathway, Ago-1and Dcr1, have high similarity with those of RNAi pathway, Ago-2 and Dcr2 [49], [56]. Since R. padi is efficient vector of BYDV-PAV and WYDV-GPV, these RNA pathways may be triggered against virus invasion [56].
Fluctuation and Comparison of Virus Titre in R. padi
Fluctuation in the virus titre in R. padi was monitored by two methods. The first method used the SD values of the raw Cq values (Cq SD) for the CP gene and RTP gene (the Cq values of the no-template controls and 0-h-feeding duration sample of these two genes were either higher than 36 or undetermined and thus not analyzed here) as calculated by BestKeeper. In WYDV-GPV and BYDV-PAV, viruliferous winged and wingless R. padi, the Cq SD values of the CP gene and RTD gene were all higher than 1, which was affirmed to be inconstant and statistically significantly different among feeding durations; but the Cq SD values of the CP gene and RTD gene of BYDV-GAV in winged and wingless R. padi were lower than 1, with no variance in virus titre during the 4-day AAP (Table S2 and S3). These results agreed with the transmission specificity of these three YDVs by R. padi, and the difference in Cq at the initial virus-acquisition duration (maximal Cq value) implied the presence of a sluggish receptor-mediated recognition process at the midgut and/or hindgut [9]. One problem with this method is that the trend for increased virus load is inversely related to the duration of virus-feeding.
The second one is the relative virus load, normalized by one reference gene or a combination of reference genes (with the same normalization as the endogenous genes). The expression levels of the CP gene and RTD gene of WYDV-GPV, BYDV-PAV and BYDV-GAV in winged (Figure 9 A–F) and wingless R. padi (Figure 10 A–F) increased more than 5-fold from the extraordinarily low value at the initial 12 h to the highest value during the last day (72 to 96 h), and the difference in expression of either the CP or RTD gene across all feeding durations was significant (all p<0.05, Figures 9 A–F and 10 A–F), regardless of the normalization setting adopted. When the duration of feeding on either WYDV-GPV-, BYDV-PAV- or BYDV-GAV-infected oat plants exceeded 48 h, the relative expression values of the CP gene and RTD gene continued to rise at a slow rate and rose and fell within a narrow range. The findings reflected accumulation but no tendency for replication of the YDV particles in R. padi and the saturability of virus load by aphid, in agreement with Gray and Gildow’s hypothesis about the nonpropagative transmission of YDVs by aphids [9]. As shown in Figures 9 and 10, the fluctuation in virus titre of the three viruses in winged and wingless R. padi at each duration within the 4-day AAP had some slight differences, relied on the method of normalization, and a good reference gene or reference gene combination estimated by GeNorm resulted in a much lower mean SE value for the sample replicates.
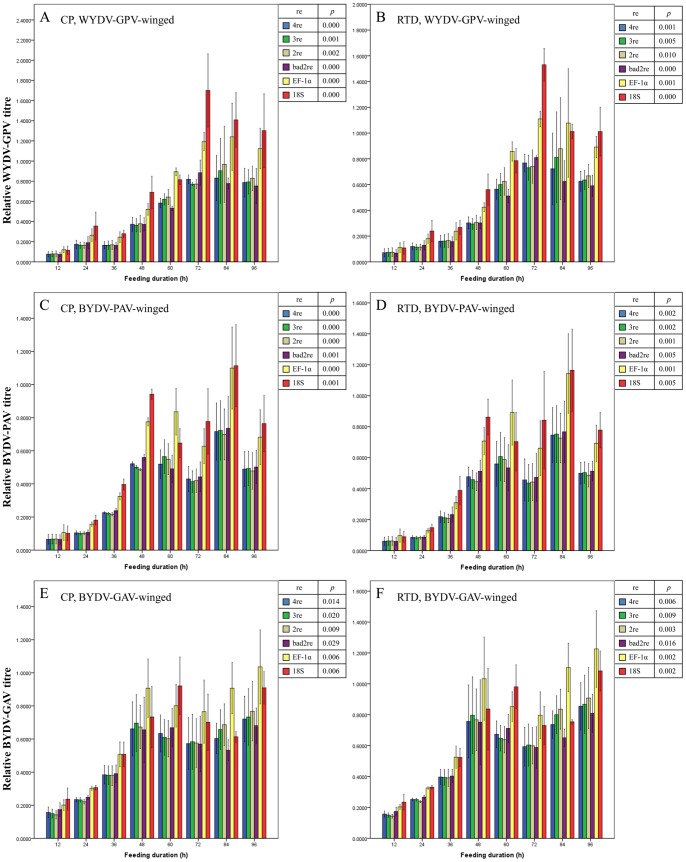
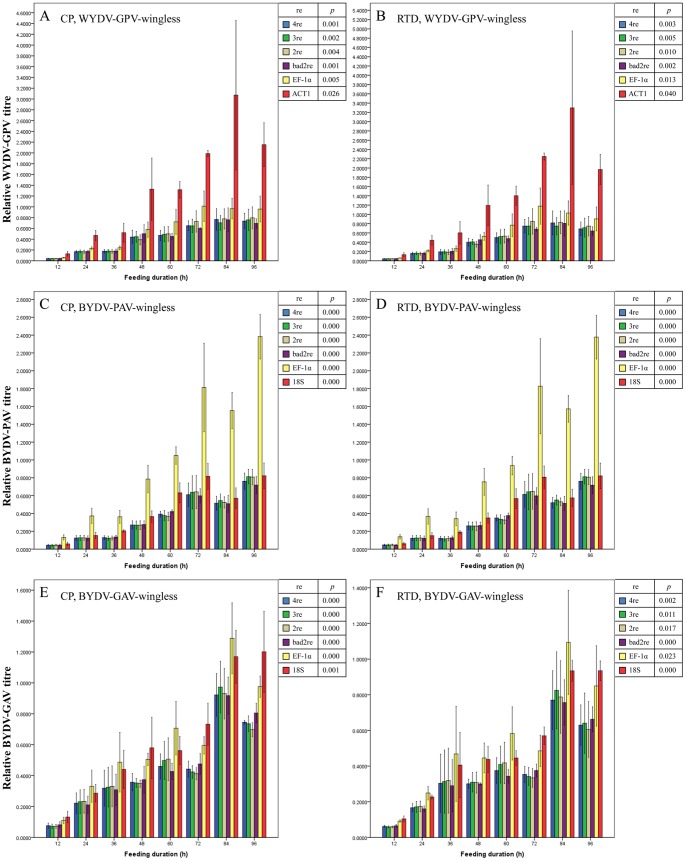
Figure 9. Mean relative titre of YDVs (± SE, n = 3) in viruliferous winged adults of Rhopalosiphum padi after different virus-feeding durations.
Virus titre is illustrated by relative expression values of the CP (A, C and E) and RTD (B, D and F) gene of YDVs. Expression values for the CP or RTD gene at each duration were normalized with the reference gene(s) selected by GeNorm and then compared with a one-way ANOVA (p) among these durations with each normalization condition. A and B: WYDV-GPV; C and D: BYDV-PAV; E and F: BYDV-GAV. 2re = 2 best reference genes; 3re = 3 best reference genes; 4re = all 4 reference genes; bad2re = 2 least stable reference genes; EF-1α = EF-1α as the reference gene; 18S = 18S rRNA as the reference gene.
Figure 10. Mean relative titre of YDVs (± SE, n = 3) in viruliferous wingless adults of Rhopalosiphum padi after different virus-feeding durations.
Virus titre is illustrated by relative expression values of the CP (A, C and E) and RTD (B, D and F) gene of YDVs. Expression values for the CP or RTD gene at each duration were normalized with the reference gene(s) selected by GeNorm and then compared with a one-way ANOVA (p) among these durations with each normalization condition. A and B: WYDV-GPV; C and D: BYDV-PAV; E and F: BYDV-GAV. 2re = 2 best reference genes; 3re = 3 best reference genes; 4re = all 4 reference genes; bad2re = 2 least stable reference genes; EF-1α = EF-1α as the reference gene; 18S = 18S rRNA as the reference gene.
We could not uncover differences in transmissibility of these three YDVs by R. padi by comparing the relative virus titre during the first 2 days of feeding with a one-way ANOVA because the relative virus titre between transmissible WYDV-PAV or BYDV-PAV and non- transmissible BYDV-GAV did not differ significantly) (all p>0.05, data not shown), even though the relative titre of WYDV-GPV and BYDV-PAV (Figures 9 A–D and 10 A–D) for the 12, 24 and 36 h of feeding was lower than that of BYDV-GAV (Figures 9E, 9F, 10E, and 10F). Perhaps the typical biological transmission assay is the most efficient and convincing measure for determining the insect vector’s transmission specificity for a particular plant virus [19].
With the extension of aphid feeding duration on infected oat plants, the relative expression values of the CP or RTD gene of all three YDVs in R. padi increased gradually from 12 h to 60 h or 72 h, but tended to stabilized after 60 h or 72 h. BYDV-PAV and WYDV-GPV once acquired are retained life of R. padi. Following ingestion during feeding, the virus moves through the alimentary canal of R. padi, invades the gut epithelial cells and then is released into the hemolymph, ultimately infects the salivary glands and is released into the salivary ducts where it can be transferred to new plants via the saliva released during feeding [57]. Virus titre continues to rise during the first 48 h of the AAP and IAP, but decreases slightly because the virus particles are transmitted into plants. As aphids continue sucking the virus-containing phloem sap, the virus load in aphids rises again. For the untransmissible one, the virus only moves through the alimentary canal and then direct excretes from body. Therefore, the virus titre increases at initial feeding duration, and then keeps a dynamic balance because of the continuous sucking [12], [58].
Conclusions
In this study, we found that in testing EF-1α, ACT1, GAPDH and 18S rRNA as reference genes, combining the first 2 or 3 genes or using all 4 genes can be used to normalize the relative expression level of endogenous and virus titre in BYDV-PAV and BYDV-GAV-viruliferous winged and wingless R. padi. Using only one reference gene, regardless of its expression stability, is not recommended for relative quantification by RT-qPCR. Importantly, for accurate and precise results, the use of different regions of a gene, including the 5′-terminus, middle and 3′-terminus, at the same in the relative RT-qPCR is strongly advised, since the different regions of one gene endure different selection pressures and may evolve some other new functions in complex life cycle of aphids. To assess relative expression and virus load in winged and wingless WYDV-GPV-viruliferous R. padi, the optimal number of combination of reference genes should be selected carefully, abandoning genes that are not useful alone or in combination and testing new ones as needed.
Materials and Methods
Virus Maintenance and Aphid Rearing
Laboratory isolates of WYDV-GPV, BYDV-PAV, and BYDV-GAV have been maintained on oat plants (Avena sativa K. Koch cv. Coast-Black Oat) in our laboratory since the 1990s [59]. A virus-free laboratory population of R. padi was reared on winter wheat (Triticum aestivum L., cv. Yangmai 158), under controlled conditions at 18–23°C. A low-density population was maintained for the wingless morph of R. padi, and a high-density population was maintained to obtain the winged morph. This R. padi population transmits BYDV-PAV (70–80% transmission rate) and WYDV-GPV (50% transmission rate) effectively, but poorly transmits BYDV-GAV (5–7% transmission rate) [60].
To confirm the morph features of wingless aphids and winged aphids, we observed different developmental stages of R. padi from first to fifth instar daily with a stereomicroscope (Olympus SZX16). Because the lifespan of the winged and wingless adult aphids (more than 5 days) is much longer than that of each nymph instar (less than 2 days) and the adult aphids are distinguishable from the nymphs, winged and wingless adults were used for the experiments.
Experimental Treatment and Sampling
Approximately 300 wingless and 300 winged adult aphids were picked from wheat with a soft brush, and then released carefully onto two oat plants infected with WYDV-GPV, BYDV-PAV or BYDV-GAV for acquisition of viruses, with insect-proof nets separating the three sets of infected plants. After each feeding duration (0, 12, 24, 36, 48, 60, 72, 84 or 96 h), 24 wingless adult aphids in each net were collected and divided randomly into 3 pools, then 8 aphids of each type were placed into an RNase-free tube. The winged adult aphids were treated the same way. The tubes were immediately placed in liquid nitrogen and stored in a −70°C freezer for RNA extraction.
Selection of Candidate Reference Genes and Target Genes and Design of Primers
Four HKGs of R. padi, including 18S rRNA, ACT1, EF-1α, and GAPDH were selected as candidate reference genes (Table 7). Based on the corresponding sequences of R. padi and A. pisum from the NCBI GenBank, primers for reverse transcription-PCR (RT-PCR) amplification of 18S rRNA and EF-1α were designed (Table 1). Primer pairs for amplifying the ACT1 and GAPDH sequences of R. padi were designed using homologous sequences of A. pisum deposited in GenBank (Table 1).
Table 7. Description of candidate reference genes for Rhopalosiphum padi and of target genes.
| Genes | Gene name | Molecular functiona | Sourceb | ||||
| Candidate referencegenes | |||||||
| 18S rRNA | 18S ribosomal RNA (18S rRNA) | Participates in protein translationof Eukaryotes | U27825 | ||||
| ACT1 | Actin1 | Cytoskeletal structural protein | NM_001126200 | ||||
| EF-1α | Elongation factor-1 alpha | Involved in protein biosynthesis | AY219719 | ||||
| GAPDH | Glyceraldehyde-3-phosphatedehydrogenase | Catalyzes the reversible oxidativephosphorylation | XM_001943014 | ||||
| Endogenous genes | |||||||
| ago-1a-1 | Argonaute-1 region1 | miRNA machinery; RISCkey component | HE585880-81 | ||||
| ago-1a-2 | Argonaute-1 region2 | miRNA machinery; RISCkey component | HE585908-09 | ||||
| ago-1a-3 | Argonaute-1 region3 | miRNA machinery; RISCkey component | HE585932-33 | ||||
| dcr1 | Dicer-1 | RNase III enzyme | HE585957-58 | ||||
| Viral genes | |||||||
| BYDV-GPV CP | Polerovirus Wheat yellowdwarf virus-GPV, Coatprotein | Encodes major structuralprotein of virus particle; aphidtransmission-related protein | FM865413 | ||||
| BYDV-GPV RTD | Polerovirus Wheat yellowdwarf virus-GPV,Readthrough domain | Encodes minor structuralprotein of virus particle; aphidtransmission-related protein | FM865413 | ||||
| BYDV-PAV CP | Luteovirus Barley yellowdwarf virus-PAV, Coatprotein | Encodes major structuralprotein of virus particle; aphidtransmission-related protein | EU332307-336 | ||||
| BYDV-PAV RTD | Luteovirus Barley yellowdwarf virus-PAV,Readthrough domain | Encodes minor structuralprotein of virus particle; aphidtransmission-related protein | EU332307-386 | ||||
| BYDV-GAV CP | Luteovirus Barley yellowdwarf virus-GAV, Coatprotein | Encodes major structuralprotein of virus particle; aphidtransmission-related protein | EU402386-391 | ||||
| BYDV-GAV RTD | Luteovirus Barley yellowdwarf virus-GAV,Readthrough domain | Encodes minor structuralprotein of virus particle; aphidtransmission-related protein | EU402386-391 | ||||
Terminology cited from Gene Ontology (GO) of NCBI.
GenBank accession number (NCBI).
Two endogenous genes of R. padi, ago-1a and dcr1, which might be involved in the miRNA pathway in aphids [48], [49], were used as target genes. The 3 packed regions of the ago-1a have been showed different selective pressures, region 1 and region 2 reflecting strong purifying selection (negative selection), but region 3 owning no positively selected sites [49]. The chosen region of dcr1 has the same selection pressure with region 3 of ago-1a [49]. For the RT-PCR of these two genes, primers (Table 1) were devised using the DNA regions for the ago-1a gene and dcr1 gene.
The CP gene (ORF3), encoding the coat protein, and RTD gene (ORF5), encoding the readthrough protein, of WYDV-GPV, BYDV-PAV and BYDV-GAV from infected oat plants were amplified by RT-PCR using their respective primers (Table 1). All primers used in this study were synthesized by Sangon Biotech (Shanghai) Co.
RNA Extraction and Purification
For plant leaves and a large number of aphids, samples were first homogenized in liquid nitrogen with a sterile mortar and pestle. Then total RNA was extracted from the resulting fine powder using the instructions for TRIzol reagent (Ambion, USA) with minor modifications. RNA was precipitated in isopropanol overnight at 4°C. The 8-aphid pooled sample was frozen with liquid nitrogen and ground with a clean glass rod for total RNA extraction in 400 µl TRIzol reagent. The concentration and purity of RNA samples were measured with a NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific, Roskilde, Denmark). The integrity was checked using electrophoresis in 1.5% agarose gels and 1× Tris-acetate EDTA (TAE). The gel was then photographed in UV light (Bio-Rad, Universal Hood II, USA).
RT-PCR of Candidate Reference and Target Genes
The total RNA isolated from plant leaves and the aphid samples of all developmental stages was reverse transcribed into first-strand cDNA only using the reverse primer of each gene. The components in each 25-µl RT reaction system included 7 µl RNase-free ddH2O, 1 µl 5 µM reverse primer and 2 µl total RNA. The mixture was briefly centrifuged and denatured at 70°C for 10 min, and then combined with 3.5 µl RNase-free ddH2O, 5 µl dNTPs (2.5 µM, TaKaRa, Dalian, China), 5 µl 5× RT reaction buffer, 1 µl M-MLV Reverse Transcriptase (200 U/µl, Promega, Madison, WI, USA), 0.5 µl Recombinant RNase Inhibitor (40 U/µl, TaKara). All reagents in the tube were centrifuged shortly and incubated at 42°C for 1 h. The RT reaction was terminated at 95°C for 5 min. The final cDNA samples were kept at −20°C.
The PCR reaction mixture for each first-strand cDNA comprised 17.8 µl ddH2O, 0.25 µl forward primer (5 µM), 0.25 µl reverse primer (5 µM), 10× LA Taq Buffer II (Mg2+ plus), 2 µl dNTPs, and 0.2 µl LA Taq (TaKaRa). The PCR program for all reactions was set as follows: pre-denaturation at 94°C for 4 min; 32 cycles of denaturation at 94°C for 1 min, annealing at 56–70°C for 1 min 30 s, and elongation at 72°C for 1 min 30 s; and 10 min at 72°C for the final elongation. RT-PCR products were purified and ligated with pMD18T simple vector (TaKaRa), and finally transformed into competent DH5α E. coli cells (TIANGEN BIOTECH, Beijing, China) according to a standard transformation method [61]. Three to five positive clones were sequenced by Sangon Biotech (Shanghai) Co., Ltd.
RT-qPCR Data Collection
Two-step RT-qPCR was used to obtain quantification cycle values (Cq, terminology from MIQE [62]) for all genes of all samples. First-stranded cDNA of each sample was synthesized according to the protocol of FastQuant RT Kit (with gDNase, TIANGEN BIOTECH). Genomic DNA (gDNA) in a 20 µl RT reaction system was first removed by adding 2 µl 5× gDNA Buffer, 1 µg total RNA, and appropriate RNase-Free ddH2O, and incubating at 42°C for 3 min. Then 2 µl 10× Fast RT Buffer, 2 µl FQ-RT Primer Mix (random hexamers and oligo-dT primers), 1 µl RT Enzyme Mix, and 5 µl RNase-Free ddH2O were added and the mixture held at 42°C for 15 min, then at 95°C for 3 min. The final cDNA was kept on ice or at −20°C for the qPCR. Subsequently, SYBR Green I dye-based qPCR was performed for all cDNA samples. Each 20 µl qPCR reaction system contained 6.4 µl RNase-free ddH2O, 10 µl 2× SuperReal PreMix Plus (SuperReal PreMix Plus kit [SYBR Green I, TIANGEN BIOTECH]), 0.6 µl forward primer (10 µM), 0.6 µl reverse primer (10 µM), 0.4 µl 50× ROX Reference Dye, 2 µl diluted cDNA (1∶30 diluted cDNA sample template) or ddH2O (as the no-template control). qPCRs of all genes were run on a 7500 real time PCR system (Applied Biosystems) with the same program: pre-denaturation at 95°C for 15 min, and 3-step amplification procedure of 40 cycles of 95°C for 10 s, 59.2°C for 32 s and 72°C for 32 s; followed by the melting curve stage of 95°C for 15 s, 60°C for 1 min, 95°C for 30 s and 60°C for 15 s. The primers used for the qPCR reactions are listed in Table 2. In each single run, the raw Cq values were automatically calculated by the 7500 real time PCR system detection software. Three replicate qPCRs were done for each cDNA dilution sample. The amplification efficiency (E) of each candidate reference gene and the endogenous genes were calculated using the Cq values for a 5-fold dilution series of the cDNA (1∶5, 1∶25, 1∶125, 1∶625, 1∶3,125, and 1∶15,625). For virus genes, E values were determined using a 2-fold dilution series of cDNA (1∶2, 1∶4, 1∶8, 1∶16, 1∶32, and 1∶64).
Sequence Analysis
The nucleotide sequences of all genes that were cloned from the cDNA were analyzed using the BLAST tool on the NCBI website (http://blast.ncbi.nlm.nih.gov/) to confirm the genes. Vector NTI Advance 10.3 package (Invitrogen) served as the alignment tool.
Algorithms and Statistical Analysis
Expression stability of 4 candidate reference genes in YDV-viruliferous R. padi after the AAPs was evaluated by three algorithms: NormFinder 0.953 [37], BestKeeper version1 [38], and GeNorm3.5 [25] respectively. Cq values were analyzed directly with BestKeeper, and then the stability of the HKGs was ordered according to standard deviation (SD) values. In addition, linear scale expression quantities (Q) were transformed from the Cq values by the delta-Cq method (ΔCq = min Cq − sample Cq; min Cq = lowest Cq value of the data set, sample Cq = Cq values of other samples) and corrected by the modified amplification efficiency (E′ = E +1) for the individual gene, according to the formula Q = E′ ΔCq, then analyzed by NormFinder and GeNorm.
To reach a reliable and unambiguous conclusion about qPCR results, we need two or more reference genes that are stably, i.e., consistently, expressed to normalize calculations [25], [62]. In addition to ranking the expression stability of a gene, GeNorm can also determine the optimal number of reference genes by pairwise variation, V, between two sequential normalization factors containing an increasing number of reference genes, whose default cut-off value of 0.15 has been accepted by many studies of finding reference genes [15], [44], [63]. When values are higher than 0.15, representing a large variation between the two sequential normalization factors, then another reference gene needs to be included to calculate a dependable normalization factor [25]. The pairwise variation starts from the first two and three (V2/3) most stable candidate reference genes, terminating with the addition of the most unstable one. As the number of reference genes increases, the best number (n) of reference genes for calculating the normalization factor will be decided, because the value of Vn/n +1 being first lower than 0.15, which means the variation in normalization factors from between n and n +1 reference genes is so small that an additional reference gene does not need to be included for calculating the normalization factor [25].
The normalization factors produced by GeNorm for the selected reference genes were used to calculate the relative virus titre and the expression value for the endogenous genes (as described in the GeNorm manual). To reach a persuasive conclusion, we used the best single reference gene (EF-1α for all experimental groups), the worst single reference gene (ACT1 for WYDV-GPV-wingless group, and 18S rRNA for the remaining 5 groups), a 4-reference gene combination (4re), 3-reference gene combination (3re), 2 best reference genes (2re) and 2 worst reference gene combination (bad2re) (according to Figure 2B) to normalize the expression levels for ago-1a and dcr1 in each group (using the serial formulas below ). The means among treatments were compared by a one-way ANOVA.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
All statistical analyses were done with the program SPSS version 19 (IBM SPSS Statistics 19 Core System, USA). Plots were graphed using SigmaPlot version 12.5 (Systat, San Jose, CA, USA).
Supporting Information
Total RNA extraction of oats and aphids. (A) Oat RNAs. H: healthy oat plants; BYDV-PAV: BYDV-PAV-infected oat plants; WYDV-GPV: WYDV-GPV-infected oat plants; BYDV-GAV-infected oat plants. (B) RNAs from mixed developmental stages of Rhopalosiphum padi; (C) RNAs from BYDV-PAV- winged or wingless adult after various feeding durations (h); (D) RNAs from WYDV-GPV- winged or wingless adult after various feeding durations (h); (E) RNAs from BYDV-GAV- winged or wingless adult after various feeding durations (h). M: DL2000 (TaKaRa).
(PDF)
RT-PCR amplification of candidate reference genes and target genes. (A) RT-PCR amplification of the CP and RTD genes from total RNAs of BYDV-PAV-, WYDV-GPV- and BYDV-GAV-infected oat plants in Figure S1 (A). H: healthy oat plants; W: ddH2O as no-template control; 1–4: four YDV-infected oat plants; (B) RT-PCR amplification of 4 candidate reference genes from total RNA in Figure S1 (B). Four lanes of each candidate gene indicated four RT-PCR products of 1/100 diluted RNA sample under different annealing temperature; (C) RT-PCR amplification of endogenous genes from total RNA in Figure S1 (B). Eight lanes of each gene indicated eight RT-PCR products of 1/10 (first four lanes) and 1/100 (last four lanes) diluted RNA sample under different annealing temperature. M: DL2000.
(PDF)
Melt curve and gel photo to check specificity and the size of RT-qPCR primer pair of each gene. NTC: no-template control; 0 h: 0 h feeding duration; selected samples: 12 randomly selected samples; M: DL2000.
(PDF)
Relative standard curves to determine amplification efficiencies of candidate reference genes and endogenous genes of the aphid Rhopalosiphum padi (diluted cDNA from virus-free winged and wingless adult aphids) and CP and RTD genes of YDVs (diluted cDNA from YDV-viruliferous winged and wingless aphids after a 12-h virus-feeding.
(PDF)
Result of best Blast search hit of 18S rRNA, ACT1, EF-1α, and GAPDH.
(PDF)
BestKeeper analysis of virus titre in YDV-viruliferous winged adults of Rhopalosiphum padi after different virus-feeding durations. n = 24: total number of samples used for analysis; Geo mean: geometric mean; Ar Mean: arithmetic mean; Min: minimun value of Cq; Max: maximum value of Cq; SD: standard deviation; CV: coefficient of variance.
(PDF)
BestKeeper analysis of virus titre in YDV-viruliferous wingless adults of Rhopalosiphum padi after different virus-feeding durations. n = 24: total number of samples used for analysis; Geo mean: geometric mean; Ar Mean: arithmetic mean; Min: minimun value of Cq; Max: maximum value of Cq; SD: standard deviation; CV: coefficient of variance.
(PDF)
Funding Statement
The Special Fund for Agro-scientific Research in the Public Interest (201303021); and the Natural Science Foundation of China (NSFC31272017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hesler LS (2005) Resistance to Rhopalosiphum padi L. (Homoptera: Aphididae) in three triticale accessions. J Econ Entomol 98: 603–610. [DOI] [PubMed] [Google Scholar]
- 2. Descamps LR, Sanchez C (2011) Population growth of Rhopalosiphum padi L. (Homoptera: Aphididae) on different cereal crops from the semiarid pampas of Argentina under laboratory conditions. Chilean Journal of Agricultural Research 71: 390–394. [Google Scholar]
- 3. Hadi BAR, Flanders KL, Bowen KL, Murphy JF, Blount AR (2012) Survey of Barley Yellow Dwarf Virus and Cereal Yellow Dwarf Virus on Three Perennial Pasture Grasses in Florida. Journal of Entomological Science 47: 35–43. [Google Scholar]
- 4. Krueger EN, Beckett RJ, Gray SM, Miller WA (2013) The complete nucleotide sequence of the genome of Barley yellow dwarf virus-RMV reveals it to be a new Polerovirus distantly related to other yellow dwarf viruses. Front Microbiol 4: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang W, Cheng Z, Xu L, Wu M, Waterhouse P, et al. (2009) The complete nucleotide sequence of the barley yellow dwarf GPV isolate from China shows that it is a new member of the genus Polerovirus. Arch Virol 154: 1125–1128. [DOI] [PubMed] [Google Scholar]
- 6.Wu BL, Blanchard-Letort A, Liu Y, Zhou GH, Wang XF, et al.. (2011) Dynamics of Molecular Evolution and Phylogeography of Barley yellow dwarf virus-PAV. Plos One 6. [DOI] [PMC free article] [PubMed]
- 7. Liu F, Wang X, Liu Y, Xie J, Gray SM, et al. (2007) A Chinese isolate of barley yellow dwarf virus-PAV represents a third distinct species within the PAV serotype. Arch Virol 152: 1365–1373. [DOI] [PubMed] [Google Scholar]
- 8.D’Arcy CJ, Domier LL (2005) Family Luteoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy 8th Report of the International Committee on Taxonomy of Viruses. San Diego, USA: Elsevier Academic Press. 891–900.
- 9. Gray S, Gildow FE (2003) Luteovirus-aphid interactions. Annu Rev Phytopathol 41: 539–566. [DOI] [PubMed] [Google Scholar]
- 10. Liu Y, Zhai H, Zhao K, Wu BL, Wang XF (2012) Two suppressors of RNA silencing encoded by cereal-infecting members of the family Luteoviridae. Journal of General Virology 93: 1825–1830. [DOI] [PubMed] [Google Scholar]
- 11. Peter KA, Gildow F, Palukaitis P, Gray SM (2009) The C Terminus of the Polerovirus P5 Readthrough Domain Limits Virus Infection to the Phloem. Journal of Virology 83: 5419–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gildow FE (1993) Evidence for receptor-mediated endocytosis regulating luteovirus aquisition by aphids. Phytopathology 83: 270–277. [Google Scholar]
- 13. Gildow FE, Gray SM (1993) The aphid salivary gland basal lamina as a selective barrier associated with vector-specific transmission of barley yellow dwarf luteoviruses. Phytopathology 83: 1293–1302. [Google Scholar]
- 14. Kliot A, Ghanim M (2013) The role of bacterial chaperones in the circulative transmission of plant viruses by insect vectors. Viruses 5: 1516–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jarosova J, Kundu JK (2010) Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol 10: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Balaji B, Bucholtz DB, Anderson JM (2003) Barley yellow dwarf virus and Cereal yellow dwarf virus Quantification by Real-Time Polymerase Chain Reaction in Resistant and Susceptible Plants. Phytopathology 93: 1386–1392. [DOI] [PubMed] [Google Scholar]
- 17. Cilia M, Tamborindeguy C, Fish T, Howe K, Thannhauser TW, et al. (2011) Genetics coupled to quantitative intact proteomics links heritable aphid and endosymbiont protein expression to circulative polerovirus transmission. J Virol 85: 2148–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seddas P, Boissinot S, Strub JM, Van Dorsselaer A, Van Regenmortel MH, et al. (2004) Rack-1, GAPDH3, and actin: proteins of Myzus persicae potentially involved in the transcytosis of beet western yellows virus particles in the aphid. Virology 325: 399–412. [DOI] [PubMed] [Google Scholar]
- 19. Wang XF, Zhou GH (2003) Identification of a protein associated with circulative transmission of Barley yellow dwarf virus from cereal aphids, Schizaphis graminum and Sitobion avenae. Chinese Science Bulletin 48: 2083–2087. [Google Scholar]
- 20. Yang XL, Thannhauser TW, Burrows M, Cox-Foster D, Gildow FE, et al. (2008) Coupling genetics and proteomics to identify aphid proteins associated with vector-specific transmission of polerovirus (Luteoviridae). Journal of Virology 82: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tamborindeguy C, Monsion B, Brault V, Hunnicutt L, Ju HJ, et al. (2010) A genomic analysis of transcytosis in the pea aphid, Acyrthosiphon pisum, a mechanism involved in virus transmission. Insect Molecular Biology 19 Suppl 2259–272. [DOI] [PubMed] [Google Scholar]
- 22. Zhang X, Wang XF, Zhou GH (2008) A one-step real time RT-PCR assay for quantifying rice stripe virus in rice and in the small brown planthopper (Laodelphax striatellus fallen). Journal of Virological Methods 151: 181–187. [DOI] [PubMed] [Google Scholar]
- 23. Liu WW, Zhao XJ, Zhang P, Mar TT, Liu Y, et al. (2013) A one step real-time RT-PCR assay for the quantitation of Wheat yellow mosaic virus (WYMV). Virology Journal 10: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, et al. (1999) Housekeeping genes as internal standards: use and limits. Journal of Biotechnology 75: 291–295. [DOI] [PubMed] [Google Scholar]
- 25. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eisenberg E, Levanon EY (2013) Human housekeeping genes, revisited. Trends in Genetics 29: 569–574. [DOI] [PubMed] [Google Scholar]
- 27. Winkles JA (2008) The TWEAK - Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nature Reviews Drug Discovery 7: 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rubie C, Kempf K, Hans J, Su TF, Tilton B, et al. (2005) Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Molecular and Cellular Probes 19: 101–109. [DOI] [PubMed] [Google Scholar]
- 29. Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al. (2000) The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- 30. You MS, Yue Z, He WY, Yang XH, Yang G, et al. (2013) A heterozygous moth genome provides insights into herbivory and detoxification. Nature Genetics 45: 220–225. [DOI] [PubMed] [Google Scholar]
- 31.Teng XL, Zhang Z, He GL, Yang LW, Li F (2012) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects. Journal of Insect Science 12. [DOI] [PMC free article] [PubMed]
- 32. Fu W, Xie W, Zhang Z, Wang SL, Wu QJ, et al. (2013) Exploring Valid Reference Genes for Quantitative Real-time PCR Analysis in Plutella xylostella (Lepidoptera: Plutellidae). International Journal of Biological Sciences 9: 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reim T, Thamm M, Rolke D, Blenau W, Scheiner R (2013) Suitability of three common reference genes for quantitative real-time PCR in honey bees. Apidologie 44: 342–350. [Google Scholar]
- 34. Bansal R, Mamidala P, Mian MAR, Mittapalli O, Michel AP (2012) Validation of Reference Genes for Gene Expression Studies in Aphis glycines (Hemiptera: Aphididae). Journal of Economic Entomology 105: 1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Canning ES, Penrose MJ, Barker I, Coates D (1996) Improved detection of barley yellow dwarf virus in single aphids using RT-PCR. J Virol Methods 56: 191–197. [DOI] [PubMed] [Google Scholar]
- 36. Ban L, Didon A, Jonsson LM, Glinwood R, Delp G (2007) An improved detection method for the Rhopalosiphum padi virus (RhPV) allows monitoring of its presence in aphids and movement within plants. J Virol Methods 142: 136–142. [DOI] [PubMed] [Google Scholar]
- 37. Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 38. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 39. Watanabe S, Greenwell AM, Bressan A (2013) Localization, concentration, and transmission efficiency of Banana bunchy top virus in four asexual lineages of Pentalonia aphids. Viruses 5: 758–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M, Zhou YW, Wang H, Jones HD, Gao Q, et al.. (2013) Identifying potential RNAi targets in grain aphid (Sitobion avenae F.) based on transcriptome profiling of its alimentary canal after feeding on wheat plants. BMC Genomics 14. [DOI] [PMC free article] [PubMed]
- 41.Bhatia V, Bhattacharya R, Uniyal PL, Singh R, Niranjan RS (2012) Host Generated siRNAs Attenuate Expression of Serine Protease Gene in Myzus persicae. Plos One 7. [DOI] [PMC free article] [PubMed]
- 42. Bansal R, Mian MAR, Mittapalli O, Michel AP (2012) Characterization of a Chitin Synthase Encoding Gene and Effect of Diflubenzuron in Soybean Aphid, Aphis Glycines. International Journal of Biological Sciences 8: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puinean AM, Foster SP, Oliphant L, Denholm I, Field LM, et al.. (2010) Amplification of a Cytochrome P450 Gene Is Associated with Resistance to Neonicotinoid Insecticides in the Aphid Myzus persicae. Plos Genetics 6. [DOI] [PMC free article] [PubMed]
- 44. Maroniche GA, Sagadin M, Mongelli VC, Truol GA, del Vas M (2011) Reference gene selection for gene expression studies using RT-qPCR in virus-infected planthoppers. Virol J 8: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Consortium TIAG (2010) Genome sequence of the pea aphid Acyrthosiphon pisum. Plos Biology 8: e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burrows ME, Caillaud MC, Smith DM, Benson EC, Gildow FE, et al. (2006) Genetic Regulation of Polerovirus and Luteovirus Transmission in the Aphid Schizaphis graminum. Phytopathology 96: 828–837. [DOI] [PubMed] [Google Scholar]
- 47. Liu SJ, Sivakumar S, Sparks WO, Miller WA, Bonning BC (2010) A peptide that binds the pea aphid gut impedes entry of Pea enation mosaic virus into the aphid hemocoel. Virology 401: 107–116. [DOI] [PubMed] [Google Scholar]
- 48. Jaubert-Possamai S, Rispe C, Tanguy S, Gordon K, Walsh T, et al. (2010) Expansion of the miRNA pathway in the hemipteran insect Acyrthosiphon pisum. Mol Biol Evol 27: 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ortiz-Rivas B, Jaubert-Possamai S, Tanguy S, Gauthier JP, Tagu D, et al. (2012) Evolutionary study of duplications of the miRNA machinery in aphids associated with striking rate acceleration and changes in expression profiles. BMC Evol Biol 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bansal R, Michel AP (2013) Core RNAi Machinery and Sid1, a Component for Systemic RNAi, in the Hemipteran Insect, Aphis glycines. International Journal of Molecular Sciences 14: 3786–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ma LM, Wang WJ, Liu CH, Yu HY, Wang ZG, et al. (2013) Selection of reference genes for reverse transcription quantitative real-time PCR normalization in black rockfish (Sebastes schlegeli). Marine Genomics 11: 67–73. [DOI] [PubMed] [Google Scholar]
- 52. Moran NA, Kaplan ME, Gelsey MJ, Murphy TG, Scholes EA (1999) Phylogenetics and evolution of the aphid genus Uroleucon based on mitochondrial and nuclear DNA sequences. Systematic Entomology 24: 85–93. [Google Scholar]
- 53. Normark BB (1999) Evolution in a putatively ancient asexual aphid lineage: recombination and rapid karyotype change (vol 53, pg 1458, 1999). Evolution 53: 2016–2016. [DOI] [PubMed] [Google Scholar]
- 54. Kim H, Lee S (2008) A molecular phylogeny of the tribe Aphidini (Insecta: Hemiptera: Aphididae) based on the mitochondrial tRNA/COII, 12S/16S and the nuclear EF1 alpha genes. Systematic Entomology 33: 711–721. [Google Scholar]
- 55. Bartel DP (2004) MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 116: 281–279. [DOI] [PubMed] [Google Scholar]
- 56. Bansal R, Michel AP (2013) Core RNAi Machinery and Sid1, a Component for Systemic RNAi, in the Hemipteran Insect, Aphis glycines. International Journal of Molecular Sciences 14: 3786–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hogenhout SA, Ammar el D, Whitfield AE, Redinbaugh MG (2008) Insect vector interactions with persistently transmitted viruses. Annual Review of Phytopathology, Vol 48 46: 327–359. [DOI] [PubMed] [Google Scholar]
- 58. Chay CA, Gunasinge UB, DineshKumar SP, Miller WA, Gray SM (1996) Aphid transmission and systemic plant infection determinants of barley yellow dwarf luteovirus-PAV are contained in the coat protein readthrough domain and 17-kDa protein, respectively. Virology 219: 57–65. [DOI] [PubMed] [Google Scholar]
- 59. Liu Y, Sun B, Wang X, Zheng C, Zhou G (2007) Three digoxigenin-labeled cDNA probes for specific detection of the natural population of Barley yellow dwarf viruses in China by dot-blot hybridization. J Virol Methods 145: 22–29. [DOI] [PubMed] [Google Scholar]
- 60. Du ZQ, Li L, Liu L, Wang XF, Zhou G (2007) Evaluation of aphid transmission abilities and vector transmission phenotypes of barley yellow dwarf viruses in China. Journal of Plant Pathology 89: 251–259. [Google Scholar]
- 61.Sambrook JF, Russell DW (2001) Molecular Cloning: A Laboratory Manual (3rd edition) Cold Spring Harbor Laboratory Press.
- 62. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622. [DOI] [PubMed] [Google Scholar]
- 63. Gopaulchan D, Lennon AM, Umaharan P (2013) Identification of reference genes for expression studies using quantitative RT-PCR in spathe tissue of Anthurium andraeanum (Hort.). Scientia Horticulturae 153: 1–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total RNA extraction of oats and aphids. (A) Oat RNAs. H: healthy oat plants; BYDV-PAV: BYDV-PAV-infected oat plants; WYDV-GPV: WYDV-GPV-infected oat plants; BYDV-GAV-infected oat plants. (B) RNAs from mixed developmental stages of Rhopalosiphum padi; (C) RNAs from BYDV-PAV- winged or wingless adult after various feeding durations (h); (D) RNAs from WYDV-GPV- winged or wingless adult after various feeding durations (h); (E) RNAs from BYDV-GAV- winged or wingless adult after various feeding durations (h). M: DL2000 (TaKaRa).
(PDF)
RT-PCR amplification of candidate reference genes and target genes. (A) RT-PCR amplification of the CP and RTD genes from total RNAs of BYDV-PAV-, WYDV-GPV- and BYDV-GAV-infected oat plants in Figure S1 (A). H: healthy oat plants; W: ddH2O as no-template control; 1–4: four YDV-infected oat plants; (B) RT-PCR amplification of 4 candidate reference genes from total RNA in Figure S1 (B). Four lanes of each candidate gene indicated four RT-PCR products of 1/100 diluted RNA sample under different annealing temperature; (C) RT-PCR amplification of endogenous genes from total RNA in Figure S1 (B). Eight lanes of each gene indicated eight RT-PCR products of 1/10 (first four lanes) and 1/100 (last four lanes) diluted RNA sample under different annealing temperature. M: DL2000.
(PDF)
Melt curve and gel photo to check specificity and the size of RT-qPCR primer pair of each gene. NTC: no-template control; 0 h: 0 h feeding duration; selected samples: 12 randomly selected samples; M: DL2000.
(PDF)
Relative standard curves to determine amplification efficiencies of candidate reference genes and endogenous genes of the aphid Rhopalosiphum padi (diluted cDNA from virus-free winged and wingless adult aphids) and CP and RTD genes of YDVs (diluted cDNA from YDV-viruliferous winged and wingless aphids after a 12-h virus-feeding.
(PDF)
Result of best Blast search hit of 18S rRNA, ACT1, EF-1α, and GAPDH.
(PDF)
BestKeeper analysis of virus titre in YDV-viruliferous winged adults of Rhopalosiphum padi after different virus-feeding durations. n = 24: total number of samples used for analysis; Geo mean: geometric mean; Ar Mean: arithmetic mean; Min: minimun value of Cq; Max: maximum value of Cq; SD: standard deviation; CV: coefficient of variance.
(PDF)
BestKeeper analysis of virus titre in YDV-viruliferous wingless adults of Rhopalosiphum padi after different virus-feeding durations. n = 24: total number of samples used for analysis; Geo mean: geometric mean; Ar Mean: arithmetic mean; Min: minimun value of Cq; Max: maximum value of Cq; SD: standard deviation; CV: coefficient of variance.
(PDF)