Abstract
DNA methylation is involved in tissue-specific and developmentally regulated gene expression. Here, we screened a novel methylation gene Sox30, whose methylation might contribute to its regulation and testis development in mice. Sox30 is a member of Sox transcription factors, and is considered to be involved in spermatogonial differentiation and spermatogenesis. However, the precise function and regulatory expression pattern remain unclear. In the present study, we found that Sox30 is highly expressed in adult testes but not in ovaries. Sox30 expression begins in early development, and in the testes, it is specifically increased coincidentally with development until adulthood. Moreover, Sox30 is expressed not only in testis germ cells, but also in sertoli cells. Sox30 is hypo-methylated in testis, epididymis and lung of adult mice, in which Sox30 is expressed. By contrast, Sox30 is hypermethylated in ovary, heart, brain, liver, kidney, spleen, pancreas, muscle, intestine, pituitary gland, blood and hippocampus of adult mice, in which the Sox30 is absent. Importantly, decreased methylation at CpG islands of Sox30 is observed in mouse developmental testes after birth, which is associated with enhanced Sox30 expression. However, the hypermethylated status of Sox30 is maintained in ovaries that does not express Sox30 during this period. Further, following demethylation treatment using 5-aza-dC, Sox30 expression is restored in GC2, TM3 and TM4 cell lines. This observation convincingly confirms that methylation really contributes to Sox30 silencing. In summary, we show that Sox30 expression is under the control of DNA methylation status, and this expression pattern is associated with testis development in mice.
Introduction
Epigenetic systems underlie the transcription factor networks, which establish gene expression in response to tissues and developmental stages [1]. In these systems, DNA methylation is one of the most major common players [2]. In mammals, formation of DNA methylation pattern is one of the important epigenetic events and is essential for development of various tissues [3]. Previous studies have revealed that DNA methylation pattern is specific to cell type, and is associated with cell differentiation [4], [5]. DNA methylation is always involved in various gene regulatory processes by silencing, switching and stabilizing genes, as well as remodeling chromatin [6]–[14].
The Sox family of transcription factors encode proteins that are characterized by a sequence-specific DNA binding HMG-box, and are a group of genes related to the mammalian testis determining factor, Sry. In general, the proteins containing an HMG domain with more than 50% similarity in the HMG domain of Sry are considered as Sox members. At present, about 20 different Sox genes have been identified in mammalian [15]. Accumulating evidence showed that Sox members play key roles in a wide variety of developmental processes, including sex determination and differentiation, testis development, male fertility maintenance and other respects [16], [17].
Sox30, a member of the Sox family, was firstly cloned from mouse (Mus musculus, location Chr 11) and human (Homo sapiens, location Chr 5), and then form Nile tilapia [18], [19]. In mouse and human, Sox30 is considered to be involved in spermatogonial differentiation and spermatogenesis [18], [20]. In the Nile tilapia, Sox30 plays roles in gonadal development [19]. However, up to now, the precise function and regulatory expression pattern of Sox30 have remained unknown.
In the present study, we first revealed that Sox30 was expressed both in mouse testis germ cells and sertoli cells, and the CpGs of Sox30 were specifically demethylated coincident with the spatiotemporal pattern expression found in mouse testis development. Further, re-expression of silenced Sox30 was found after treatment of cell lines with 5-aza-dc, a pharmacological inhibitor of DNA methylation, substantiating the role of methylation in the silencing of Sox30. The data demonstrate that DNA methylation plays key role in transcriptional regulation of Sox30 in testis development of mice.
Materials and Methods
Cell lines
The GC2 (Spermatocyte, GC-2spd), TM3 (Leydig cell) and TM4 (Sertoli cell) cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), cultured in DMEM or Ham's F12/DMEM (HyClone) supplemented with 10% fetal bovine serum or 5% horse serum/2.5% fetal bovine serum (FBS) (HyClone), and incubated in an atmosphere of 5% CO2 in air at 37°C.
Animals
BALB/c mice of both sexes were obtained from the animal center of Daping Hospital, Third Military Medical University (Chongqing, China). Mice were housed in cages, at a constant temperature (25°C), with a relative humidity of 50–60%, and a 12 h dark/light cycle. Food and water were given ad libitum. All experiments conformed to the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Committee of Laboratory Animal Experimentation at Third Military Medical University, China.
Sertoli and germ cells separation and purification
Sertoli cells were separated from 18 days old male mouse testes according to previous procedure [21]–[24]. Trypsin (Hyclone) and collagenase IV (Sigma) were used to digest the testis tissue for 30 min at 37°C separately. Dulbecco's Modified Eagle's Medium/Ham's Nutrient Mixture F12 (DMEM/F12, 1:1) medium (Hyclone) containing 10% FBS (Hyclone) was used to finish the digestion. The enzyme-digested product was sieved under 80 meshes. Primary separated cells were cultured in 6 centimeters diameter dishes (Corning) with DMEM/F12, 10% FBS and 1× Penicillin-Streptomycin (Beyotime, China) in a humidified atmosphere of 5% CO2 at 35°C. Germ cells (medium) were collected to another dish for culture [25]–[29]. Then adherent cells were treated with 20 mM Tris-HCl (pH 7.4) for 2 minutes after 48 hr of culture.
Genomic DNA extraction
DNA was extracted from the various tissues and cell lines with a Promega DNA Purification Wizard kit (Promega) according to the manufacturer's instructions. The samples were collected into lysis buffer with proteinase K, digested at 55°C overnight, and then incubated at 95°C for 10 min. The DNA was then precipitated with ethanol, resuspended in 50 µl deionized water and stored at −20°C until analysis.
Total RNA extraction
Total RNA was isolated from various tissues (mixed tissues from five individuals) and cell lines using Trizol reagent (Invitrogen) according to the manufacturer's instructions. The RNA (2.0 µg) was treated with DNase I to eliminatethe genomic DNA contamination. Then cDNAs were synthesized using M-MLV First Strand Kit (Invitrogen), and stored at −20°C.
Analysis of Sox30 expression by reverse transcription-polymerase chain reaction (RT-PCR)
RT-PCR was performed according to methods described previously [30]. The templates were set with Sox30 plasmid DNA and deionized water for positive and negative controls, respectively. The β-actin was amplified as internal control. A series of PCRs with different cycles (from 18 to 36, with an interval of 2) were performed to determine the linear phase using 1 µl cDNA. Based on these pilot experiments, 20 cycles for β-actin and 32 cycles for Sox30 were chosen. The primers used for RT-PCR are listed in Table 1.
Table 1. Sequence of primers used in the present study.
| Primer | Sequence | Purpose |
| M-Sox30F | GCCTCGTCCAGGGAAGCGAA | Expression analysis (RT-PCR) |
| M-Sox30R | CCGAAGGGAGCCTATGAATGTC | |
| MSox30F(M) | GGAGAGAGTTAGGTCGGAGTTATC | MSP analysis |
| MSox30R(M) | CGACTTCACTTACAACAAACGTC | |
| MSox30F(U) | TGGAGAGAGTTAGGTTGGAGTTATT | |
| MSox30R(U) | TTCCAACTTCACTTACAACAAACAT | |
| MSox30F(B) | TTTTTTTTATGGAGAGAGTTAGGT | BSP analysis |
| MSox30R(B) | AAAACAACAACAACACCTATTCC | |
| Sox30F | CCCATTCCACACTCACACGTCTA | Expression analysis (Real Time PCR) |
| Sox30R | AACCAAGACATTCTGGCATTGAACT | |
| β-actin-F | GGAGATTACTGCTCTGGCTCCTA | Internal control |
| β-actin-R | GACTCATCGTACTCCTGCTTGCTG |
Analysis of Sox30 expression by RT- quantitative PCR (RT-qPCR)
The cDNAs were prepared specific for RT-qPCR using PrimeScript RT reagent Kit With gDNA Eraser (Perfect Real Time) (Takara, RR047A). Real-time PCR was carried out using an iQ5 real-time detection system (Bio-Rad Laboratories) and SYBR Premix Ex TaqII (Perfect Real Time) (Takara, RR820A) according to manufacturer's instructions. The mRNA levels were normalized to β-actin and the 2−ΔΔt method was used to analyze the relative levels of expression. The primers used for RT-qPCR are listed in Table 1.
Western blotting (WB) analysis
Protein (80 micrograms) extracted from samples was run on 10% SDS–PAGE and transferred to PVDF membrane (Millipore). The membrane was blocked for 2 hr at room temperature, and incubated overnight at 4°C with primary antibody (Sox30; 1∶1000; Santa Cruz Biotechnology, sc-20104). The membranes were then washed, incubated with secondary antibody (1∶4000, Jackson ImmunoResearch Laboratories) and developed with SuperSignal West Pico Chemiluminescent substrate (Pierce). The same membrane was stripped and incubated with β-actin antibody (Sigma), serving as an internal control.
Immunohistochemistry (IHC) analysis
The paraffin-embedded tissue sections were baked at 60°C for 2 h, deparaffinized in xylene, rehydrated through graded alcohol to water, immersed in citrate buffer (pH 6.0), and antigen retrieval accomplished at 95°C for 15 min. The sections were blocked with 0.3% H2O2, treated with 10% normal goat serum for 15 min, and incubated at 4°C citrate buffer overnight with Sox30 antibody (1∶50; Santa Cruz Biotechnology). The reaction of antigen–antibody was visualized using streptavidin-horseradish peroxidase conjugated with diaminobenzidine after reaction with a biotinylated secondary antibody. The section incubated with PBS instead of primary antibody was used as a negative control.
Methylation analysis of Sox30 by methylation-specific polymerase chain reaction (MSP) and bisulfite sequencing PCR (BSP) sequencing
DNA samples were modified using EZ DNA Methylation-Gold Kit (Zymo Research, D5005). The MSP, BSP and sequencing were performed as previously reported [31]. Two pairs of MSP primers were designed (one pair in the promoter region and other pair in Exon 1 region), but only one pair of MSP primers in Exon 1 region was amplified successfully. Primers for MSP and BSP are listed in Table 1.
Treatment of cell lines with 5-aza-2-deoxycytidine (5-aza-dC)
To assess whether the expression of Sox30 was restored by 5-aza-dC treatment, demethylation experiments were performed. Briefly, GC2, TM3 and TM4 cells with Sox30 hypermethylation were exposed to 5-aza-dC (10 µM, Sigma) for three days, with media and drugs being replaced daily. The controls were treated in parallel with DMSO at each time point. DNA and total RNA were extracted, and the methylation status and mRNA expression of Sox30 were detected. The primer sequences used are shown in Table 1.
Statistical analysis
Statistical analyses were performed with the SPSS version 16.0 software. The results were expressed as the mean ± standard error (S.D). The differences between groups were analyzed by the Chi-square test and the Student's t-test. All p values are 2-sided, and p values less than 0.05 were taken as statistically significant.
Results
Sox30 was highly expressed in testes of adult mice
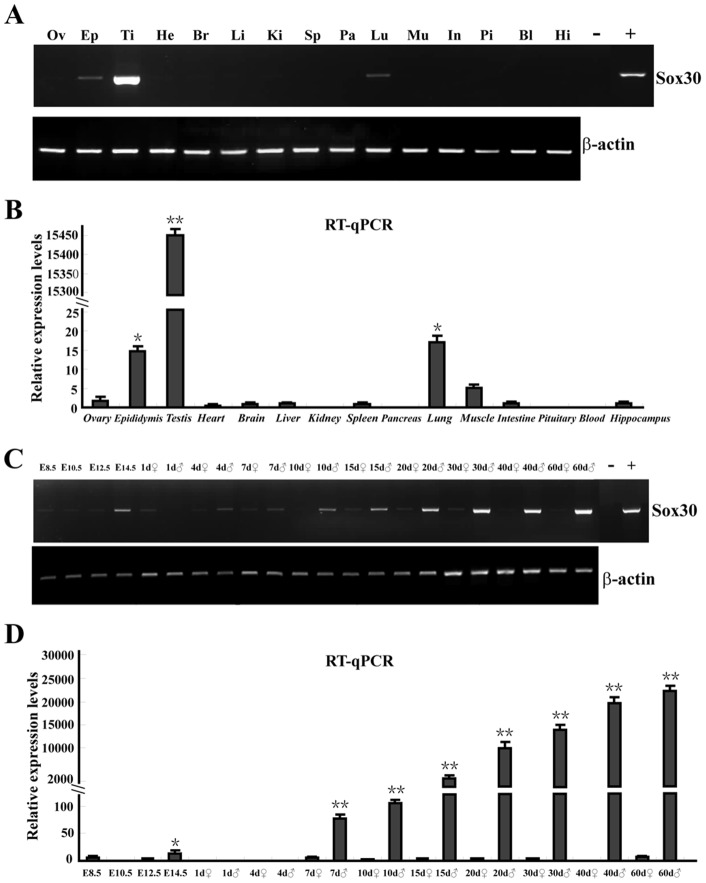
Tissue distribution analysis using RT-PCR and RT-qPCR revealed that Sox30 was expressed in adult testis, epididymis and lung, but was absent from ovary, heart, brain, liver, kidney, spleen, pancreas, muscle, intestine, pituitary, blood and hippocampus. The expression of Sox30 in testes was much higher (about 1000-fold) than that in epididymis and lung (Figure 1A, 1B).
Figure 1. Expression patterns of Sox30 in various adult tissues and developmental stages of the gonads.
(A) Expression analysis of Sox30 from various tissues of adults by RT-PCR. Ov, ovary; Ep, epididymis; Ti, testis; He, heart; Br, brain; Li, liver; Ki, kidney; Sp, spleen; Pa, pancreas; Lu, lung; Mu, muscle; In, intestine; Pi, pituitary; Bl, blood; Hi, hippocampus; −, negative control (deionized water); +, positive control (Sox30 plasmid DNA in place of the template); β-actin was used as the internal control. (B) Expression of Sox30 in various tissues of adults by RT-qPCR. Error bars indicate standard deviation (S.D). (n = 3); * <0.05; **<0.01. (C) Ontogeny of Sox30 expression in mouse gonads by RT-PCR. E8.5, embryonic day 8.5; 1d, 1 day post partum; ♀, female; ♂, male. (D) Ontogeny of Sox30 expression in mouse gonads by RT-qPCR. Error bars indicate S.D. (n = 3); * <0.05; **<0.01. Each sample of adult tissue and developmental stage gonad is a mix tissue from five individuals.
Sox30 was specifically increased coincidentally with testis development
The expression of Sox30 was detected in the whole embryos at E8.5 (embryonic day 8.5), E10.5, E12.5 and E14.5, and the gonads at 1d (1 day post partum), 4d, 7d, 10d, 15d, 20d, 30d, 40d and 60d of mice by RT-PCR, RT-qPCR and WB. Sox30 expression was detected at low levels in the whole embryos at E8.5, E10.5, E12.5 and gonads at 1d, while at relatively high level in the whole embryos at E14.5 (Figure 1C, 1D). Thereafter, Sox30 expression in the testes was specifically increased coincidentally with growing developmental stages until adult (4d, 7d, 10d, 15d, 20d, 30d, 40d and 60d), and a sharp increase of Sox30 expression was observed between 15d and 20d. However, the low expression status of Sox30 was maintained in ovaries during this period (Figure 1C, 1D and Figure 2A).
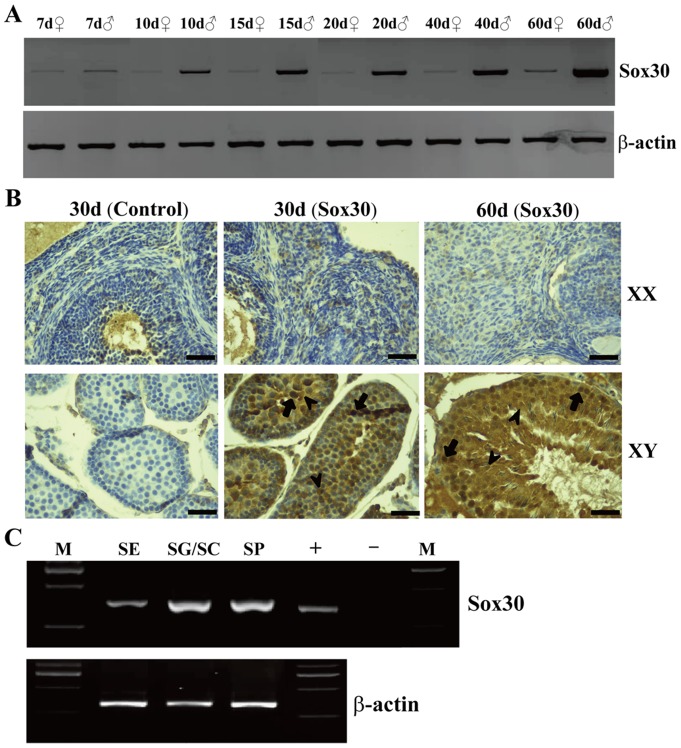
Figure 2. Expression of Sox30 in developmental gonads and cell types of testes.
(A) Ontogeny of Sox30 protein expression in mouse testes and ovaries (mix tissues from five individuals) by WB. 7d, 7 day post partum; ♀, female; ♂, male; β-actin was used as the internal control. (B) IHC analysis of Sox30 expression in gonads at 30d and 60d. XX, female; XY, male; Control, negative control (PBS in place of a primary antibody); Arrow, signals of Sox30; Scale bars are 50 µm. (C) Sox30 expression in germ cells and sertoli cells of male gonads by RT-PCR; SE, sertoli cells; SG, spermatogonia; PS, primary spermatocyte; SP, spermatid; −, negative control; +, positive control; M, marker; β-actin was used as the internal control.
Sox30 was expressed in germ cells and sertoli cells of mouse testes
To ascertain which population of cells expresses Sox30, IHC was performed using mouse gonads at 30d and 60d. Specific signals were observed in germ cells including spermatogonia and spermatocytes, and sertoli cells of the testes. However, no signal was detected in the ovaries (Figure 2B). To further confirm this result, germ cells and sertoli cells were separated and purified from 18 days old mouse testes. RT-PCR analysis showed that Sox30 was indeed expressed in germ cells (high level) and sertoli cells (lower level) of the testes in mice (Figure 2C).
The tissue-specific expression of Sox30 was associated with its methylation in adult mice
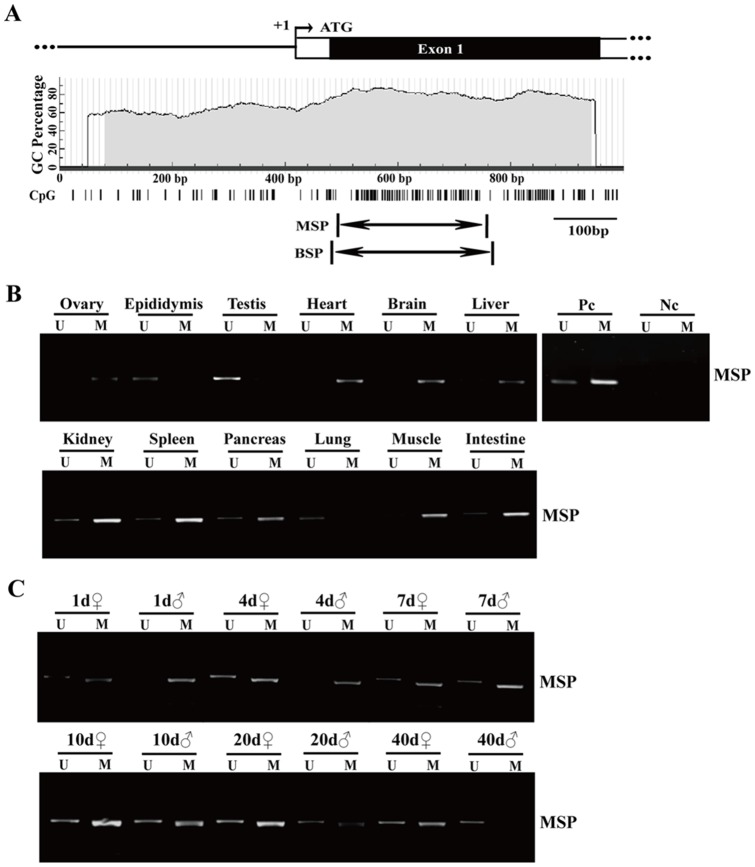
To determine whether tissue-specific expression of Sox30 was correlated with its methylation, the methylation patterns of Sox30 were detected. Pairs of MSP and BSP primers were showed in Figure 3A. The MSP analysis showed that hyper-methylation of Sox30 was detected in ovary, heart, brain, liver, kidney, spleen, pancreas, muscle, intestine, pituitary, blood and hippocampus, but was absent from the testis, epididymis and lung (Figure 3B). Consequently, Sox30 was silenced in the ovary, heart, brain, liver, kidney, spleen, pancreas, muscle, intestine, pituitary, blood and hippocampus, but was expressed in the testis, epididymis and lung (Figure 1A, 1B). These data suggested that Sox30 expression was likely associated with its methylation state in various tissues of adult mice.
Figure 3. Methylation status of Sox30 in various tissues and developmental gonads.
(A) Schematic representation of the mouse Sox30. Open boxes, non-coding regions; Closed boxes, coding regions; Arrow, the transcriptional start site (+1); ATG, start codon; Vertical bars, CpG sites; Arrows below the CpG sites, the regions subjected to MSP and BSP. (B) The methylation status was analyzed by MSP in various tissues of adult mice. U, unmethylated-specific primers; M, methylation-specific primers; Pc, positive control, including fully unmethylated control (blood genomic DNA sample) and fully methylated control (blood genomic DNA sample treated with SssI-methylase); Nc, negative control (deionized water). (C) The methylation analysis by MSP was performed in developmental gonads of mice. U, unmethylated-specific primers; M, methylation-specific primers; ♀, female; ♂, male.
The stage-specific expression of Sox30 was correlated with its methylation in developmental testes
To investigate whether the relationship between Sox30 methylation and its expression also existed in mouse developmental gonads, the differential and developmental stage-specific methylation of Sox30 was studied in gonads. Hypermethylation of Sox30 was maintained in ovaries from 1d to 40d. By contrast, decreased methylation was observed in testes from 1d to 40d (Figure 3C). Taking the ontogeny results into consideration, Sox30 expression was correlated to its methylation state in developmental gonads, and this decreased methylation in developmental testes was associated with enhanced expression of Sox30.
De-methylation treatment with 5-aza-dC restored Sox30 expression
To further confirm the methylation being associated with inhibition of Sox30 expression, we detected Sox30 mRNA level and methylation status in GC2, TM3 and TM4 cell lines that was incubated with/without 5-aza-dC (a DNA methylation inhibitor). Hypermethylation and decreased expression of Sox30 were detected in the three cell lines (Figure 4A, 4B). After 5-aza-dC treatment, methylated alleles of Sox30 were not found in any of the cell lines (Figure 4A). As expected, the cell lines, which initially showed hypermethylation of Sox30 and loss of its expression, were re-expressed Sox30 after 5-aza-dC treatment (Figure 4B, 4C). These data strongly suggested that methylation of Sox30 directly contributed to the regulation of Sox30 expression.
Figure 4. De-methylation analysis of Sox30 in cell lines from gonad.
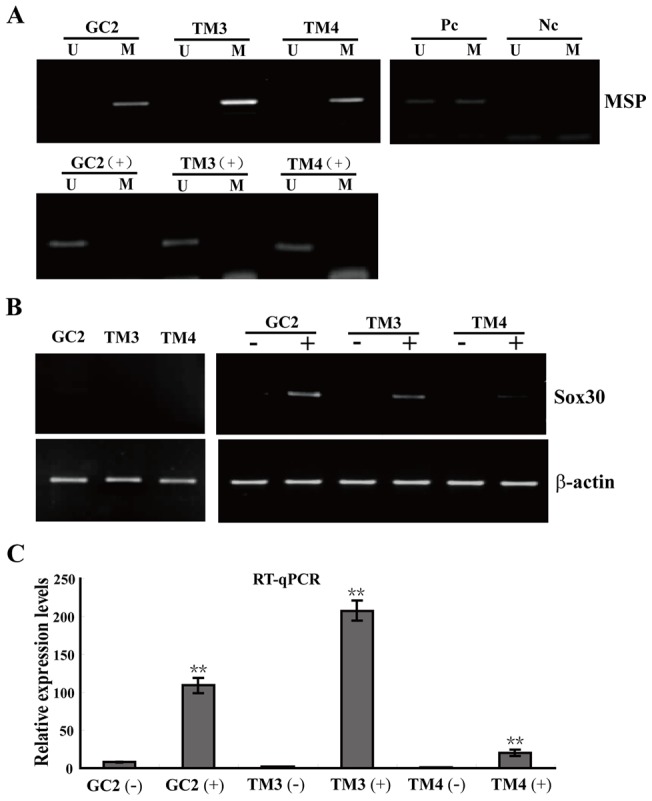
(A) The electrophoresis results of MSP products from GC2, TM3 and TM4 cells. U, unmethylated-specific primers; M, methylation-specific primers; Pc, positive control; Nc, negative control; (+), Cells treated with 5-aza-dC (10 µM). (B) Sox30 expression was analyzed in cell lines by RT-PCR. Sox30 expression significantly increased after treatment with 5-aza-dC; (+), treated with 5-aza-dC; (−), treated with DMSO and without 5-aza-dC; β-actin was used as an internal control; (C) The expression of Sox30 transcripts significantly increased after 5-aza-dC treatment as shown by RT-qPCR. Error bars indicate S.D. (n = 3).
The MSP results were confirmed by BSP sequencing
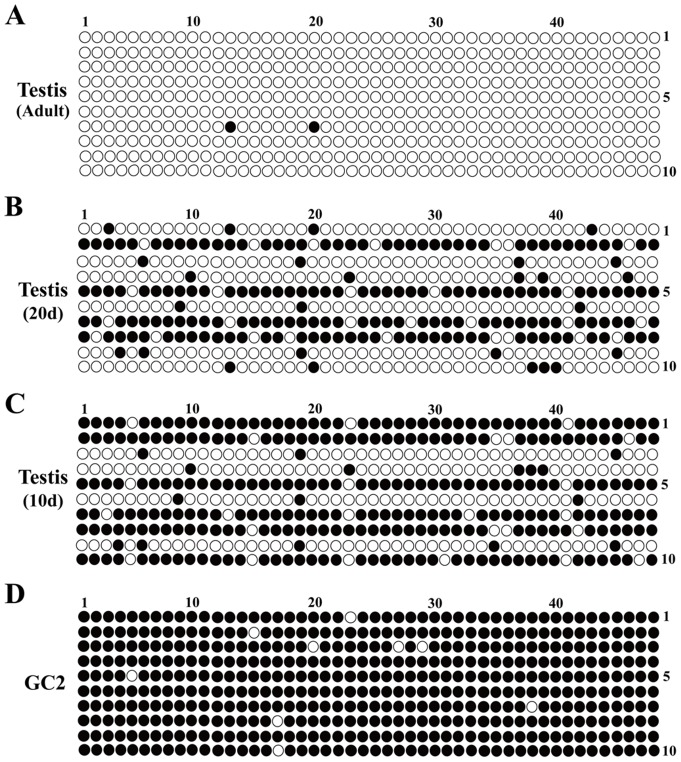
In order to make the results more reliable, the MSP results above were further validated by BSP sequencing. Amplified fragments were cloned into a specific vector, and 10 clones were sequenced for each amplification product. The sequencing results were consistent with those of MSP, in which dense methylation was found in methylated GC2 cells and testis tissues at 10d and 20d but not in unmethylated adult testis tissues (Figure 5A–5D). Moreover, decreased methylation level of Sox30 was observed in testes from 10d, 20d and adult mouse (60d) (Figure 5A–5C).
Figure 5. Methylation status of Sox30 was confirmed by BSP sequencing.
BSP sequencing of Sox30 was performed to confirm MSP results in 60d old adult testis (A), 20d old testis (B), 10d old testis (C) tissues and GC2 cells (D). Number of rows, number of clones; Number of columns, number of CpG; Solid circles, methylated CpG sites; Open circles, unmethylated CpG sites.
Discussion
Tissue distribution analysis showed that Sox30 was expressed in the testis, epididymis and lung of adult mice, and was highly expressed only in the testes. Moreover, the expression levels of Sox30 in the testes were nearly 1000-fold greater than that found in the epididymis and the lung. Thus, Sox30 was expressed exclusively in adult testes. However, high expression of Sox30 in extra-gonadal tissues in other developmental stages still cannot be completely excluded.
Previous reports showed that Sox30 was expressed in normal adult testis and specifically in germ cells of mice [18], [20]. In our present study, Sox30 expression was specifically increased coincident with the growing developmental stages until adult in the testes, and it was not only expressed in germ cells, but was also expressed in sertoli cells of mouse testes. These results indicated that Sox30 was involved in the testis development and spermatogenesis in mice. In particular, a sharp increase of Sox30 expression observed between 15d and 20d may be correlated with meiosis, which normally starts around 10d and continues thereafter in mice [32]. While the precise function of Sox30 remains obscure and should be revealed in a future study by gene knockout animal model.
DNA methylation plays key roles in many biological processes and is particularly important for development [32]. In mice, transcriptional activation of genes is always preceded by a decrease in DNA methylation as a result of demethylation [33], [34]. It is also known that mammalian tissues are heterogeneous in their methylation state, which is hypothesized to be the driving force of the orchestrated specific gene expression in response to tissue specification [35]. Thus, the methylation and demethylation dynamics is an important mechanism in regulating gene expression and development. In our study, it was observed that dynamic changes in the expression and methylation of Sox30 occurred in various tissues and gonads throughout mouse development. The changes in methylation were associated with opposite changes in the expression of Sox30, which indicated that hypermethylation might negatively contribute to regulation of Sox30 expression. Further, demethylation treatment with the reagent 5-aza-dc, a pharmacological inhibitor of DNA methylation, could restore the expression of Sox30 in GC2, TM3 and TM4 cells. The results indicated that hypermethylation of Sox30 indeed mediate its transcriptional silencing directly in mouse testes. In addition, the hypermethylated and hypomethylated Sox30 MSP products were both present in kidney, spleen, pancreas and intestine, which might show that both methylated and unmethylated primer-binding sites exist. This might be due to the defect of the MSP method, which is not quantitative.
Increasing evidence suggests that DNA methylation is rather “orderly” with a clear deterministic pattern throughout development. This orderly pattern is preserved over evolutionary time, and can be thought of as evolutionary “memory”, that represents a mechanism to maintain stable cellular identities [36]–[38]. On the other hand, the DNA methylation state proceeds over embryonic and postnatal development, and it is hypothesized as a driving force to direct life events, including shaping development, defining the stages of life, and determining cell fate for tissue specification [39]. Recently, new findings revealed that DNA methylation is not random, nor fixed, but is an orchestrated event during development [39]. It is spatiotemporally programmed in growing embryo, and impeding this program is found to disrupt developmental progression leading to defects [39]. Our study showed that the methylation and demethylation dynamics of Sox30 are unveiled in various adult tissues and testes at different developmental stages. This expression and DNA methylation patterns of Sox30 is likely to be an orchestrated event, which is essential for testis development in mice. However, it remains unknown whether or not impeding this program will disrupt developmental progression and subsequently leads to testis deficits in mice. Detailed studies will be required to answer this question.
Previous study had revealed that DNA hypermethylation is maintained until birth when germ cells resume their cycle during spermatogenesis, and the DNA methylation decreased along with successive divisions corresponding to demethylation after birth [32], [40]–[42]. In the present study, DNA methylation of Sox30 had decreased between 10d and adult testes, which may be due to the decreased expression and activity of Dnmt1 (a DNA methyltransferase) in mouse testes between 6d and the adult stage of development [43], [44]. Moreover, both de novo gene methylation and demethylation events also usually occur after completion of DNA replication during meiotic prophase [45]. These results suggested that DNA methylation of Sox30 was needed for spermatogenesis in mice. In addition, many testis-specific genes are demethylated and expressed in the testis, and are methylated in non-expressing somatic tissues [32]. This methylation and expression pattern is also suitable for Sox30.
Conclusions
Our data demonstrated that Sox30 expression was under the control of an epigenetic mechanism mediated by DNA methylation in testis development of mouse for the first time.
Acknowledgments
We thank MedSci for language editing.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are included in the manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Nos.81030052; 81172714). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yagi S, Hirabayashi K, Sato S, Li W, Takahashi Y, et al. (2008) DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res 18: 1969–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirabayashi K, Shiota K, Yagi S (2013) DNA methylation profile dynamics of tissue-dependent and differentially methylated regions during mouse brain development. BMC Genomics In press. [DOI] [PMC free article] [PubMed]
- 3. Shiota K, Yanagimachi R (2002) Epigenetics by DNA methylation for development of normal and cloned animals. Differentiation 69: 162–166. [DOI] [PubMed] [Google Scholar]
- 4. Shiota K, Kogo Y, Ohgane J, Imamura T, Urano A, et al. (2002) Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells 7: 961–969. [DOI] [PubMed] [Google Scholar]
- 5. Ohgane J, Hattori N, Oda M, Tanaka S, Shiota K (2002) Differentiation of trophoblast lineage is associated with DNA methylation and demethylation. Biochem Biophys Res Commun 290: 701–706. [DOI] [PubMed] [Google Scholar]
- 6. Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6: 597–610. [DOI] [PubMed] [Google Scholar]
- 7. Reik W, Dean W, Walter J (2001) Epigenetic reprogramming in mammalian development. Science 293: 1089–1093. [DOI] [PubMed] [Google Scholar]
- 8. Li E, Bestor TH, Jaenisch R (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926. [DOI] [PubMed] [Google Scholar]
- 9. Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, et al. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393: 386–389. [DOI] [PubMed] [Google Scholar]
- 10. Eden S, Hashimshony T, Keshet I, Cedar H, Thorne AW (1998) DNA methylation models histone acetylation. Nature 394: 842. [DOI] [PubMed] [Google Scholar]
- 11. Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. [DOI] [PubMed] [Google Scholar]
- 12. Cho JH, Kimura H, Minami T, Ohgane J, Hattori N, et al. (2001) DNA methylation regulates placental lactogen I gene expression. Endocrinology 142: 3389–3396. [DOI] [PubMed] [Google Scholar]
- 13. Imamura T, Ohgane J, Ito S, Ogawa T, Hattori N, et al. (2001) CpG island of rat sphingosine kinase-1 gene: tissue-dependent DNA methylation status and multiple alternative first exons. Genomics 76: 117–125. [DOI] [PubMed] [Google Scholar]
- 14. Li E (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 3: 662–673. [DOI] [PubMed] [Google Scholar]
- 15. Schepers GE, Teasdale RD, Koopman P (2002) Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell 3: 167–170. [DOI] [PubMed] [Google Scholar]
- 16. Jiang T, Hou CC, She ZY, Yang WX (2013) The SOX gene family: function and regulation in testis determination and male fertility maintenance. Mol Biol Rep 40: 2187–2194. [DOI] [PubMed] [Google Scholar]
- 17. Sarkar A, Hochedlinger K (2013) The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12: 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Osaki E, Nishina Y, Inazawa J, Copeland NG, Gilbert DJ, et al. (1999) Identification of a novel Sry-related gene and its germ cell-specific expression. Nucleic Acids Res 27: 2503–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han F, Wang Z, Wu F, Liu Z, Huang B, et al. (2010) Characterization, phylogeny, alternative splicing and expression of Sox30 gene. BMC Molecular Biology 11: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ballow D, Meistrich ML, Matzuk M, Rajkovic A (2006) Sohlh1 is essential for spermatogonial differentiation. Dev Biol 294: 161–167. [DOI] [PubMed] [Google Scholar]
- 21. Lydka M, Bilinska B, Cheng CY, Mruk DD (2012) Tumor necrosis factor alpha-mediated restructuring of the Sertoli cell barrier in vitro involves matrix metalloprotease 9 (MMP9), membrane-bound intercellular adhesion molecule-1 (ICAM-1) and the actin cytoskeleton. Spermatogenesis 2: 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicholls PK, Harrison CA, Gilchrist RB, Farnworth PG, Stanton PG (2009) Growth Differentiation Factor 9 Is a Germ Cell Regulator of Sertoli Cell Function. Endocrinology 150: 2481–2490. [DOI] [PubMed] [Google Scholar]
- 23. Shi Y, Song Y, Wang Y, Liang X, Hu Y, et al. (2009) p,p'-DDE induces apoptosis of rat Sertoli cells via a FasL-dependent pathway. J Biomed Biotechnol 2009: 181282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Su L, Cheng CY, Mruk DD (2010) Adjudin-mediated Sertoli-germ cell junction disassembly affects Sertoli cell barrier function in vitro and in vivo. Int J Biochem Cell Biol 42: 1864–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, et al. (2003) Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 69: 612–616. [DOI] [PubMed] [Google Scholar]
- 26. Izadyar F, Spierenberg GT, Creemers LB, den Ouden K, de Rooij DG (2002) Isolation and purification of type A spermatogonia from the bovine testis. Reproduction 124: 85–94. [PubMed] [Google Scholar]
- 27. Dirami G, Ravindranath N, Pursel V, Dym M (1999) Effects of stem cell factor and granulocyte macrophage-colony stimulating factor on survival of porcine typeA spermatogonia cultured in KSOM. Biol Reprod 61: 225–230. [DOI] [PubMed] [Google Scholar]
- 28. Hamra FK, Chapman KM, Nguyen DM, Williams-Stephens AA, Hammer RE, et al. (2005) Self renewal, expansion, and transfection of rat spermatogonial stem cells in culture. Proc Natl Acad Sci U S A 102: 17430–17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ogawa T, Ohmura M, Tamura Y, Kita K, Ohbo K, et al. (2004) Derivation and morphological characterization of mouse spermatogonial stem cell lines. Arch Histol Cytol 67: 297–306. [DOI] [PubMed] [Google Scholar]
- 30. Zhou LY, Wang DS, Kobayashi T, Yano A, Paul-Prasanth B, et al. (2007) A novel type of P450c17, lacking the lyase activity is responsible for C21-steroid biosynthesis in the fish ovary and head kidney. Endocrinology 148: 4282–4291. [DOI] [PubMed] [Google Scholar]
- 31. Liu WB, Han F, Jiang X, Yang LJ, Li YH, et al. (2012) ANKRD18A as a novel epigenetic regulation gene in lung cancer. Biochem Biophys Res Commun 429: 180–185. [DOI] [PubMed] [Google Scholar]
- 32. Marchal R, Chicheportiche A, Dutrillaux B, Bernardino-Sgherri J (2004) DNA methylation in mouse gametogenesis. Cytogenet Genome Res 105: 316–324. [DOI] [PubMed] [Google Scholar]
- 33. Oswald J, Engemann S, Lane N, Mayer W, Olek A, et al. (2000) Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10: 475–478. [DOI] [PubMed] [Google Scholar]
- 34. Mayer W, Niveleau A, Walter J, Fundele R, Haaf T (2000) Demethylation of the zygotic paternal genome. Nature 403: 501–502. [DOI] [PubMed] [Google Scholar]
- 35. Tawa R, Ono T, Kurishita A, Okada S, Hirose S (1999) Changes of DNA methylation level during pre- and postnatal periods in mice. Differentiation 45: 44–48. [DOI] [PubMed] [Google Scholar]
- 36. Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21. [DOI] [PubMed] [Google Scholar]
- 37. De Carvalho DD, You JS, Jones PA (2010) DNA methylation and cellular reprogramming. Trends Cell Biol 20: 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou FC (2012) DNA methylation program during development. Front Biol (Beijing) 7: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coffigny H, Bourgeois C, Ricoul M, Bernardino J, Vilain A, et al. (1999) Alterations of DNA methylation patterns in germ cells and Sertoli cells from developing mouse testis. Cytogenet Cell Genet 87: 175–181. [DOI] [PubMed] [Google Scholar]
- 41. Bernardino J, Lombard M, Niveleau A, Dutrillaux B (2000) Common methylation characteristics of sex chromosomes in somatic and germ cells from mouse, lemur and human. Chromosome Res 8: 513–525. [DOI] [PubMed] [Google Scholar]
- 42. Bernardino-Sgherri J, Chicheportiche A, Niveleau A, Dutrillaux B (2002) Unusual chromosome cleavage dynamic in rodent neonatal germ cells. Chromosoma 111: 341–347. [DOI] [PubMed] [Google Scholar]
- 43. Trasler JM, Alcivar AA, Hake LE, Bestor T, Hecht NB (1992) DNA methyltransferase is developmentally expressed in replicating and non-replicating male germ cells. Nucleic Acids Res 20: 2541–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benoit G, Trasler JM (1994) Developmental expression of DNA methyltransferase messenger ribonucleic acid, protein, and enzyme activity in the mouse testis. Biol Reprod 50: 1312–1319. [DOI] [PubMed] [Google Scholar]
- 45. Trasler JM, Hake LE, Johnson PA, Alcivar AA, Millette CF, et al. (1990) DNA methylation and demethylation events during meiotic prophase in the mouse testis. Mol Cell Biol 10: 1828–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are included in the manuscript.