Abstract
Glutathione S-transferases (GSTs) form a superfamily defined by their ability to catalyze the conjugation of glutathione with electrophilic substrates. These enzymes are proposed to play a critical role in protection of cellular components from damage mediated by reactive metabolites. Twenty-two cytosolic GSTs, grouped into seven families, are recognized in mice. This complexity hinders the assignment of function to a subset or family of these genes. We report generation of a mouse line in which the locus encoding three GST gene families is deleted. This includes the four Gstt genes spanning 65 kb on chromosome 10 and the seven Gstm genes found on a 150 kb segment of DNA chromosome 3. In addition, we delete two Gstp genes on chromosome 19 as well as a third related gene located 15 kb telomeric to Gstp1 and Gstp2, which we identify as a potential new member of this gene family. We show that, despite the loss of up to 75% of total GST activity in some tissues from these animals, the mice are healthy and fertile, with normal life expectancy. The normal development and health of these animals make them an appropriate model for defining the role of these families in redox homeostasis and metabolism of drugs and environmental pollutants.
Introduction
Glutathione S-transferases (GSTs) catalyze the conjugation of glutathione (GSH) with electrophilic molecules (Hayes et al., 2005). These enzymes facilitate this catalysis through proton extraction from GSH. Various GST isoforms have now been identified in a variety of complex organisms, with over 20 present in the mouse alone. The GST enzymes are generally grouped into one of three classes based on their subcellular localization: membrane-bound microsomal, mitochondrial, and cytoplasmic. Membrane-bound microsomal GSTs include enzymes such as PTGES (prostaglandin E synthase) in which glutathione transfer plays a critical role in the synthesis of prostaglandins (Trebino et al., 2003). Mammals express a single, evolutionarily divergent mitochondrial GST, which was presumed because of its mitochondrial localization to have a critical role in redox processes. However, mice lacking the gene encoding this mitochondrial GST are relatively healthy, with glomerular changes in males being the only pathology reported (Blackburn et al., 2011). The largest and most diverse group of GSTs encompasses those present in the cytosol. Cytosolic GSTs are further grouped into the following families based on substrate specificity and, more importantly, sequence similarity: alpha (A), mu (M), pi (P), theta (T), sigma (S), zeta (Z), and omega (O). The families vary in size from one to seven genes, and when there are multiple family members, the genes encoding these are linked in loci that suggest expansion from a primordial founding gene. Thus, for example, the seven mouse Gstm genes are located in a single locus on chromosome 1 (chr 1).
Significant progress has been made in understanding the function of some GST families, especially those consisting of a single enzyme. This progress has resulted from extensive biochemical studies as well as from studies using knockout mice in which individual GST genes have been inactivated by homologous recombination (Henderson et al., 1998; Engle et al., 2004; Lim et al., 2004; Fujimoto et al., 2006, 2007). For example, GSTZ plays an important role in the catabolism of aromatic amino acids, as it catalyzes the isomerization of maleylacetoacetate to fumarylacetoacetate. Generation of mice in which the gene encoding GSTZ1-1 had been inactivated by targeted recombination showed that this enzyme plays an important role in this pathway, as mice lacking this enzyme are very sensitive to any increase in tyrosine or phenylalanine in their diet (Lim et al., 2004). The high conservation between human and mice of the encoded protein (>80%) and the fact that a single GSTZ1 gene is present in both species allows extrapolation of the result from this experimental system to humans.
Unlike the GSTZ gene, however, many GSTs belonging to larger families display considerable interspecies variation in family size as well as in the expression pattern and substrate preferences of various family members. A role for the members of the various GST families in protection of the cell against endogenous metabolites has not been ascertained. Although the relative importance of the GST families in detoxification has not been established, a central role for these enzymes in this process is suggested by their unique capacity for conjugation of a tremendous variety of reactive intermediates. As phase II enzymes, the GSTs conjugate glutathione to xenobiotic electrophiles that are often produced by cytochrome P450-mediated reactions, increasing solubility and thereby facilitating export from cells or enzymatic degradation and subsequent secretion as mercapturic acid.
Mice lacking individual GST genes have provided some functional insight into the role of these enzymes in drug metabolism and risk from environmental carcinogens, especially in the case of GSTP (Henderson et al., 2000; Conklin et al., 2009). However, generating a panel of mice lacking each of the 22 cytosolic mouse GSTs to assess their role in development and normal physiology as well as biotransformation of drugs and xenobiotics would be an arduous undertaking. In addition, the documented variation between the human and mouse isoforms of these genes would call into question the relevance to human health of any experimental findings regarding the function of these GSTs.
As a first step in addressing these obstacles, we have generated mouse lines in which all members of the GSTT, GSTP, or GSTM family, respectively, are completely deleted. Each of these lines was generated from a corresponding embryonic stem (ES) cell line in which all members of a given GST family were excised in a single recombination event. Furthermore, we have interbred the three mouse lines carrying the individual GST family deletions to generate mice lacking these three gene families.
Materials and Methods
Generation of Mouse Lines.
Replacement type targeting vectors were constructed from the following 129 derived bacterial artificial chromosomes: Gstp: bMQ415B6, Gstt: bMQ379m08, Gstm: bMQ403h10, and bMQ75m22, Gstm1: bMQ386j24. The region of the mouse genome to be deleted was replaced with a phosphoglycerate kinase–driven neomycin resistance gene. 129S6-derived ES cells were cultured using standard methods, and DNA was introduced by electroporation (Mohn and Koller, 1995). Cells were selected in G418 (Geneticin) and were evaluated by polymerase chain reaction (PCR) and Southern blot analysis using methods previously detailed elsewhere (Koller et al., 1991). Correctly targeted cells were introduced into C57BL/6 blastocysts, which were returned to B6D2F1/J foster mothers to complete their development. All mice were maintained in specific pathogen-free housing in ventilated caging. The 129S6 mice were purchased from Taconic (Hudson, NY) and bred to chimeras to maintain the deleted loci on this genetic background. Heterozygous animals were intercrossed to obtain mice homozygous for the deletion as well as 129S6 mice. These animals were used to establish mutant breeding colonies and an in-house 129S6 colony. The mutant mice were age and sex matched at weaning and housed together until used in an experiment. All mice used in these studies were coisogenic for the locus under study. The mice were genotyped using the described primer sets (Supplemental Methods). All studies were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals as well as the Institutional Animal Care and Use Committee guidelines of the University of North Carolina–Chapel Hill.
Expression Analysis.
Total RNA was isolated from tissue using RNA isolation solvent (RNA-Bee; Tel-Test, Friendswood, TX) according to the manufacturer’s protocol. Northern blot analysis was conducted using a 32P-labeled probe prepared from the cDNA probes described in the legend for Fig. 3. Reverse transcription of RNA to cDNA for quantitative reverse transcription-polymerase chain reaction (RT-PCR) was performed using a high-capacity cDNA archive kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Primers and probes were purchased from Applied Biosystems. DNA was amplified with FastStart Universal Probe Master (Roche Applied Science, Indianapolis, IN). All samples were run in duplicate, and relative expression was determined by normalizing the samples to 18S RNA (ΔCT). To facilitate use of this highly expressed internal standard, samples were diluted to 10 pg/µl for analysis with this primer set, compared with a dilution of 2.5 ng/µl for all other probe sets. Data were analyzed using the comparative CT method as described by Applied Biosystems. In some experiments, samples from mutant animals were normalized to values obtained from cohoused control animals (ΔΔCT).
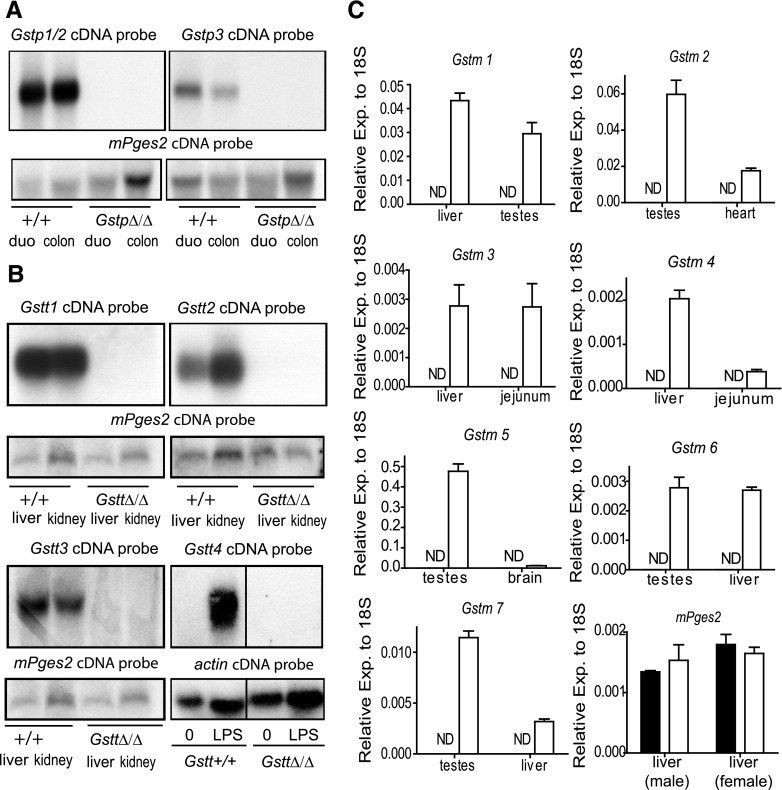
Fig. 3.
Expression of the Gstp, Gstm, and Gstt genes in the mice homozygous for the deleted loci. (A) RNA was prepared from the duodenum (duo) and colon of two GstpΔ/Δ mice and their controls and was analyzed by Northern blot using the a full-length cDNA probe. The Gstp1 cDNA does not discriminated between Gstp1 and Gstp2 expression because of the high level of homology between these genes (99.3% by Wilbur-Lipman DNA alignment). As expected, a strong signal was observed in the lanes corresponding to wild-type mice but not in the lanes loaded with RNA from GstpΔ/Δ animals. A band is also detected in wild-type RNA from these tissues when analyzed with a full-length cDNA probe corresponding to Gstp3. Although GSTP3 has 71.4% (Lipman-Pearson protein alignment) homology at the protein level with GSTP1/2, this cDNA probe has only 65% similarity index (Wilbur-Lipman DNA alignment) with Gstp1 and Gstp2 and thus will not hybridize to these transcripts in the conditions used in this study. Consistent with a deletion that encompasses this telomeric Gstp gene, this band is absent in lanes corresponding to the GstpΔ/Δ mice. To verify the quality and quantity of the bound RNA, filters were probed with mPges2, a gene that is expressed at moderate levels throughout the intestinal tract. (B) RNA was isolated from liver, kidney, and bone marrow derived macrophages of wild-type and GsttΔ/Δ mice. Northern blot analysis using cDNA probes specific for Gstt1 and Gstt2 under stringent hybridization conditions showed high expression of both genes in these tissues. Gstt3 expression, detected using a full-length cDNA, was also observed in both liver and kidney, although longer exposure of the film was required. No signal was observed using the three Gstt probes in RNA prepared from GsttΔ/Δ mice. Gstt4 is expressed at low levels in most tissues, including bone marrow-derived macrophages. However, we identified robust expression of this gene after treatment of macrophages for 16 hours with LPS using a full-length cDNA probe specific for this gene. No signal was observed in lanes corresponding to RNA isolated from LPS-treated cells derived from GsttΔ/Δ mice. (C) Total RNA was prepared from the indicated tissues of GstmΔ/Δ mice and cohoused sex- and age-matched controls. Expression of the Gstm gene indicated in each of the panels from tissues known to express each family member was determined by qPCR using gene-specific primers obtained from Applied Biosciences. Expression levels were normalized to 18S RNA. Data were analyzed using the comparative CT method (ΔCT) as described by Applied Biosystems. No signal (ND: not detected) was observed in all cases in samples prepared from mice homozygous for the GstmΔ/Δ locus. Values represent mean of three animals ± S.E.M.
Hematologic and Clinical Chemistry.
Blood samples were collected in EDTA (5 mM) from deeply anesthetized animals by cardiac puncture and analyzed using Heska’s animal blood counter (Heska, Loveland, CO). Clinical chemistry was performed on plasma prepared from the blood using an Automatic Chemical Analyzer (V350; Johnson & Johnson, New Brunswick, NJ).
Fluorescence-Activated Cell Sorting Analysis.
Cells from whole blood (white blood cells), spleen, thymus, and lymph nodes isolated from ΔPMT or control mice 6–8 weeks of age were stained with markers specific for T cells (CD3, CD4, CD8), B cells (B220), myeloid cells (CD11, GR-1). Fluorescence-activated cell sorting (FACS) was performed using CyAn ADP Analyzer (Beckman Coulter, Brea, CA) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Measurement of GST Activities in Mouse Tissues.
Mice were transcardially perfused with phosphate-buffered saline (PBS), pH 7.4, to remove red blood cells from the tissue. 150 mg of tissue was homogenized in 1 ml of cold buffer (100 mM potassium phosphate, pH 7.0, 2 mM EDTA) and centrifuged at 10,000g for 15 minutes at 4°C to enrich for cytosolic and microsomal GSTs. To further enrich for cytoplasmic GSTs, the supernatant was transferred to new tubes and centrifuged at 10,000g for 60 minutes at 4°C. GST activity in the supernatant was then determined spectrophotometrically as previously described (Habig et al., 1974). Substrates used in this analysis include ethacrynic acid (TCI, Portland, OR); 1,2-epoxy-3-(4-nitrophenoxy) propane (EPNP), (Oakwoods Chemical, West Columbia, SC); 1,2-dichloro-4 nitrobenzene (DCNB) (Sigma-Aldrich, St. Louis, MO); 1-chloro-2,4-dinitrobenzene (CDNB) (Sigma-Aldrich).
Statistical Analysis.
The method used for the evaluation of data are detailed in each figure legend.
Results
General Strategy.
To create a more streamlined system for the analysis of GST function, we have generated a mouse line lacking all members of the Gstm, Gstt, and Gstp gene families. Because the members of each of these families form a cluster on a different mouse chromosome, each family was deleted separately in mouse ES cells by a single targeted recombination event. The ES cell lines carrying the deleted GST loci were used to generate three mouse lines, each carrying a deletion of one GST gene family. These lines were in turn interbred to produce the final line lacking all three gene families. Each deletion removes all coding sequences as well as regulatory elements that might modulate expression of genes introduced into the loci in the future (Fig. 1). By performing assays of GST function in the mouse line carrying all three deletions, it should be possible to determine relatively quickly whether any of the members of these families contributes to the function being assayed. Once a functional deficit is identified in this mouse line, it should be possible to identify the cause of the deficit more narrowly by repeating the assay in each of the lines lacking a single GST family. Because the structures of the deleted loci that we have created are amenable to restoring function by insertion of single or multiple GST genes, it should be possible in the future to engineer mouse lines for pinpointing the GSTs responsible for particular functions. In addition, restoration of function using human GST genes should provide a system for performing functional assays that are more relevant to human physiology.
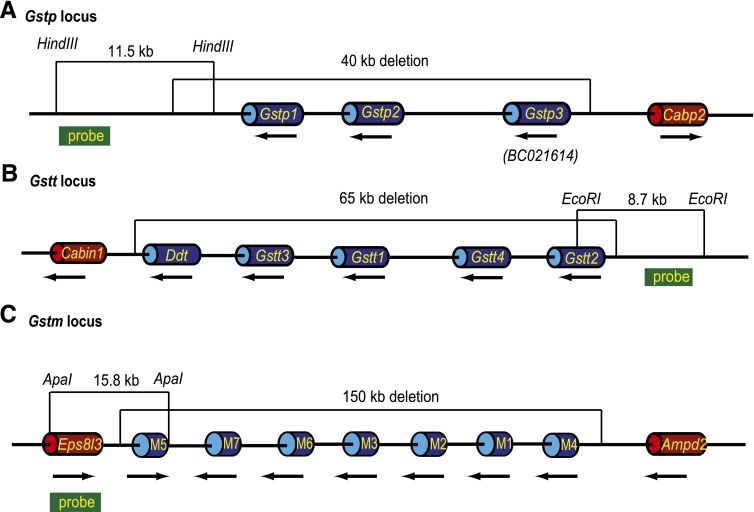
Fig. 1.
Schematic for generation of mice lacking the mouse Gstp, Gstt, and Gstm loci. All loci are shown with the proximal (centromere) on the left and distal (telomere) to the right. Genes are shown as cylinders, and the 5′ to 3′ orientation of each is indicated by a black arrow below. Intergenetic DNA is shown as a thin black line. Distances are approximately proportional but not to scale. The region deleted is indicated above each locus. Genes in the deleted region are shown as blue cylinders; genes outside the deleted region are shown as red cylinders. The restriction fragment and the probe used for initial verification of the targeting event by Southern blot analysis are shown for each locus. (A) The mouse Gstp locus extends over ~40 kb and is located between the Cabp2 and Ndufv1 (not shown) genes on chr 19. It includes the two well-characterized and highly homologous (91.4% protein homology) genes Gstp1 and Gstp2. Located telomeric to Gstp2 we identified an additional GST-like gene with the current designation BC021614, which we here refer to as Gstp3 based on sequence analysis. The deletion at this locus encompasses chr19:4,028,404–4,067,695 and includes not only the coding regions of the three genes but also approximately 8.5 kb of DNA 5′ of the mouse Gstp3 and about 7 kb of DNA 3′ of the Gstp1. (B) The mouse Gstt locus extends over ~65 kb on chr 10 between Ddt and Mif (not shown) and includes Gstt1, Gstt2, Gstt3, and Gstt4. Because of the close proximity of Ddt to Gstt3 and the possibility that they share regulatory elements, the gene was included in the Gstt deletion, which extends from chr10:75,234,558–75,299,968. (C) Seven Gstm genes are located in a ~150 kb segment extending from the Eps8l3 gene to the Ampd2 on mouse chr 3. Removal of the entire family of genes and relevant 5′ and 3′ regions required deletion of chr3:107,697,426–107,849,488.
Generation of Mice Lacking the Gstp Locus.
The two mouse Gstp genes, Gstp1 and Gstp2, which lie in close proximity (~2.4 kb) to one another on chr 19 between Cabp2 and Ndufv1, have been characterized. However, during the design of a vector to remove these two genes and the upstream regulatory elements, we identified an additional GST-like gene in this region, designated BC021614 (Fig. 1A). Sequence analysis shows that this gene is most closely related to the mouse and human Gstp genes. A blast search of the human genome with the sequences from BC021614 identifies GSTP1 as the only human ortholog. Comparison of the protein structure encoded by this sequence to mouse and human GSTP further supports the contention that BC021614 represents a previously unrecognized mouse Gstp gene.
We aligned the known GSTP protein sequences and examined that extrapolated from the sequence of BC021614 for the presence of residues conserved between species and/or shown, using a mutagenesis approach, to play a role in enzyme activity (Fig. 2A) (discussed in Vasieva, 2011). Tyr 8 is critical for many aspects of GSTP function including folding and structural mobility, interactions with other proteins, substrate binding, and catalysis (Oakley et al., 1998; Pettigrew et al., 1999). Trp 39, Lys 45, and Gln 52 contribute to the GSH binding site (Oakley et al., 1998). Cys 48 and Tyr 80 are involved in binding and regulatory activities such as heterodimerization (Ralat et al., 2006). Chemical modification studies of porcine lung pi class GST suggest that Tyr 109 may be located near or at the binding site (Pettigrew et al., 1999), while Tyr 154 has been shown to contribute to the hydrophobic staple motif (Stenberg et al., 2000b). Tyr 50 is a key residue at the dimer interface and has been implicated in the cooperativity of the enzyme (Stenberg et al., 2000a). All these residues are conserved in mGSTP1, mGSTP2, hGSTP1, and the predicted protein encoded by BC021614 except for Tyr 50, which is replaced by Phe in the BC021614 protein. Interestingly, this substitution has been shown in human GSTP1 to result in higher thermal stability and catalytic efficiency (Stenberg et al., 2000a). BC021614 is an expressed gene, with transcripts identified in a number of tissues including the liver. In contrast with the conservation of functionally important amino acids among the three mouse GSTP proteins, expression of BC021614 does not display the well-characterized sexual dimorphism of Gstp1 and Gstp2. Unlike the expression of these genes, expression of BC021614 is similar in the male and female liver in both young and sexually mature animals (Fig. 2B).
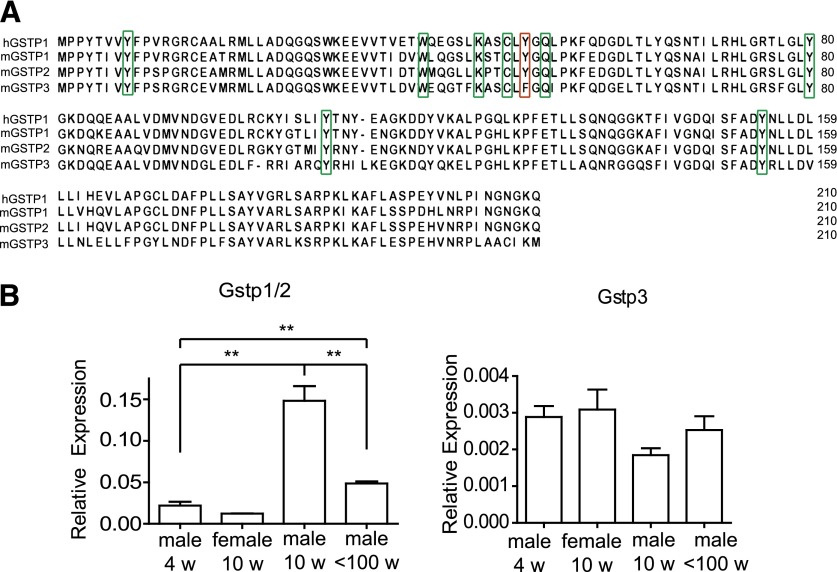
Fig. 2.
Comparison of human GSTP1 and mouse GSTP1 and GSTP2 with BC021614 (mGSTP3). (A) The primary structure of human GSTP1 is compared with mouse GSTP1 and GSTP2 and the deduced primary structure of the protein encoded by BC021614 (GSTP3). Conserved residues that have been implicated in GSTP function are marked by green rectangles. Tyr 50, a residue at the dimer interface implicated in the cooperativity of the enzyme, is marked with a red rectangle. This residue is replaced by Phe in GSTP3, a substitution that has been shown in human GSTP1 to result in higher thermal stability and catalytic efficiency. Numbering includes the AUG encoded methionine as recommended by the human genome variation society (www.hgvs.org). (B) Comparison of expression of BC021614 (mGstp3) and Gstp1/2 in liver of male and female mice. mRNA was isolated from liver of 4-week-old male and female mice, sexually mature male mice (10 weeks), and males that were no longer fertile (100 weeks), and the expression of Gstp1/2 and Gstp3 was compared. The qPCR probes do not distinguish between transcripts originating from Gstp1 and Gstp2. Expression was normalized to that of 18s RNA. Expression of Gstp1/2 is significantly higher in the 10-week-old males compared with all other groups. In comparison, no statistically significant increase is seen in Gstp3 expression between males of different ages (n = 3 for all groups). **P < 0.05 determined by unpaired t test.
Together, our findings suggest that it is likely that BC021614 represents an additional member of the mouse GST pi family. Although clearly assignment of BC021614 to the Gstp gene family awaits demonstration that the encoded protein catalyzes the conjugation of glutathione with electrophilic substrates, for simplicity, we refer to this gene as Gstp3 throughout this study. Importantly, this analysis indicates that generation of a mouse lacking all GSTP activity might require removal of this gene, which lies approximately 15 kb telomeric to the Gstp1/2 genes. Thus, a targeting vector was designed that removes ~40 kb of DNA. This includes the three Gstp genes and intergenic DNA as well as ~8.5 kb of DNA upstream of Gstp3 and 7 kb of DNA downstream of Gstp1 that may be involved in regulation of the locus. ES cells in which this targeting construct integrated by homologous recombination were identified by PCR analysis and verified by Southern blot analysis (Supplemental Fig. 1A). Mice homozygous for the deleted locus (GstpΔ/Δ) were generated, and DNA and RNA were analyzed to confirm the loss of the segment of DNA carrying the three genes. DNA was probed with full-length cDNA probes corresponding to Gstp1 and Gstp3. No binding of either probe was observed in DNA obtained from the GstpΔ/Δ mice (Supplemental Fig. 2A). These cDNA probes were also used to confirm loss of expression of the genes. As expected, a Gstp1/2-specific cDNA probe hybridized to RNA prepared from the duodenum and colon of wild-type mice. The high homology between these two genes (99.3% by Wilbur-Lipman DNA alignment) does not allow generation of a probe that distinguishes between the two transcripts. Gstp3 expression was also detected by Northern blot analysis of total RNA prepared from the gut by hybridization with a full-length cDNA probe corresponding to Gstp3. Neither probe detected Gstp-derived transcripts in RNA prepared from GstpΔ/Δ mice (Fig. 3A).
Generation of Mice Lacking the Gstt Locus.
The theta locus on mouse chr 10 contains four Gstt genes as well as Ddt (d-dopachrome tautomerase) clustered between Cabin1 (calcineurin-binding protein 1) and Mif (macrophage migration inhibitory factor). Ddt is located in close proximity to Gstt3 (Fig. 1B). Because of the possible overlap in the regulatory elements controlling Ddt and Gstt3, we included this gene in the deletion, thereby removing mouse genes between Cabin1 and Mif. Targeted ES cells were identified by PCR and Southern blot and were used to generate a mouse line homozygous for the deleted Gstt locus (GsttΔ/Δ), (Supplemental Fig. 1B). The correct deletion of the locus was verified by analysis of DNA and RNA from homozygous GsttΔ/Δ mice. Analysis of DNA with a number of probes unique to the 65 kb Gstt locus failed to bind to DNA prepared from these animals (Supplemental Fig. 2B). RNA was prepared from liver and kidney and was analyzed with cDNA probes specific for Gstt1, Gstt2, and Gstt3. Gstt4 is expressed at low levels in most tissue. To verify deletion of this gene, we generated macrophages from bone marrow. We found that Gstt4 was induced after stimulation with lipopolysaccharide (LPS) at sufficient levels to allow detection by Northern blot. No expression was observed in cells derived from GsttΔ/Δ animals (Fig. 3B).
Generation of Mice Lacking the Entire Gstm Locus.
The mouse Gstm locus spans 150 kb and contains seven Gstm genes (Fig. 1C). Targeted ES cells were identified by PCR and Southern blot analysis (Supplemental Fig. 1C), and these cells were used to generate a GstmΔ/Δ mouse line. DNA and RNA from these mice were used to verify the deletion of the locus, including all seven Gstm genes. In this case, because of the complexity and size of the deletion, we generated a probe that would allow a comprehensive evaluation of the deletion by Southern blot analysis (Fig. 4). DNA from the homozygous GstmΔ/Δ mice was digested with a number of enzymes and subjected to Southern blot analysis using a probe corresponding to the region of DNA extending from the middle of exon 3 to just downstream of exon 4 of Gstm6. Genome analysis showed that this fragment of DNA shares sufficient homology to allow detection by Southern blot analysis of eight regions in the Gstm gene cluster, with only minimum homology to four other regions in the mouse genome (Supplemental Table 1). An example of evaluation of such an analysis is shown (Fig. 4).
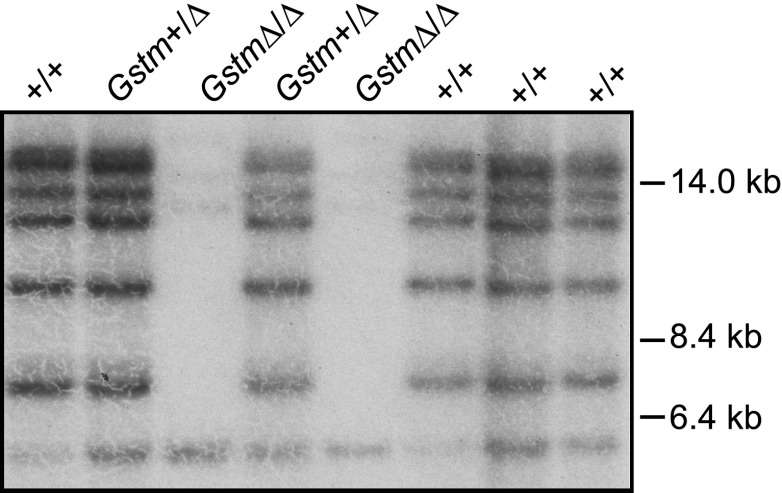
Fig. 4.
Verification of deletion of the seven mouse Gstm genes by Southern blot analysis. DNA was prepared from tail biopsy samples obtained from pups generated by the intercross of GstmΔ/+ mice. DNA was digested with BglI and analyzed by Southern blot using a probe that binds to multiple fragments of carrying Gstm genes located on chr 1. These bands were not observed in DNA obtained from pups Gstm Δ/Δ. The few bands remaining correspond to cross-reacting loci present on other chromosomes. For example, the 5.5 kb fragments observed in all lanes correspond to a segment of DNA on chr 10. See Supplemental Table 1 for assigning all fragments to chromosome and locus.
The hybridization pattern of the probe with DNA from two of the eight pups derived from the intercross of Gstm+/Δ mice is consistent with the absence of all the Gstm genes in these animals. We also verified the loss of expression of each of the seven genes. Based both on our own studies and published reports, we identified tissues with the highest expression for each of the seven genes. RNA was prepared from these tissues from wild-type mice as well as mice homozygous for the GstmΔ/Δ locus and was analyzed by quantitative PCR (qPCR). No expression could be detected for any of these genes, even in tissue in which high levels can be observed in wild-type animals (Fig. 3C).
As an additional control for future studies assigning function to various Gstm genes, we generated a mouse line lacking only Gstm1. We chose to generate a line lacking this individual gene because our expression analysis as well as published studies (Knight et al., 2007) indicated that expression of this gene dominates in many tissues, and thus direct comparison of the GstmΔ/Δ mice and Gstm1−/− mice would be important in future studies assigning function to other members of this large gene family. The 18.6 kb segment of the Gstm locus encoding GSTM1 was deleted, and loss of the locus and lack of expression of Gstm1 were verified by Southern blot and qPCR analysis of homozygous Gstm1−/− animals (Supplemental Fig. 3). The deletion includes the entire gene as well as promoter elements, thus mimicking the deletion event that resulted in the null GSTM1*0 allele present in humans. We verified that this deletion does not result in loss of expression of the neighboring two genes, particularly Gstm2 and Gstm4 (data not shown).
Generation of Mice Lacking Gstp, Gstt, and Gstm Genes.
The null allele for each of the three loci was maintained on the 129S6 genetic background by mating the chimeras generated from the three ΔES cell lines directly with 129S6 female mice. This makes it possible to generate coisogenic 129 mice in which the only genetic difference between experimental animals and control animals is the absence of these three loci. All DNA flanking these mutations will be of 129S6 origin. We first generated a mouse line lacking both the Gstt and Gstp loci. This line was then intercrossed with the GstmΔ/Δ mice to generate the mice lacking all three gene families. These GstpΔ/Δ/GstmΔ/Δ/GsttΔ/Δ,(ΔPMT), mice appeared normal and could not be visually distinguished from 129S6 mice. The weight of the three mouse lines did not differ significantly when examined through the first 8 weeks from control 129S6 animals (Supplemental Table 2).
Morphologic evaluation of tissues showed no gross abnormalities, including in size of organs, nor did histologic evaluation reveal an overt impact of loss of these 14 genes on development (data not shown). For example, histologic evaluation of liver, lung, and kidneys indicated normal development of these organs, and changes indicating increased cell death and/or inflammation were not observed. Staining with Oil Red revealed no fatty changes to the liver; an increase in fibrosis, evaluated by staining sections with Masson’s trichrome, was not observed in the ΔPMT animals. Changes consistent with ongoing activation of the immune system were not apparent on evaluation of the leukocyte composition of the blood, thymus, spleen, and lymph nodes (Supplemental Table 3). A decrease in the circulating number of B cells was observed; however, the change was small, and normal B cell numbers were observed in both the spleen and nodes.
Biochemical analyses showed no difference in hematocrit, red cell, or platelet numbers between ΔPMT mice and 129S6 controls (Supplemental Table 4). Normal blood urea, nitrogen, and creatinine indicated normal kidney function, and normal alanine aminotransferase and aspartate aminotransferase levels indicated that loss of these three Gst gene families was not sufficient to induce liver disease in the animals (Supplemental Table 4). The only significant difference observed was a slight decrease in the levels of alkaline phosphatase in the ΔPMT mice. The values, however, were still well within the range observed for mouse (Foreman et al., 2005).
Both female and male mice were fertile and raised litters within the normal size range for the 129S6 mouse strain. Taken together, this indicates that these 14 Gst genes are not required for normal development, maturation, and health in a controlled environment. Thus, this line will provide a useful tool for the study of the role of these genes in disease models as well as for the study of their role in metabolism of drugs and xenobiotics.
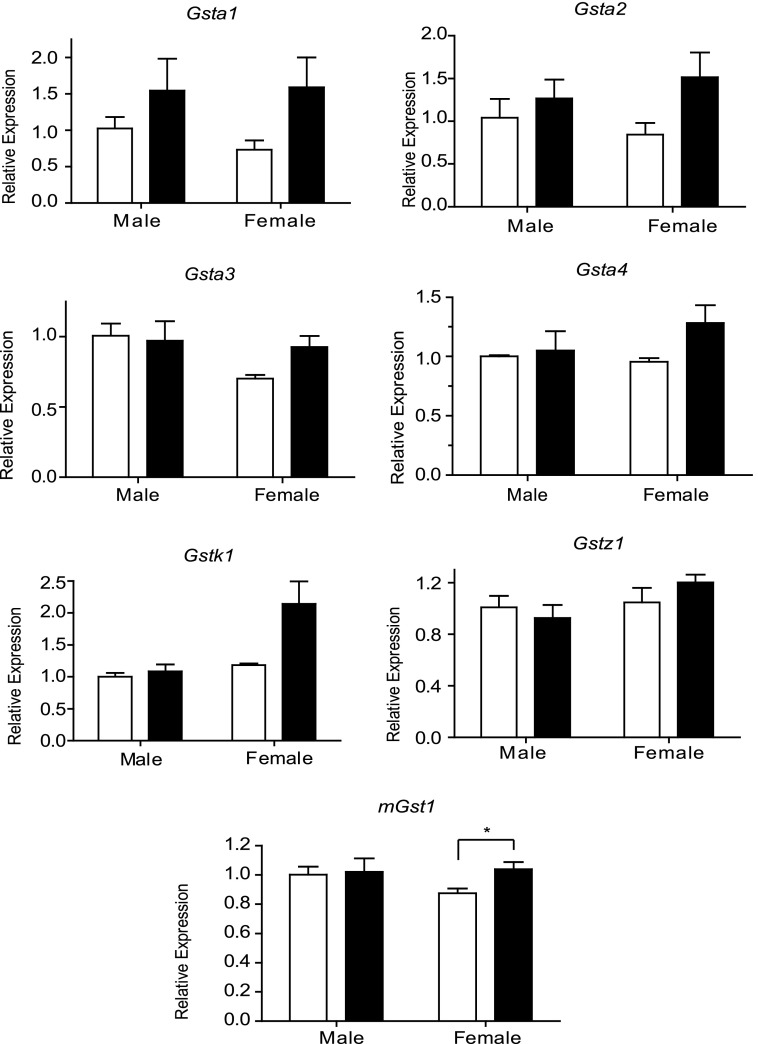
Gene Expression in ΔPMT Mice.
We next asked whether the absence of these three gene families resulted in a compensatory change in the expression of remaining GSTs. Expression of four mouse Gsta1-4 genes, Gstk, Gstz1, and mGst1 was examined in the liver of both male and female ΔPMT animals. Although there was a trend toward increased expression of Gsta genes, particularly in females, none of the changes achieved statistical significance. Also a trend toward increased expression of the microsomal enzyme mGst1 and the peroxisome enzyme Gstk1 was observed in females; however, these increases were small and only achieved statistical significance in the case of mGst1. Overall loss of these three gene families had minimal impact on the expression of the remaining GST genes whose expression was examined (Fig. 5). Increase in the expression of antioxidant enzymes can provide evidence of ongoing oxidative stress. To determine whether such changes were present in the ΔPMT mice, the transcript levels of superoxide dismutase and glutathione peroxidase were examined (Supplemental Fig. 4). This evaluation indicated that, under normal dietary conditions, loss of the Gstp, Gstm, and Gstt gene families did not result in significant oxidative stress in the liver.
Fig. 5.
Relative abundance of GST transcripts in the liver of ΔPMT mice. Expression of the gene indicated in each panel was determined by qPCR of RNA prepared from liver of male and female ΔPMT mice (solid bars) and coisogenic 129S6 controls (open bars). Expression levels were normalized to 18S RNA. Data were analyzed using the comparative CT method as described by Applied Biosystems. In all cases, the expression is shown as the fold change relative to levels in male 129S6 livers. Values represent the mean from three animals. *P < 0.05 determined by unpaired t test.
Metabolism of GST Substrates by ΔPMT Mice.
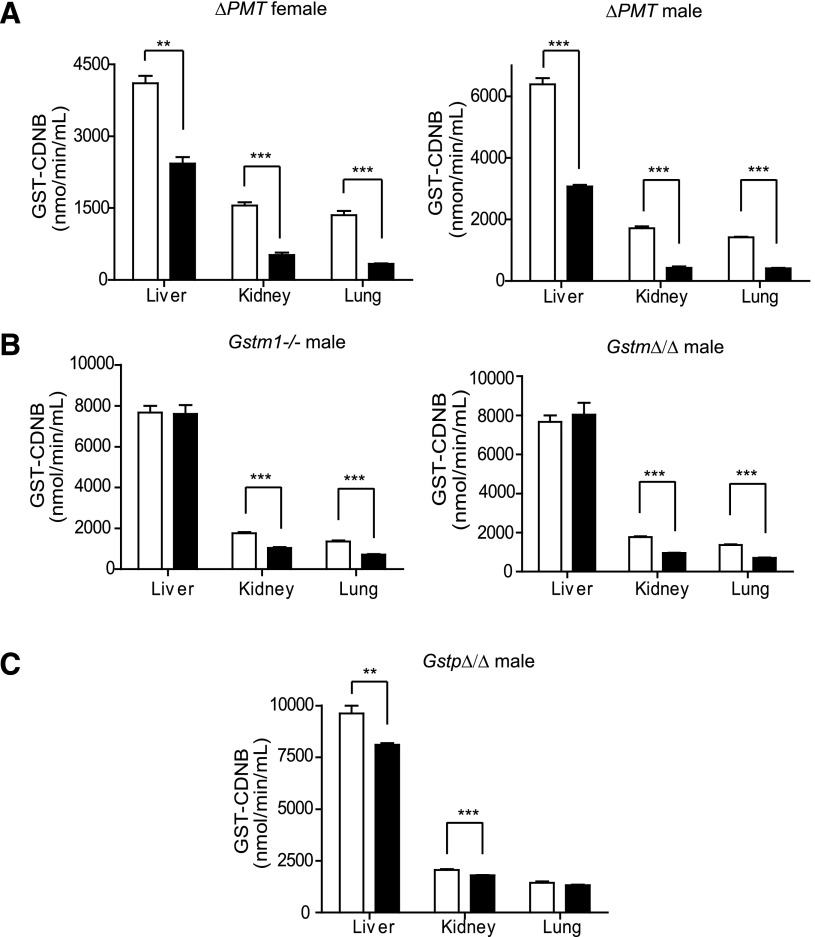
GST-CDNB activity is commonly used as an indicator of total GST activity, and we therefore determined the impact of loss of these 14 genes on the activity toward this substrate. Cytoplasmic fractions were prepared from the liver, lung, and kidney of the male and female ΔPMT mice and GST-CDNB activity measured (Fig. 6). A dramatic decrease in CDNB metabolism was seen in all three tissues in samples prepared from both male and female ΔPMT mice, with levels of activity in the mutant mice reduced by ~50% to 75% (Fig. 6A). Previous studies using tissue from mouse lines with mutations in Gstp1 and Gstp2 have demonstrated decreases in the CDNB activity varying from 10% to 40%, depending on the study, the tissue examined, and the sex of the animals from which the tissue was harvested. A small decrease in CDNB activity was also reported in mice with a mutant Gstm1 locus. Therefore, we next determined whether the large reduction in the CDNB activity could be attributed to the combined loss of Gstm1 and the Gstp locus. We observed a small but significant decrease in the activity of CDNB in the cytosolic extracts from the liver and kidney of the GstpΔ/Δ mice (Fig. 6C). We however did not observe a difference between these groups in the CDNB activity in lung extracts as reported for the Gstp1/2-deficient mice. In all cases the magnitude of the change in activity was small, representing less than 25% of the total CDNB activity. Similar to previous studies, we observed a decrease in the CDNB activity in extracts from the Gstm1−/− mice. Therefore, as expected, a decrease was also detected in the GstmΔ/Δ animals. However, no significant difference in the magnitude of the change in CDNB-GST activity was observed between these two groups, suggesting that the CDNB activity in this family is attributable primarily to GSTM1 (Fig. 6B). The large decrease in GST-CDNB activity cannot be accounted for by loss of the Gstt locus. Consistent with the reported inability of GSTT to metabolize CDNB, no change in CDNB activity was observed in extracts from the GsttΔ/Δ mice (data not shown). Together our results indicate that the loss of all three loci has dramatic impact on total CDNB activity that cannot be accounted for by the loss of the GSTM1 and GSTP alone.
Fig. 6.
Comparison of GST-CDNB activity in tissues from ΔPMT mice and mice lacking individual GST gene families. Cytoplasmic fractions were prepared from liver, kidney, and lung of wild-type (open bars), ΔPMT, GstpΔ/Δ, GsttΔ/Δ, GstmΔ/Δ, and Gstm1−/− mice (solid bars), and activity toward CDNB was determined spectrophotometrically. (A) The activity toward CDNB is reduced by 41%, 67%, and 75% in the liver, kidney, and lung, respectively, in the ΔPMT male mice. In female animals, the reductions were of similar magnitude (52% in liver, 75% in kidney, and 71% in lung). (B) A small but significant reduction in activity toward CDNB was observed in the cytoplasmic preparations from the kidney and lung of the Gstm1−/− and GstmΔ/Δ mice. No difference was observed in this activity when similar subcellular fractions were prepared from the liver of these animals. (C) A statistically significant decrease in activity toward CDNB is also observed in cytosol prepared from GstpΔ/Δ liver and kidney (15% and 12%, respectively). Values are the mean of four animals ± S.E.M. **P < 0.01, ***P < 0.001, determined by unpaired t test. All mice were 129S6 coisogenic between the ages of 8 and 16 weeks.
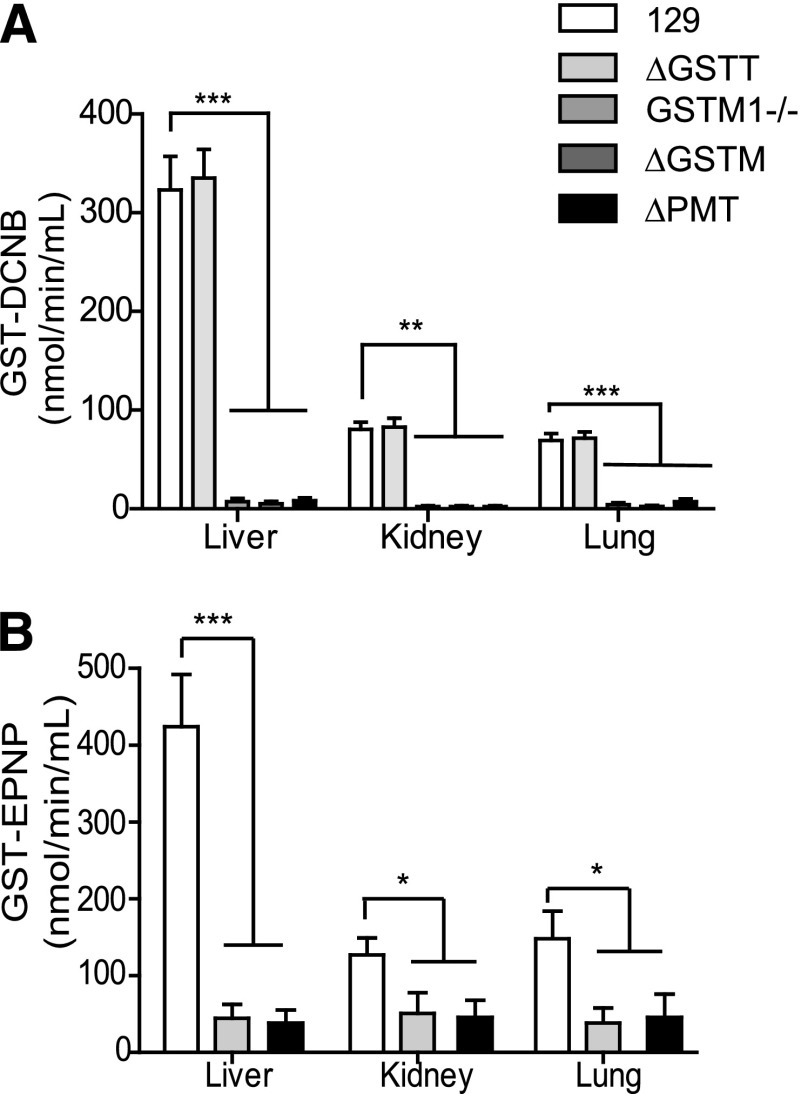
We also examined activity of cytosolic extracts prepared from the ΔPMT mice toward DCNB as a substrate preferentially metabolized by GSTM (Fig. 7A). A 90% reduction in activity was observed in extracts from the liver, kidney, and lung of ΔPMT mice. Although no change in activity toward this substrate is observed in extracts from GsttΔ/Δ mice, a decrease of similar magnitude is observed in the GstmΔ/Δ mice, assigning the GST-DCNB activity to this family. Evaluation of mice lacking only GSTM1 assigns all GST-DCNB activity to this family member, as the magnitude of the decrease in GST-DCNB activity in the cytosolic fractions from the Gstm1−/− animals was not significantly different than the activity of the GstmΔ/Δ tissues.
Fig. 7.
Enzymatic activity of liver, kidney, and lung cytosol of ΔPMT mice toward DCNB and EPNP. (A) Activity of cytosolic preparations from the three tissues was determined spectrophotometrically. The genotype represented by each bar is indicated in the panel. (B) Activity of hepatic cytosol toward EPNP was determined spectrophotometrically. Values represent the mean of four animals ± S.E.M. *P < 0.05, **P < 0.01, ***P < 0.001, determined by unpaired t test.
The activity toward EPNP is also dramatically reduced in tissues from the ΔPMT mice (Fig. 7B). EPNP is reported to be metabolized primarily by mouse GSTT1. Consistent with this report, comparison of the activity toward this substrate in cytosolic fractions from GsttΔ/Δ mice and ΔPMT mice assigned all activity to this locus. Similarly, activity toward ethacrynic acid, a substrate for GSTP, was reduced in liver extracts from ΔPMT and GstpΔ/Δ animals (data not shown).
These experiments using substrates metabolized primarily by GSTT, GSTM, and GSTP demonstrate the facility of using the ΔPMT in combination with the mice lacking individual loci and individual genes to assign the metabolism of compounds for which metabolic pathways have not been established, first generally to these GST families, and then specifically to a family of these enzymes.
Discussion
We have successfully generated mice lacking the Gstp, Gstt, and Gstm gene families, as confirmed by analysis of the structure of the DNA from the deleted loci as well as evaluation of the expression of the 14 genes located in these three major Gst families. In all cases, the loss of expression of the Gst families is the result of the deletion of the entire locus, including DNA flanking the telomeric and centromeric boundaries of the gene clusters. Thus, it is not surprising that we detect no expression of any Gstp, Gstt, or Gstm family members in mice homozygous for each of the deleted loci, even when tissue that normally displays the highest level of expression is chosen for study.
When choosing the boundaries for each of the three deletions, the organization and structure of the genes adjacent to the deleted loci were considered. In silico analysis of the regions to be deleted was performed to identify conserved, noncoding sequences of possible regulatory importance as well as transcribed regions of unknown function. Although the deleted Gst gene clusters were flanked in most cases by poorly conserved, intergenic DNA, an exception was the region between Gstt3 and Ddt, the gene encoding d-dopachrome decarboxylase, at the centromeric boundary of the Gst locus. In humans, the DDT gene is located within the GSTT cluster (Coggan et al., 1998). The name of this gene describes its enzymatic activity, conversion of d-dopachrome into 5,6-dihydroxyindole (Odh et al., 1993). However, d-dopachrome is not present in vivo, and more recent studies suggest that DDT, similar to migration inhibitory factor (MIF), binds CD74 (Merk et al., 2011). This activity suggests that, like MIF, DDT may in fact be a cytokine involved in regulation of immune responses. However, because potential regulatory regions of the Gstt3 extend into the Ddt gene, this gene was destroyed during the deletion event, as we decided it was prudent to remove all sequences in this region with the potential to influence expression of GSTT or Gstt genes introduced into the delta Gstt locus in future experiments. Should the ΔPMT mice display a phenotype that is subsequently attributed to the Gstt locus through study of the GsttΔ/Δ mice, we can assign the phenotype to one of the human or mouse Gstt genes using mice generated from ΔPMT ES cells reconstituted with various segments of the mouse Gstt or human GSTT locus. A line restored with only the Ddt gene would serve as a control.
The close chromosomal proximity of the Gstp1 and Gstp2 genes in the mouse has facilitated the use of gene targeting in a previous study to generate mice in which a mutation is introduced that disrupts expression of both genes (Henderson et al., 1998). We noted, however, on examination of this locus, the presence of a gene that, based on its primary structure, appears to represent a third member of the Gstp gene family. There are a number of intriguing features of this gene that we believe warrant its further study. First, the dramatic increase in the expression of Gstp1/2 in male mice observed at puberty (Conforto and Waxman, 2012) is not conserved in Gstp3. Second, although we have observed lower transcript levels of BC021614 relative to Gstp1/2 in the tissues examined to date, this may be compensated for by the fact that the predicted protein encoded by this gene includes an amino acid substitution that has been shown to increase the stability of human GSTP. Final assignment of this gene to the Gstp family awaits determination of whether the protein encoded by BC021614 (Gstp3) has GST activity.
Evaluation of GST activity in the four mouse lines we have generated toward molecules known to be preferred substrates for members of the GSTP, GSTT, or GSTM family, respectively, demonstrates the utility of these lines in defining the role these GST families play in the metabolism of a particular substrate. Comparison of the results of these evaluations with similar evaluations in mice lacking single genes can subsequently be used to determine the contribution of individual family members to the metabolism of a particular substrate. For example, comparison of the activity of extracts from GstmΔ/Δ mice and Gstm1−/− mice indicates that Gstm2 through Gstm7 do not contribute significantly to the metabolism of DCNB, even though the expression of Gstm2, Gstm3, Gstm6, and Gstm4 in the liver has been reported (Knight et al., 2007). Interestingly, metabolism of DCNB by human GSTM1 has not been supported in comparative studies using extracts from individuals carrying the null GSTM1 allele (Arakawa et al., 2012). Therefore, this substrate may be highly specific for a single murine Gstm1 gene, calling into question the evolutionary and functional relationship between the mouse and human Gstm isoforms (Board, 2007).
The GstpΔ/Δ mice demonstrated only a small decrease in activity toward CDNB in the liver and kidney, and no change in metabolism of this substrate could be measured in extracts from the lung. This differs in some important aspects from results reported for the Gstp1/2 mice. In an initial study of these animals, Henderson et al. (1998) reported that the activity toward CDNB was reduced in lung cytosolic fractions but not in the fractions prepared from the kidney and liver. However, a later study reported reduced activity toward CDNB in cytosolic extracts prepared from liver, lung, and kidney of this same mouse line, with the most dramatic reduction observed in the liver (Conklin et al., 2009). A possible explanation for these disparate findings is the different genetic background of the GSTP-deficient mice. Our studies were conducted with coisogenic 129S6 animals whereas the initial studies of the Gstp1/2−/− mice were performed with F2 mice carrying 129 and MF1 alleles; the subsequent studies used mice in which the mutation had been introduced into the C57BL/6 background by successive rounds of breeding. Strain differences in mouse liver GST activity have been previously noted (Egaas et al., 1995). Differences might reflect different environmental conditions, including biologic differences in the microflora colonizing skin, intestinal, and/or nasal passages as well as physical differences such as bedding and diet.
Perhaps the most surprising finding of our study of GST activity present in the cytosolic fractions prepared from ΔPMT mice was the robust and easily repeatable decrease in the metabolism of CDNB by cytosolic fractions prepared from all three tissues, particularly the lung. This change could not be assigned to the Gstt genes, as the GsttΔ/Δ mice showed no change in activity toward CDNB, consistent with other studies. Furthermore, although some of the activity toward CDNB could be assigned to the Gstm1 gene, the contribution from this gene represented a small fraction of the total activity. As discussed earlier, only a small change in activity toward CDNB was observed in the GstpΔ/Δ mice, and this was limited to the liver and kidney. No decrease was seen in the lung, the tissue with the largest decrease in activity toward CDNB in the ΔPMT animals. Taken together, the decreases in activity observed in the GstpΔ/Δ and GstmΔ/Δ mice cannot account for the dramatic decrease in the Δ PMT animals. One possible explanation is that there is compensatory up-regulation of genes in the GstpΔ/Δ and GstmΔ/Δ mice. Compensation for loss of a Gst gene was reported for Gsta4−/− mice, where increased expression of Gsta2, Gsta3, and Gstm1 was observed (Engle et al., 2004). In comparison, we observed no change in Gsta, Gstz1, or Gstk1 expression and only a small increase in mGst1 in female ΔPMT mice. However, we did not examine tissue from the GstpΔ/Δ mice or the GstmΔ/Δ mice for changes in gene expression. A possible role for compensatory changes in gene expression between the Gstp and the Gstm loci can be tested by the generation of mice lacking only these two loci.
The mice lacking the 14 Gst genes are remarkably healthy and fertile, with no apparent increase in morbidity. Evaluation of blood chemistry from three of the oldest available animals indicated no significant loss of liver or kidney function. The normal development and good health of these animals suggests that these enzymes are not required to maintain redox homeostasis under normal conditions. This interpretation is supported by our failure to detect an increase in the expression of antioxidant enzymes, a signature of oxidative stress. This suggests that the Gstm, Gstp, and Gstt genes function primarily in response to changes in environmental stress factors that are largely limited or controlled in a vivarium. Such an interpretation would be consistent with the significant differences between humans and rodents in the number, substrate specificity, and organization of these gene families, as species differences would reflect the very different environmental pressures driving evolution of these genes in humans and mice. Mice in which the deleted loci are restored with individual mouse genes or, alternatively, with their proposed human orthologs should provide models for pharmacologic and toxicologic studies of the role of these phase II enzymes in the metabolism of drugs and xenobiotics.
Supplementary Material
Acknowledgments
The authors thank Rebecca Dye, William Barker, and Syed Jaffery, for assistance in breeding mouse lines and genotyping.
Abbreviations
- CDNB
1-choloro-2,4-dinitrobenzene
- chr
chromosome
- DCNB
1,2-dichloro-4 nitrobenzene
- EPNP
1,2-epoxy-3-(4-nitrophenoxy)propane
- ES
embryonic stem
- G418
Geneticin
- Gst
glutathione S-transferase
- kb
kilo base pair(s)
- LPS
lipopolysaccharide
- PCR
polymerase chain reaction
- qPCR
quantitative PCR
Authorship Contributions
Participated in research design: Snouwaert, Nguyen, Koller.
Conducted experiments: Xiang, Snouwaert, Nguyen, Repenning, Latour, Kovarova, Cyphert, Koller.
Wrote or contributed to writing of manuscript: Snouwaert, Nguyen, Kovarova, Koller.
Footnotes
This work was supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant HL107780] and [Grant HL098941].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Arakawa S, Fujimoto K, Kato A, Endo S, Fukahori A, Shinagawa A, Fischer T, Mueller J, Takasaki W. (2012) Evaluation of hepatic glutathione transferase Mu 1 and Theta 1 activities in humans and mice using genotype information. Drug Metab Dispos 40:497–503 [DOI] [PubMed] [Google Scholar]
- Blackburn AC, Coggan M, Shield AJ, Cappello J, Theodoratos A, Murray TP, Rooke M, Larter CZ, Koina ME, Dahlstrom JE, et al. (2011) Glutathione transferase kappa deficiency causes glomerular nephropathy without overt oxidative stress. Lab Invest 91:1572–1583 [DOI] [PubMed] [Google Scholar]
- Board PG. (2007) The use of glutathione transferase-knockout mice as pharmacological and toxicological models. Expert Opin Drug Metab Toxicol 3:421–433 [DOI] [PubMed] [Google Scholar]
- Coggan M, Whitbread L, Whittington A, Board P. (1998) Structure and organization of the human theta-class glutathione S-transferase and D-dopachrome tautomerase gene complex. Biochem J 334:617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforto TL, Waxman DJ. (2012) Sex-specific mouse liver gene expression: genome-wide analysis of developmental changes from pre-pubertal period to young adulthood. Biol Sex Differ 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin DJ, Haberzettl P, Lesgards JF, Prough RA, Srivastava S, Bhatnagar A. (2009) Increased sensitivity of glutathione S-transferase P-null mice to cyclophosphamide-induced urinary bladder toxicity. J Pharmacol Exp Ther 331:456–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egaas E, Falls JG, Dauterman WC. (1995) A study of gender, strain and age differences in mouse liver glutathione-S-transferase. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol 110:35–40 [DOI] [PubMed] [Google Scholar]
- Engle MR, Singh SP, Czernik PJ, Gaddy D, Montague DC, Ceci JD, Yang Y, Awasthi S, Awasthi YC, Zimniak P. (2004) Physiological role of mGSTA4-4, a glutathione S-transferase metabolizing 4-hydroxynonenal: generation and analysis of mGsta4 null mouse. Toxicol Appl Pharmacol 194:296–308 [DOI] [PubMed] [Google Scholar]
- Foreman JE, Blizard DA, Gerhard G, Mack HA, Lang DH, Van Nimwegen KL, Vogler GP, Stout JT, Shihabi ZK, Griffith JW, et al. (2005) Serum alkaline phosphatase activity is regulated by a chromosomal region containing the alkaline phosphatase 2 gene (Akp2) in C57BL/6J and DBA/2J mice. Physiol Genomics 23:295–303 [DOI] [PubMed] [Google Scholar]
- Fujimoto K, Arakawa S, Shibaya Y, Miida H, Ando Y, Yasumo H, Hara A, Uchiyama M, Iwabuchi H, Takasaki W, et al. (2006) Characterization of phenotypes in Gstm1-null mice by cytosolic and in vivo metabolic studies using 1,2-dichloro-4-nitrobenzene. Drug Metab Dispos 34:1495–1501 [DOI] [PubMed] [Google Scholar]
- Fujimoto K, Arakawa S, Watanabe T, Yasumo H, Ando Y, Takasaki W, Manabe S, Yamoto T, Oda S. (2007) Generation and functional characterization of mice with a disrupted glutathione S-transferase, theta 1 gene. Drug Metab Dispos 35:2196–2202 [DOI] [PubMed] [Google Scholar]
- Habig WH, Pabst MJ, Jakoby WB. (1974) Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem 249:7130–7139 [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. (2005) Glutathione transferases. Annu Rev Pharmacol Toxicol 45:51–88 [DOI] [PubMed] [Google Scholar]
- Henderson CJ, Smith AG, Ure J, Brown K, Bacon EJ, Wolf CR. (1998) Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc Natl Acad Sci USA 95:5275–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CJ, Wolf CR, Kitteringham N, Powell H, Otto D, Park BK. (2000) Increased resistance to acetaminophen hepatotoxicity in mice lacking glutathione S-transferase pi. Proc Natl Acad Sci USA 97:12741–12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight TR, Choudhuri S, Klaassen CD. (2007) Constitutive mRNA expression of various glutathione S-transferase isoforms in different tissues of mice. Toxicol Sci 100:513–524 [DOI] [PubMed] [Google Scholar]
- Koller BH, Kim HS, Latour AM, Brigman K, Boucher RC, Jr, Scambler P, Wainwright B, Smithies O. (1991) Toward an animal model of cystic fibrosis: targeted interruption of exon 10 of the cystic fibrosis transmembrane regulator gene in embryonic stem cells. Proc Natl Acad Sci USA 88:10730–10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CE, Matthaei KI, Blackburn AC, Davis RP, Dahlstrom JE, Koina ME, Anders MW, Board PG. (2004) Mice deficient in glutathione transferase zeta/maleylacetoacetate isomerase exhibit a range of pathological changes and elevated expression of alpha, mu, and pi class glutathione transferases. Am J Pathol 165:679–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merk M, Zierow S, Leng L, Das R, Du X, Schulte W, Fan J, Lue H, Chen Y, Xiong H, et al. (2011) The d-dopachrome tautomerase (DDT) gene product is a cytokine and functional homolog of macrophage migration inhibitory factor (MIF). Proc Natl Acad Sci USA 108:E577–E585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn AR, Koller BH. (1995) Genetic manipulation of embryonic stem cells, In: Mammalian Systems, pp 143–184, Oxford University Press, New York [Google Scholar]
- Oakley AJ, Lo Bello M, Ricci G, Federici G, Parker MW. (1998) Evidence for an induced-fit mechanism operating in pi class glutathione transferases. Biochemistry 37:9912–9917 [DOI] [PubMed] [Google Scholar]
- Odh G, Hindemith A, Rosengren AM, Rosengren E, Rorsman H. (1993) Isolation of a new tautomerase monitored by the conversion of d-dopachrome to 5,6-dihydroxyindole. Biochem Biophys Res Commun 197:619–624 [DOI] [PubMed] [Google Scholar]
- Pettigrew NE, Moyer-Myers M, Colman RF. (1999) Affinity labeling of pig lung glutathione S-transferase pi by 4-(fluorosulfonyl)benzoic acid. Arch Biochem Biophys 364:107–114 [DOI] [PubMed] [Google Scholar]
- Ralat LA, Manevich Y, Fisher AB, Colman RF. (2006) Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase pi with activity changes in both enzymes. Biochemistry 45:360–372 [DOI] [PubMed] [Google Scholar]
- Stenberg G, Abdalla AM, Mannervik B. (2000a) Tyrosine 50 at the subunit interface of dimeric human glutathione transferase P1-1 is a structural key residue for modulating protein stability and catalytic function. Biochem Biophys Res Commun 271:59–63 [DOI] [PubMed] [Google Scholar]
- Stenberg G, Dragani B, Cocco R, Mannervik B, Aceto A. (2000b) A conserved “hydrophobic staple motif” plays a crucial role in the refolding of human glutathione transferase P1-1. J Biol Chem 275:10421–10428 [DOI] [PubMed] [Google Scholar]
- Trebino CE, Stock JL, Gibbons CP, Naiman BM, Wachtmann TS, Umland JP, Pandher K, Lapointe JM, Saha S, Roach ML, et al. (2003) Impaired inflammatory and pain responses in mice lacking an inducible prostaglandin E synthase. Proc Natl Acad Sci USA 100:9044–9049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasieva O. (2011) The many faces of glutathione transferase pi. Curr Mol Med 11:129–139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.