Abstract
Intratumoral hypoxia has been proposed to create a “mutator” phenotype through downregulation of DNA repair, leading to increased genomic instability and drug resistance. Such downregulation of DNA repair has been proposed to sensitize hypoxic cancer cells to DNA-damaging agents and inhibitors of DNA repair. Here, we showed that prostate cancer cells with mutant p53 were resistant to the poly(ADP-ribose) polymerase inhibitor, veliparib (2-[(2R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide, dihydrochloride; Abbott Laboratories, Abbott Park, IL), and the DNA-damaging topoisomerase I inhibitor camptothecin-11 (CPT-11) or SN38 (7-ethyl-10-hydroxycamptothecin) under hypoxia. Upregulation of Rad51 by E2F1 upon DNA damage under hypoxia contributed to such resistance, which was reversed by either inhibiting RAD51 transcription with small interfering RNA or by expressing wild-type p53 in the p53 null prostate cancer line. Accumulation of endogenous p53 but not E2F1 and suppressed RAD51 transcription was observed in prostate cancer line with wild-type p53 after DNA damage under hypoxia. Combining veliparib with CPT-11 significantly enhanced DNA damage and apoptosis under both hypoxic and normoxic culture conditions. Such enhanced DNA damage and antitumor activities were seen in the presence of Rad51 upregulation and confirmed in vivo with PC3 mouse xenografts. These data illustrate a dynamic regulation of Rad51 by E2F1 and p53 in prostate cancer cells’ response to hypoxia and DNA damage. The veliparib and CPT-11 combination can be further explored as a treatment of metastatic castration-resistant prostate cancers that have frequent p53 mutations and enriched genomic instability.
Introduction
With the recent success of treatment of BRCA1 or BRCA2 mutated cancers with the poly(ADP-ribose) polymerase (PARP) inhibitor (Fong et al., 2009; Tutt et al., 2010), there has been increasing interest in exploring synthetic lethality in cancers with defective DNA repair pathways (Helleday, 2010; Yap et al., 2011). This could potentially offer a unique therapeutic opportunity to directly target the aggressive cancer cells that obtain genomic instability through diminished DNA repair. Although two of the most common genetic alterations in prostate cancer, ETS gene rearrangement and loss of PTEN (phosphatase and tensin homolog), have been linked to sensitivity to PARP inhibition in preclinical studies (Mendes-Pereira et al., 2009; Brenner et al., 2011), neither of them was associated with antiprostate cancer activities—time to disease progression, prostate-specific antigen response rate, or decline in circulating tumor cells—in a phase 1 study with the PARP inhibitor niraparib (Sandhu et al., 2013). Among the 23 prostate cancer patients in this trial, only one had a documented BRCA mutation, and nine had stable disease for a median duration of 254 days. Developing biomarkers to identify this subgroup of prostate cancer, which is sensitive to drug-induced DNA damage, and improving the therapeutic index of the PAPR inhibitor with novel combinations are unmet challenges.
Intratumoral hypoxia has been proposed to create a “mutator” phenotype with increased genomic instability and drug resistance (Bristow and Hill, 2008). This hypothesis is supported by observations that DNA repair proteins are frequently downregulated in hypoxic cancer cells, including prostate cancer cells (Bindra et al., 2004; Bindra and Glazer, 2007; Chan et al., 2010). Downregulation of Rad51 expression, in particular, has been reported in lung, breast, colon, prostate, and cervical cancer cell lines grown under chronic hypoxic conditions (Bindra et al., 2004; Meng et al., 2005; Chan et al., 2010). Rad51 is an essential protein in homologous recombination repair, an error-free pathway for DNA double-strand break repairs (Moynahan and Jasin, 2010). Although mutations in the RAD51 open-reading frame are rare in cancer, overexpression of Rad51 has been reported in a wide variety of cancers, especially those harboring p53 mutations (Klein, 2008). Rad51 overexpression can lead to resistance to both drug- and radiation-induced DNA damage and has been shown to compensate for the homologous recombination defects caused by BRCA1 or BRCA2 deficiency (Martin et al., 2007; Brown and Holt, 2009; Lee et al., 2009; Yang et al., 2012).
Using cell lines derived from metastatic lesions of prostate cancer patients with nonfunctional p53 (DU145, mutant p53; PC3, p53 null) as well as wild-type p53 (LNCaP), we found that the p53 status determined the sensitivity of prostate cancer cells to DNA-damaging drugs under hypoxia. Prostate cancer cells with nonfunctional p53 were resistant to PARP inhibitor and topoisomerase I inhibitor under hypoxia, and such resistance was mediated by upregulation of Rad51 by E2F1. The RAD51 transcription was suppressed by p53 in LNCaP cells, and expressing wild-type p53 in PC3 cells reversed their resistance to DNA damage under hypoxia. Combining the PARP inhibitor veliparib (2-[(2R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide, dihydrochloride) with camptothecin-11 (CPT-11) overcame such resistance in p53 mutant prostate cancer cells and showed synergistic antitumor activities both in vitro and in vivo.
Materials and Methods
Cell Culture and Drugs.
Human prostate cancer cell lines PC3 (p53 null), DU145 (mutant p53), LNCaP (p53 wild type), and Vcap (mutant p53) were obtained from the American Type Culture Collection (Manassas, VA) and were maintained in culture media as instructed by American Type Culture Collection. For hypoxia experiments, cells were incubated in a hypoxic chamber (Biospherix, New York, NY) with constant 0.2% oxygen. CPT-11/irinotecan and its active metabolite SN38 (7-ethyl-10-hydroxycamptothecin) were purchased from Sigma-Aldrich (St. Louis, MO). Unless otherwise specified in the figures, the doses of SN38 were 1 μM for PC3, 0.1 μM for DU145, and 0.5 μM for LNCaP . The PARP inhibitor veliparib was kindly provided by Abbott Laboratories (Abbott Park, IL), and 1 μM was used in all the in vitro data shown in the figures.
Western Blot Analysis.
Protein lysate preparation and immunoblotting were performed as described previously elsewhere (Zhang et al., 2004). Antibodies to PARP, E2F1, E2F4, p53, Rad51, poly(ADP) ribose, γ-H2AX, β-actin, and tubulin were purchased from Cell Signaling Technology (Boston, MA), Santa Cruz Biotechnology (Santa Cruz, CA), Trevigen (Gaithersburg, MD), Millipore (Billerica, MA), and Sigma-Aldrich. Immunoreactive protein was detected using enhanced chemiluminescence reagents (Roche, Indianapolis, IN) according to the manufacturer's instructions.
Flowcytometry.
Cells were collected and analyzed for apoptosis with propidium iodide and annexin V (BD Biosciences, San Jose, CA). The stained cells were sorted by FACSCAN1 (BD Biosciences), and the data were analyzed with Flowjo software (Treestar, Inc., Ashland, OR).
Real-Time Reverse-Transcription Polymerase Chain Reaction.
Total RNA was extracted using RNeasy Mini kit (Qiagen, Valencia, CA); 2 μg total RNA was reversely transcribed using SuperScript II Reverse Transcriptase (Applied Biosystems, Grand Island, NY) according to the manufacturer’s instructions. Sequences of RAD51 primers were as follows: GCTGCGGACCGAGTAATG (forward) and CCAGCTTCTTCCAATTTCTTC AC (reverse). Amplification reaction assays contained 1× SYBR green polymerase chain reaction (PCR) Mastermix (Applied Biosystems) at the optimal concentrations, and amplification was performed using an ABI PRISM 7000 SDS thermal cycler (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase was used as the reference gene for normalization.
Gene Silencing with Small Interfering RNA.
RAD51 small interfering RNA (siRNA) pools (three target-specific siRNAs) were purchased from Santa Cruz Biotechnology. E2F1 siRNA Smartpools (four target-specific siRNAs) were purchased from Dharmacon (Pittsburgh, PA). PC3 or DU145 cells were seeded in six-well plates and transfected with 5-μM siRNAs. After 6 hours, the medium with siRNA was removed, and the cells were incubated with fresh medium overnight before further treatment. A mock siRNA (Pittsburgh, PA) was used as the control.
Reporter Assays.
The RAD51 promoter PGL3 luciferase reporter construct and the construct with a mutated E2F1-binding site at the RAD51 promoter were kindly provided by Dr. Peter Glazer. Cells were seeded in 96-well culture plates and transfected with 2 ng/well Renilla luciferase construct along with 0.2 μg/well of each RAD51 promoter firefly luciferase reporter construct in triplicate. Firefly and Renilla luciferase activities were measured with the Dual-Luciferase Reporter Assay kit (Promega, Fitchburg, WI).
Chromatin Immunoprecipitation Assay.
The chromatin immunoprecipitation (ChIP) experiments were performed with the Chip-IT Express Kit (Active Motif, Carlsbad, CA). Briefly, cells were cross-linked with formaldehyde and incubated with 1× lysis buffer with a protease-inhibitor mixture, and sonicated to generate 200–500 bp DNA fragments. After incubation with 5 μg of anti-p53 antibody or anti-E2F1 antibody, the cross-linking was reversed. The bound DNA was obtained by phenol chloroform extraction and ethanol precipitation, and resuspended in 50 μl of H2O. PCR was performed with 5 μl of immunoprecipitated target DNA. Input material corresponding to 1% of total sample was recovered before immunoprecipitation, and PCR was performed with 1 μl of purified DNA. Primer sets used were the following: 5′-CCTCGAACTCCTAGGCTCAGA-3′, 5′-CCGCGTCGACGTAACGTAT-3′, for the p53 binding sites on the RAD51 promoter; and 5′-TAGGAGGCTCAGAGCGACCA-3′, 5′-GTCCGCCAGCGGCTTTCAGAA-3′, for the E2F1/E2F4 binding site on the proximal RAD51 promoter.
Adenoviral Infection.
Recombinant adenovirus containing wild-type p53 and green fluorescent protein (GFP), p53/GFP adenovirus 5, and the empty vector GFP adenovirus 5 were purchased from Vector Biolabs (Philadelphia, PA). PC3 cells were infected with 200 pfu/cell of either GFP adenovirus 5 or p53/GFP adenovirus 5. Infection efficiency was monitored by observation under fluorescent microscope. The plates with sufficient infection efficiency were used (>90%).
Neutral Comet Assay.
The neutral comet assay was used to detect DNA double-strand breaks and was performed based on the manufacturer’s instructions (Trevigen). Comets were visualized with the Olympus BX51 fluorescence microscope (Olympus Corporation, Tokyo, Japan).
Clonogenic Assay.
Cells were seeded in six-well plates to reach 70–80% densities and cultured for at least 24 hours before treatment with SN38, veliparib, or their combination for 16 hours. Treated and untreated cells (n = 1000 each) were seeded separately in six-well plates for 14 days to form colonies. After they were washed with phosphate-buffered saline, the colonies were fixed with 100% methanol, dried, stained with 0.5% crystal violet (Sigma-Aldrich), and counted manually.
Immunofluorescence Staining and Confocal Microscopy.
Cells were seeded onto Laboratory TekII chamber slides (Fisher Scientific, Hampton, NH). The immunofluorescence assay was performed as described previously elsewhere (Zhang et al., 2004).
Mouse anti-Rad51 (Abcam, Cambridge, MA) and rabbit anti-BRCA1 (Santa Cruz Biotechnology) were used as primary antibodies. The Alexa Fluor 488 goat anti-mouse antibody (Invitrogen, Carlsbad, CA) and the Alexa Fluor 594 goat anti-rabbit antibody (Invitrogen) were used as secondary antibodies. The coverslips were mounted with 4′,6-diamidino-2-phenylindole containing Vectashield (Vector Laboratories, Burlingame, CA). Samples were viewed with a Leica DMI6000 inverted microscope, TCS SP5 confocal scanner, and a 63×/1.4 numerical aperture Plan Apochromat oil immersion objective (Leica Microsystems, Buffalo Grove, IL). Images were captured with photomultiplier detectors and prepared with the LAS AF software version 1.6.0 build 1016 (Leica Microsystems).
Immunohistochemistry.
Immunohistochemistry (IHC) analyses for pimonadazole (Hydroxyprobe, Burlington, MA), γ-H2AX (Millipore), and Rad51 (Abcam) on formalin-fixed paraffin-embedded xenograft sections were performed at Moffitt Cancer Center’s Tissue Core with the standard antigen retrieval method. Consecutive xenograft sections were used to closely match the pimonadazole positivity with the γ-H2AX and Rad51 stains.
Xenograft Studies.
The xenograft study protocol was approved by the institutional animal care and use committees. Xenografts were established by subcutaneous injection of 5.0 × 105 PC3 cells 1:1 mixed with matrigel (BD Biosciences) to the flank area of male NOD.CB17-Prkdcsid/NcrCrl mice. After the xenografts reached ∼200 mm3, mice were randomized to treatment groups of saline control, CPT-11, veliparib, or veliparib combined with CPT-11. Male mice were weighed, and the xenograft size was measured twice per week. The mice were euthanized when the xenografts reached 1000 mm3. At day 101, all of the remaining mice underwent euthanasia regardless of their xenograft size. To highlight the hypoxic regions, pimonadazole was injected before the mice were euthanized. The mice received 1 day of assigned treatment before euthanizing if the xenografts grew to the 1000 mm3 threshold while being off treatment. This allowed us to assess the effects of treatment on DNA damage and DNA repair with these xenograft samples.
Statistical Analysis.
Statistical analysis was performed using the GraphPad Prism 5 software (GraphPad Software, San Diego, CA). For one-way analysis of variance, Tukey’s multiple comparison test was used. A t test was used for two-group comparisons. Data derived from at least three independent experiments are shown as mean ± S.E.M. Log-rank test was used for survival rate analysis in the mouse xenograft experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
Results
Upregulation of Rad51 Contributes to the Resistance of Prostate Cancer Cells to Drug-Induced DNA Damage under Hypoxia.
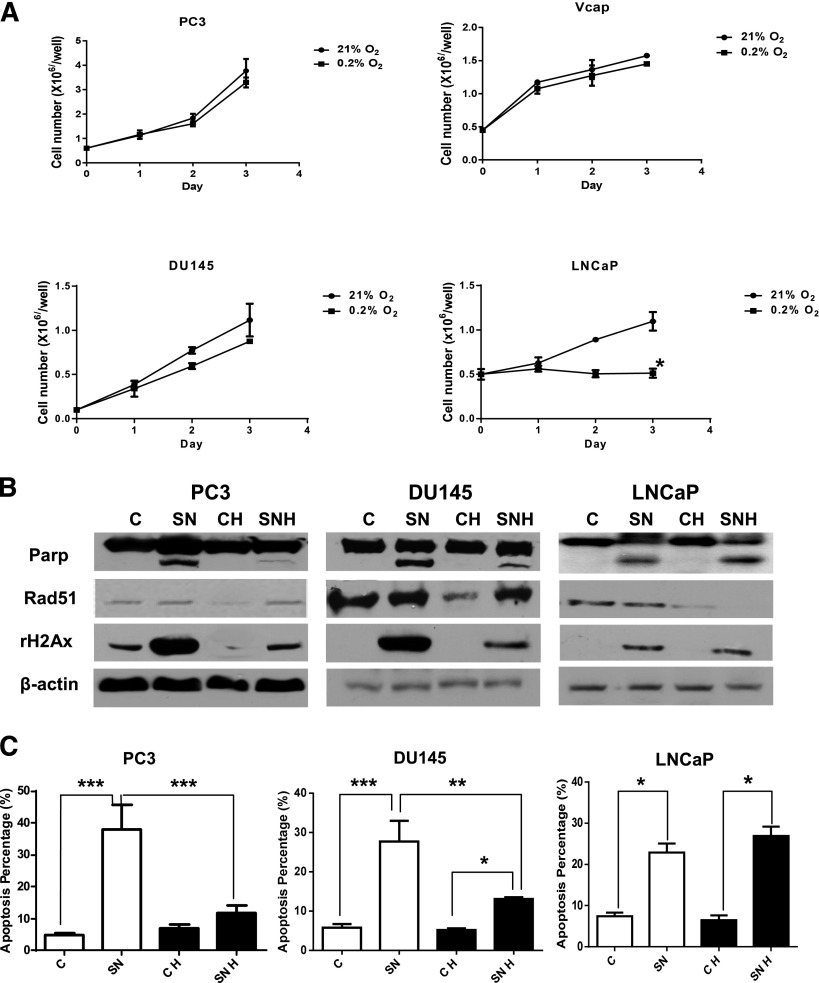
Unlike the LNCaP cells (wild-type p53), the PC3, Vcap, and DU145 cells (p53 null or mutant) continued to proliferate under chronic hypoxia (0.2% oxygen) (Fig. 1A). Although Rad51 was initially decreased under hypoxia, upregulation of Rad51 was observed in PC3 and DU145 cells after a 3-day treatment with the topoisomerase I inhibitor SN38 (Fig. 1B). Compared with PC3 and DU145 cells treated with SN38 under normoxia, the upregulation of Rad51 after SN38 treatment under hypoxia was associated with fewer DNA double-strand breaks as detected by γ-H2AX levels on Western blot (Fig. 1B); and less apoptosis as measured by PARP cleavage on Western blot (Fig. 1B) and propidium iodide-annexin V–positive cells on flow cytometry (Fig. 1C). No upregulation of Rad51 was observed in LNCaP after treatment with SN38 under hypoxia. Compared with LNCaP cells grown under normoxia, LNCaP cells grown under hypoxia remained sensitive to SN38-induced DNA damage and apoptosis (Fig. 1, B and C).
Fig. 1.
The resistance of prostate cancer cells to drug-induced DNA damage and apoptosis under hypoxia is associated with upregulation of Rad51. (A) Growth curves of prostate cancer cells under hypoxic (0.2% oxygen) and normoxic (21% oxygen) culture. The mean and S.E.M. of three independent experiments were plotted. (B) Western blots comparing levels of PARP cleavage, Rad51, and γ-H2AX in untreated control versus 3-day SN38 treatment under normoxia and hypoxia. Increased levels of Rad51 were seen in DU145 and PC3 cells but not LNCaP cells after SN38 treatment under hypoxia (SNH versus CH). (C) Percentage of apoptosis as detected by flow cytometry with or without SN38 treatment of 3 days. Each column represents the mean and S.E.M. of three independent experiments. White column: 21% oxygen; black column: 0.2% oxygen; C, untreated control; CH, untreated under hypoxia; SN, SN38; SNH, SN38 treated under hypoxia. β-Actin was used as the loading control. *P < 0.05; **P < 0.01; ***P < 0.001.
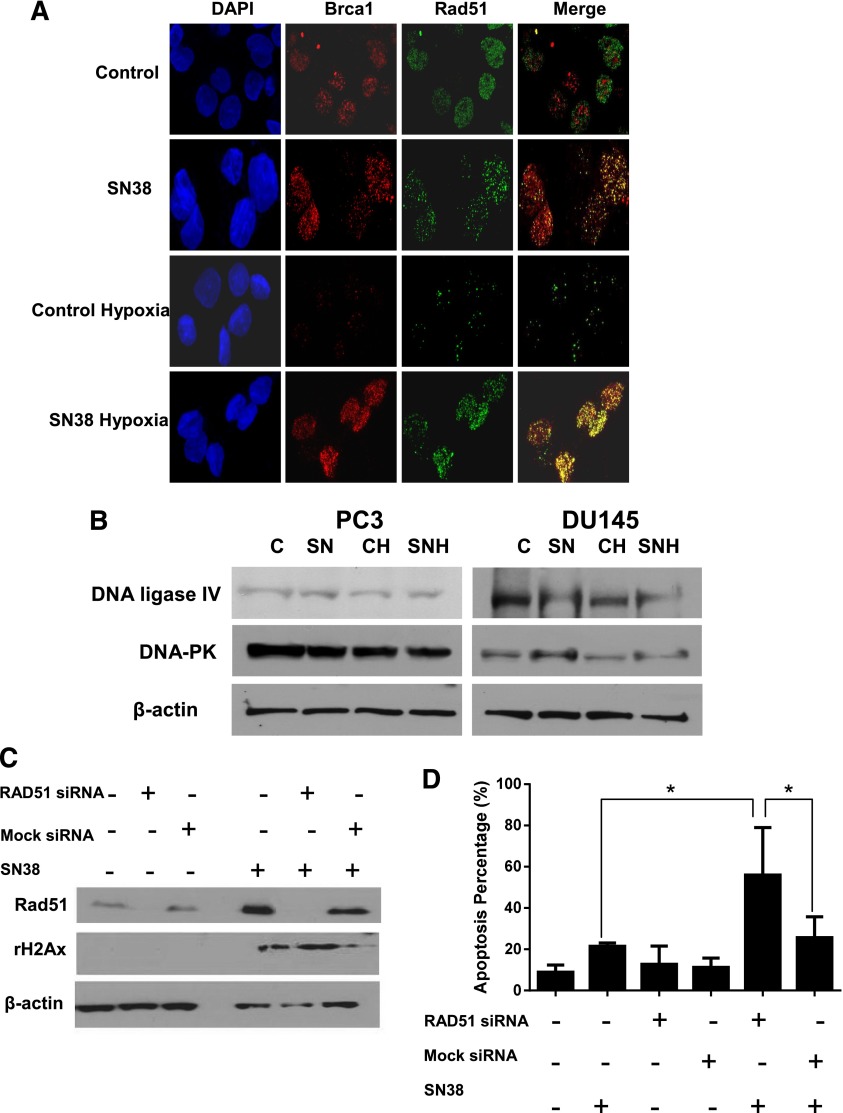
Formation of Rad51 and BRCA1 nuclear foci on immunofluorescence was used to reflect the activation of the homologous recombination repair (Scully et al., 1997). Compared with untreated PC3 cells under normoxia, there was less nuclear staining of Rad51 and BRCA1 under hypoxia (Fig. 2A). This is consistent with less DNA damage caused by oxidative stress (Fig. 1B) and, thus, less demand for DNA-repair proteins, such as Rad51 and BRCA1, under hypoxia. No changes in the levels of key proteins in the nonhomologous end-joining pathway were observed when SN38-treated cells were compared with untreated cells under either normoxia or hypoxia (Fig. 2B).
Fig. 2.
Blocking Rad51 upregulation resensitizes prostate cancer cells to drug-induced DNA damage and apoptosis under hypoxia. (A) Nuclear foci formation of Rad51 and BRCA1 as detected by immunofluorescence in normoxic and hypoxic PC3 cells either untreated (control) or treated with 0.1 µM SN38 for 4 hours. (B) Western blots of DNA ligase IV and DNA-dependent protein kinase (DNA-PK) in untreated controls and SN38-treated cells under normoxic and hypoxic conditions. (C) Western blots comparing the Rad51 and γ-H2AX levels with and without SN38 treatment in untransfected, mock siRNA-transfected and RAD51 siRNA-transfected PC3 cells under hypoxia. (D) Percentage of apoptosis as detected by flow cytometry in siRNA-transfected as well as untransfected PC3 cells under hypoxia. β-Actin was used as the loading control. Three replicates were performed for each experiment. Each column represents the mean and S.E.M. of three independent experiments. *P < 0.05.
Whether blocking Rad51 expression could resensitize metastatic castration-resistant prostate cancer cells to the topoisomerase I inhibitor under hypoxia was then studied. As shown in Fig. 2C, Rad51 protein levels can be effectively decreased with siRNA. Compared with untreated PC3 cells under hypoxia, higher levels of Rad51 were observed after SN38 treatment in untransfected and mock siRNA transfected PC3 cells, but not RAD51 siRNA transfected PC3 cells. DNA damage/γ-H2AX levels (Fig. 2C) and apoptosis (Fig. 2D) after SN38 treatment under hypoxia were significantly increased after blocking Rad51 upregulation with siRNA. These data indicate that the resistance of prostate cancer cells to drug-induced DNA damage under hypoxia requires upregulation of Rad51 and Rad51-mediated DNA repair.
PARP Inhibitor Veliparib Overcomes the Resistance of p53 Mutant Prostate Cancer Cells to Topoisomerase I Inhibitor under Hypoxia and Enhances Its Antitumor Activities.
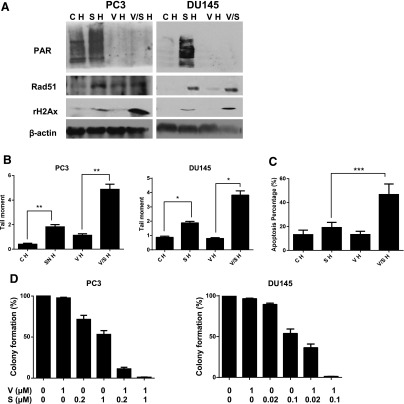
Accumulating data support the synergy between PARP inhibitor and topoisomerase I inhibitor under normoxia (Smith et al., 2005; Zhang et al., 2011; Patel et al., 2012). We therefore tested whether adding veliparib can resensitize p53 mutant prostate cancer cells to SN38 under hypoxia. As shown by the diminished PAR level, 1 μM of veliparib was sufficient to inhibit PARP activity under hypoxia (Fig. 3A). When DNA damage was assessed via the γ-H2AX levels in Western blot and tail moment in a neutral comet assay, veliparib as a single agent caused minimum DNA damage. Combining veliparib with SN38 significantly enhanced the DNA damage induced by SN38 under hypoxia (Fig. 3, A and B).
Fig. 3.
Veliparib enhances the antitumor effects of SN38 under hypoxia. (A) Western blots comparing levels of poly(ADP) ribose (PAR) and γ-H2AX in treated and untreated cells under hypoxia (H). CH, untreated control; SH, SN38; VH, veliparib; V/SH, veliparib plus SN38. (B) Neutral comet assay comparing DNA damage/tail moment in treated and untreated cells under hypoxia. The tail moment (y-axis) incorporated measurements of both the smallest detectable size of migrating DNA (reflected in the comet tail length) and the number of relaxed/broken pieces (represented by the intensity of DNA in the tail). Each column represents the mean and S.E.M. of tail moments of 50 cells. (C) Percentage of apoptosis as detected by flow cytometry in treated and untreated PC3 cells under hypoxia. (D) Clonogenic assay comparing the effects of hypoxia and 16-hour drug treatment on colony formation of PC3 and DU145 cells. The colony numbers in untreated controls were set as 100%. The colony numbers of each treatment group were divided by those of untreated control, and the percentages were reflected in y-axes. β-Actin was used as the loading control. Three replicates were performed for each experiment. Each column represents the mean and S.E.M. of three independent experiments. C, untreated control; H, hypoxia; S, SN38; V, veliparib; V/S, veliparib plus SN38. *P < 0.05; **P < 0.01; ***P < 0.001.
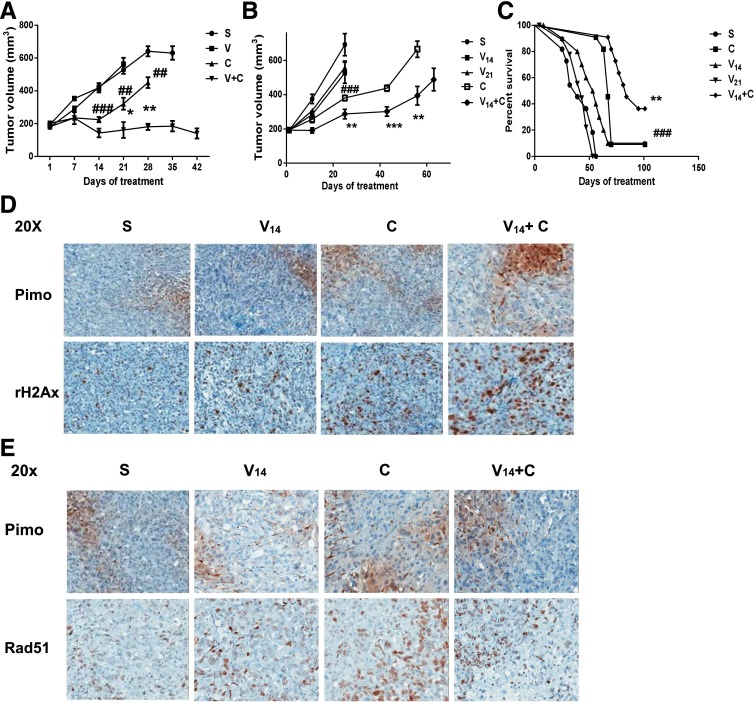
The antitumor activities of veliparib, SN38, and their combination were then assessed in vitro and in vivo. Although veliparib had no single-agent antitumor activities, its combination with SN38 significantly enhanced apoptosis (Fig. 3C) and decreased colony formation (Fig. 3D) in PC3 and DU145 cells compared with those treated with the single agent SN38. Compared with untreated cells, a dose-dependent decrease in colony formation after SN38 treatment was also observed (Fig. 3D). Based on published xenograft studies with CPT-11, we initially used a 60-mg/kg intraperitoneal injection on day 1 and day 5 of every 21-day cycle. Although adding veliparib oral gavages at 12.5 mg/kg on weekdays to CPT-11 led to decreased tumor volume (Fig. 4A), this schedule (schedule A) was not well tolerated. Two of the eight mice in the combination treatment group were excluded in the analysis after their early death on day 35 (day 14 of cycle 2). We then modified the schedule (schedule B) based on the phase I study combining veliparib with CPT-11 in nonprostate tumors (LoRusso et al., 2011). In Fig. 4B, CPT-11 was given on days 1 and 8 of every 21-day cycle. Two veliparib schedules were tested: twice daily (V21 in Fig. 4) and twice daily for 14 days of each 21-day cycle (V14 in Fig. 4). Compared with the growth curves and survival curves of the saline control, the single agent CPT-11 significantly inhibited the growth of PC3 xenografts (Fig. 4B) and prolonged the survival of the mice (Fig. 4C). Consistent with the in vitro data, veliparib had minimal single-agent activities at the doses and schedules tested. Adding valiparib to CPT-11 significantly enhanced the growth-inhibition and survival benefits of CPT-11 (Fig. 4, B and C).
Fig. 4.
Veliparib enhances DNA damage and the antitumor effects of CPT-11 in vivo. (A) Growth curves for PC3 mouse xenografts treated with schedule A as described in Results. C, CPT-11; S, saline control; V, veliparib; V + C, veliparib plus CPT-11 combination. For each treatment group, the growth curve terminated on the day when the first xenograft in the group reached the 1000-mm3 size. Each tumor volume data point represents the mean and S.E.M. of eight mice. Significant data points were labeled with # in the CPT-11 versus saline comparison and labeled with * in the valiparib + CPT-11 versus CPT-11 comparison. (B) Growth curves for the PC3 mouse xenografts treated with schedule B as described in Results. Each tumor volume data point represents the mean and S.E.M. of 10 mice. Significant data points were labeled with # in the CPT-11 versus saline comparison and labeled with * in the valiparib + CPT-11 versus CPT-11 comparison. (C) Survival curve for the PC3 mouse xenografts treated with schedule B. The median survival of each treatment group was compared, and the log-rank test was used for survival rate analysis. (D) Comparison of IHC positivity and distributions of γ-H2AX (brown staining, lower panels) and hypoxia/pimonadazole (brown staining, upper panels) among PC3 xenograft sections treated with schedule B. (E) Rad51 (brown staining, lower panels) was detected in both pimonidazole-positive (brown staining, upper panels) and pimonidazole-negative areas on the PC3 xenograft IHC sections treated with schedule B. **/##P < 0.01; ***/###P < 0.001.
The expression of markers for intratumor hypoxia (pimonadazole), DNA damage (γ-H2AX), and repair (Rad51) were then studied with IHC on the PC3 xenograft sections. As shown by the IHC image of γ-H2AX positivity and intensity, adding veliparib enhanced the DNA damage caused by CPT-11 (Fig. 4D). Most of the DNA damage and γ-H2AX staining in the CPT-11–treated xenograft were in the normoxic/pimonadazole negative area, whereas the DNA damage and γ-H2AX staining were seen in both the hypoxic/pimonadole-positive and normoxic/pimonidazole-negative areas in the valiparib and CPT-11 treatment group (Fig. 4D, C versus V14 + C). Moreover, positive Rad51 staining was noted in both the hypoxic and normoxic regions in the valiparib and CPT-11 treatment groups (Fig. 4E). This is consistent with our in vitro finding that adding veliparib overcomes the resistance of p53 mutant prostate cancer cells to a topoisomerase I inhibitor under hypoxia in the presence of elevated Rad51.
Transcriptional Regulation of Rad51 by E2F1 upon DNA Damage under Hypoxia.
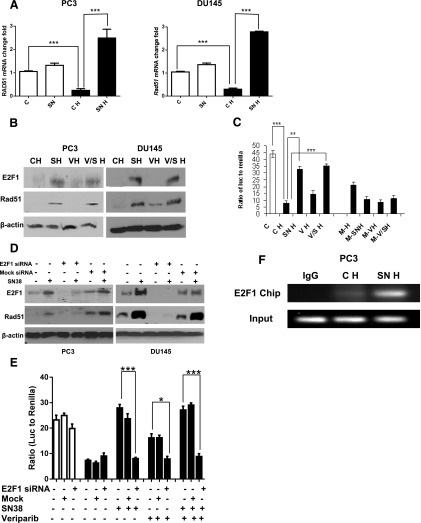
Among the E2F family of transcription factors, E2F4 has been shown to suppress the transcription of RAD51 under hypoxia by binding to the E2F4 site in the proximal promoter of the RAD51 gene (Bindra and Glazer, 2007). Because E2F1 shares a consensus binding sequence with E2F4 and both E2F1 and E2F4 can bind to the RAD51 promoter (Kachhap et al., 2010), we then studied the role of E2F1 in regulating Rad51 expression under hypoxia. Compared with untreated cells under hypoxia, more than 8-fold upregulation of RAD51 RNAs was observed after a 16-hour treatment with SN38 under hypoxia (Fig. 5A, SNH versus CH). The increase of Rad51 after treatments with SN38 and SN38 plus veliparib was associated with elevated levels of E2F1 (Fig. 5B). Consistent with reduced RAD51 mRNA and protein under hypoxia, RAD51 promoter activity was significantly suppressed in untreated PC3 cells under hypoxia (Fig. 5C). This promoter activity increased more than 3-fold after treating hypoxic PC3 cells with SN38 or veliparib plus SN38 (Fig. 5C, SNH and V/SH versus CH). Such increase in RAD51 promoter activity was blocked when the E2F1 and E2F4 consensus binding site on the RAD51 promoter was mutated (Fig. 5C, M-SNH versus SNH and M-V/SH versus V/SH). Consistent with E2F4’s role in downregulating the RAD51 promoter (Bindra and Glazer, 2007), mutating the E2F1 and E2F4 consensus binding site led to increased RAD51 promoter activity in untreated PC3 cells under hypoxia (Fig. 5C, M-H versus CH). Of note, veliparib treatment also increased RAD51 promoter activity and protein under hypoxia, and this increase was blocked after the E2F binding site was mutated.
Fig. 5.
Transcriptional regulation of Rad51 by E2F1. (A) Quantitative real-time PCR comparing RAD51 mRNA levels in treated and untreated PC3 and DU145 cells under hypoxia (black bars) and normoxia (white bars). The relative abundance of RAD51 mRNA was calculated using glyceraldehyde 3-phosphate dehydrogenase as internal control. (B) Western blots comparing E2F1 and Rad51 levels after 1 day of drug treatment under hypoxia. (C) Dual luciferase reporter assay show the enhanced RAD51 promoter activities in PC3 cells after 16 hours of drug treatment under hypoxia (black bars); such upregulated promoter activities were blocked after mutating the E2F1-binding site on the RAD51 promoter (M-SNH versus SNH and M-V/SH versus V/SH). Ratios of firefly luciferase versus Renilla luciferase activities are shown. (D) E2F1 siRNA decreased E2F1 expression and blocked the increase in RAD51 protein after SN38 treatment under hypoxia (E) Dual luciferase reporter assay comparing RAD51 promoter activities in siRNA transfected or untransfected PC3 cells under different treatments and oxygen. White bars, normoxia; black bars, hypoxia. (F) ChIP assays showing increased E2F1 occupancy at the RAD51 proximal promoter after SN38 treatment under hypoxia. C, untreated control; CH, control untreated under hypoxia; M, mutated E2F consensus binding site on the RAD51 promoter; M-H, untreated cells with mutated E2F consensus binding site on the RAD51 promoter; SN, SN38; SNH, SN38 treated under hypoxia; VH, veliparib treated under hypoxia; SNH, SN38 treated under hypoxia; V/SH, veliparib plus SN38 under hypoxia. β-Actin was used as the loading control. Three replicates were performed for each experiment. Each column represents the mean ± S.E.M. of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
E2F1 siRNA was then used to study the regulation of Rad51 by E2F1 in PC3 and DU145 cells under hypoxia. Compared with untransfected or mock siRNA transfected cells, E2F1 protein was significantly reduced by siRNA against E2F1 (Fig. 5D). Transfection with siRNA against E2F1 blocked the upregulation of RAD51 promoter activity (Fig. 5E) and Rad51 protein (Fig. 5D) upon treatment of hypoxic cells with SN38, veliparib, or their combination. To study the role of endogenous E2F1 in regulating Rad51, we performed a ChIP assay with an anti-E2F1 pull down after PC3 cells had been treated with SN38 under hypoxia. Compared with untreated cells under hypoxia, enhanced E2F1 binding to the RAD51 promoter was detected after SN38 treatment (Fig. 5F). These data support the role of E2F1 in transactivating the RAD51 promoter upon drug-induced DNA damage under hypoxia.
Wild-Type p53 Suppressed RAD51 Transcription and Sensitized Prostate Cancer Cells to Drug-Induced DNA Damage under Hypoxia.
Unlike p53 null PC3 and p53 mutant DU145 cells, p53 wild-type LNCaP cells were sensitive to SN38-induced DNA damage and apoptosis (Fig. 1). Similar to the results observed with LNCaP, lack of Rad51 upregulation after SN38 treatment under hypoxia was observed in other cell lines with wild-type p53 (A549, HCT116, and MCF7) (data not shown).
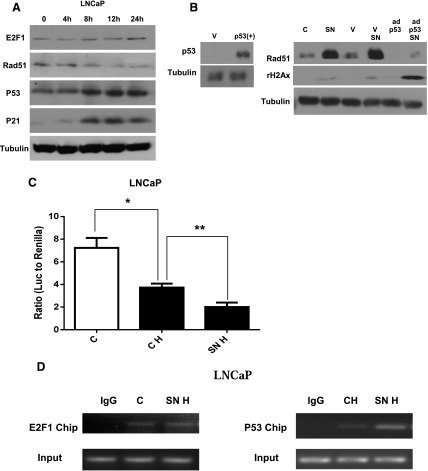
To delineate the role of endogenous p53 and E2F1 in regulating Rad51 expression, the expression of E2F1, Rad51, p53 and its downstream effector p21 were compared at different time points after SN38 treatment in LNCaP cells under hypoxia. Increasing levels of wild-type p53 and its downstream effector p21 were associated with decreasing Rad51 levels whereas no significant changes in E2F1 levels were observed (Fig. 6A). Wild-type p53 was then ectopically expressed in the p53 null PC3 cells. Compared with untransfected or vector transfected PC3 cells, restoration of p53 expression suppressed the upregulation of Rad51 and increased DNA damage after SN38 treatment under hypoxia (Fig. 6B).
Fig. 6.
p53 suppresses Rad51 transcription and sensitizes prostate cancer cells to drug-induced DNA damage. (A) Western blots showing levels of E2F1, Rad51, p53, and p21 at different time points (0, 4, 8, 12, and 24 hours) after SN38 treatment in LNCaP cells under hypoxia. (B) Western blots show the restoration of wild-type p53 in PC3 cells after infection with p53/GFP adenovirus 5 (ad-p53). Levels of rH2Ax and Rad51 in PC3 cells with or without SN38 treatment under hypoxia were compared among cells that were uninfected or infected with GFP adenovirus 5 vector (V) or p53/GFP adenovirus 5 (ad-p53) infected cells. (C) Dual luciferase reporter assays of RAD51 promoter activities in LNCaP with or without 16 hours of SN38 treatment under hypoxia. (D) ChIP assays comparing RAD51 promoter occupancy by E2F1 and p53 with or without SN38 treatment under hypoxia. C, untreated control; CH, control untreated under hypoxia; SN, SN38; SNH, SN38 treated under hypoxia. Tubulin was used as the loading control. Three replicates were performed for each experiment. Each column represents the mean and S.E.M. of three independent experiments. *P < 0.05; **P < 0.01.
Reporter and ChIP assays were then performed to study the regulation of RAD51 by p53 under hypoxia. Consistent with the decreasing Rad51 protein observed in Fig. 6A, RAD51 promoter activity was suppressed after SN38 treatment compared with untreated hypoxic LNCaP cells (Fig. 6C). When the occupancy of RAD51 promoter by E2F1 and p53 were studied with the ChIP assays in hypoxic LNCaP cells, increased binding of RAD51 promoter by p53, but not E2F1, was observed after SN38 treatment (Fig. 6D). These data indicate that wild-type p53 plays a dominant negative role in regulating RAD51 transcription in response to drug-induced DNA damage under hypoxia.
Discussion
Despite recent advances in androgen-deprivation therapy, immunotherapy, radiopharmaceuticals, and chemotherapy, metastatic castration-resistant prostate cancer remains an incurable disease (Liu and Zhang, 2013). Several mechanisms of therapy resistance have been studied (Seruga et al., 2011), and intratumoral hypoxia has been proposed to create a “mutator” phenotype with increased genomic instability and drug resistance (Bristow and Hill, 2008). Studies with p53 wild-type lung (A549, H460), breast (MCF7), and colorectal (HCT116, RKO) cell lines have shown that downregulation of DNA repair proteins under hypoxia sensitize these cells to the PARP inhibitor and DNA-damaging agents (Chan et al., 2010). Here, we show that prostate cancer cells with nonfunctional p53 are resistant to veliparib and the topoisomerase I inhibitor under hypoxia through transactivating Rad51 expression by E2F1. Our data support that hypoxia contributes to drug resistance and that such resistance in p53 mutant cells is not due to diminished but rather more effective DNA repair when there is a lack of Rad51 suppression by p53 and a lack of additional DNA damage from reactive oxygen species generated under normoxia. Our preliminary data with molecular karyotyping also have shown that 3 weeks of hypoxia culture and veliparib treatment do not enhance the genomic instability of PC3 or DU145 cells compared with their untreated counterparts grown under normoxia (J. Zhang, unpublished data). This lack of additional chromosomal copy number changes under hypoxia and PARP inhibitor treatment is at least partly due to the preservation of the DNA-damage repair response in these p53 mutant cells. Our data, therefore, provide a mechanism other than genomic instability to explain the resistance of p53 mutant cancer cells to drug-induced DNA damage under hypoxia.
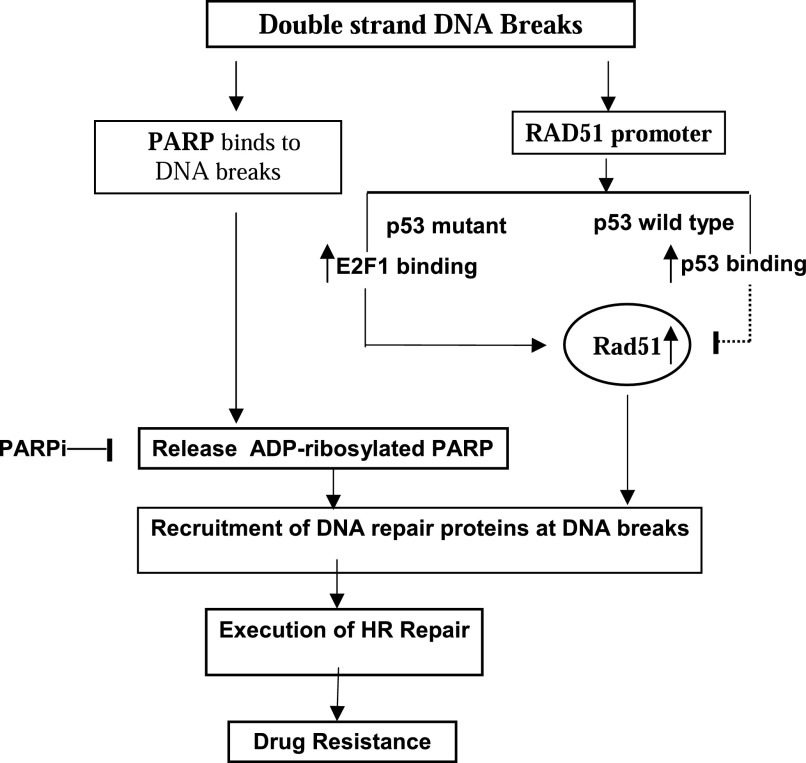
As shown in our proposed model (Fig. 7), there is a dynamic regulation of RAD51 promoter activity by p53 and E2F1 after drug-induced double-strand DNA breaks. In p53 wild-type prostate cancer cells, both E2F1 and p53 can bind to the RAD51 promoter under hypoxia. Upon drug-induced DNA damage, increased binding of endogenous p53 to the RAD51 promoter abolishes the positive regulation of RAD51 by E2F1. Without Rad51, the cells cannot execute homologous recombination repair and will die from unrepaired DNA damage. In p53 mutant prostate cancer cells, enhanced binding of E2F1 to the RAD51 promoter facilitated Rad51 mediated homologous recombination repair and led to drug resistance. Of note, Rad51 overexpression was frequently observed in cancers harboring mutant p53 (Klein, 2008). The suppression of RAD51 transcription by p53 provides a likely explanation.
Fig. 7.
Schematic diagram of the response of prostate cancer cells to drug-induced double-strand DNA breaks under hypoxia. Downregulation of the RAD51 promoter by p53 undermines homologous recombination repair and leads to cell death in cells with wild-type p53 under hypoxia. Upregulation of the RAD51 promoter by E2F1 in p53 mutant cells facilitates DNA repair and drug resistance. Such resistance can be overcome by adding PAPR inhibitor (PARPi), which traps PARP at DNA breaks and blocks access of DNA-repair proteins, such as Rad51. PAPRi by itself was not sufficient to cause detrimental DNA double-strand breaks to prostate cancer cells under hypoxia.
Adding veliparib to SN38 or CPT-11 reversed the resistance of p53 mutant prostate cancer cells to DNA damage and led to cell death in vitro and inhibition of xenograft growth in vivo. Veliparib has been shown to sensitize human colon cancer, lung cancer, and glioma cells as well as B16F10 melanoma and MX-1 breast cancer xenografts to topoisomerase I inhibitors (Smith et al., 2005; Zhang et al., 2011; Patel et al., 2012). Enhanced DNA damage and antitumor activities were also reported in phase I studies with the veliaprib-topotecan combination (Kummar et al., 2011) and the veliparib-irinotecan combination (LoRusso et al., 2011). Of note, neither phase I studies included prostate cancer patients. To the best of our knowledge, our study is the first to report such activities under hypoxic condition.
Several studies have explored the mechanisms underlying the synergies between PARP inhibitors and topoisomerase I inhibitors. Using a panel of DNA repair-deficient Chinese hamster ovary cells, Smith et al. (2005) reported that the PARP inhibitor AG14361 [1-(4-dimethyl-aminomethyl-phenyl)-8,9-dihydro-7H-2,7,9a-benzo[cd]azulen-6-one] significantly potentiated CPT-mediated cytotoxicity in all cells except in the base excision repair-deficient EM9 cells. PARP-1–dependent base excision repair was thought to be involved in the repair of DNA damage caused by the topoisomerase I inhibitor. In contrast, Patel et al. (2012) recently reported that transfecting catalytically inactive PARP-1 (E988K) or transfecting the PARP-1 DNA-binding domain alone to PARP1−/− mouse embryonic fibroblasts sensitized cells to topoisomerase I inhibitor, with cells not further sensitized by veliparib. PARP inhibition was therefore proposed to convert PARP-1 into a protein that binds to topoisomerase I–induced DNA damage, preventing its normal repair. This model of PARP inhibitor–induced trapping of PARP at damaged DNA was also supported by two other recent publications (Murai et al., 2012, 2014). Given that Rad51 remained elevated in PC3, DU145 cells, and PC3 xenografts treated with veliparib and topoisomerase I inhibitor, failure to release PARP after PARP inhibition can block the access of Rad51 to damaged DNA and is the likely explanation for the enhanced DNA damage observed in p53 mutant prostate cancer cells with this combination (Fig. 7).
As a key DNA damage checkpoint gene, loss of functional p53 is relatively common in both primary and metastatic prostate cancer (Barbieri et al., 2012; Grasso et al., 2012). The exomic sequencing data reported by Grasso et al. (2012) reported two point mutations and two frame shift mutations in 11 treatment-naïve high-grade primary prostate cancers (36% mutation frequency) and 14 point mutations and 5 frame shift mutations in 50 heavily treated lethal castration-resistant prostate cancers (38% mutation frequency). Copy number loss of p53 was also observed in 9 of these 50 lethal cases. Thus, prostate cancer cells with mutant p53 not only can evade apoptosis but likely will have more effective homologous recombination repair due to lack of suppression of RAD51 transcription by wild-type p53. Although none of the DNA-damaging chemotherapy agents has been approved for the treatment of prostate cancer, a platinum-based chemotherapy regimen demonstrated a 16-month median overall survival in a phase 2 study of 120 patients who met the predefined criteria of anaplastic prostate cancers (Aparicio et al., 2013). Anaplastic prostate cancer shares several features of small-cell prostate cancer and is likely enriched with cell cycle checkpoint alterations, based on its rapid proliferation. Our data highlight the importance of p53 in determining the sensitivity of prostate cancer cells to DNA damage and provide the preclinical rationale for testing the PARP inhibitor-irinotecan combination in selected patients with prostate cancer that has mutant p53 and anaplastic features.
Acknowledgments
The authors thank Abbott Laboratories for providing veliparib, and Dr. Peter Glazer at Yale University for providing the RAD51 promoter PGL3 luciferase reporter constructs.
Abbreviations
- AG14361
1-(4-dimethyl-aminomethyl-phenyl)-8,9-dihydro-7H-2,7,9a-benzo[cd]azulen-6-one
- ChIP
chromatin immunoprecipitation
- CPT-11
camptothecin-11
- GFP
green fluorescent protein
- IHC
immunohistochemistry
- PARP
poly(ADP-ribose) polymerase
- PCR
polymerase chain reaction
- siRNA
small interfering RNA
- SN38
7-ethyl-10-hydroxycamptothecin; veliparib, 2-[(2R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide, dihydrochloride
Authorship Contributions
Participated in research design: Zhang, Wu, Pienta.
Conducted experiments: Wu, Wang, Mcgregor.
Performed data analysis: Wu, Zhang.
Wrote or contributed to the writing of the manuscript: Zhang, Wu.
Footnotes
This work was supported by the National Institutes of Health National Cancer Institute Moffitt Cancer Center Support Grant for new faculty recruitment (P30-CA076292-11S5); and the Conquer Cancer Foundation 2010 Young Investigator Award of American Society of Clinical Oncology (EG ID 1083).
References
- Aparicio AM, Harzstark AL, Corn PG, Wen S, Araujo JC, Tu SM, Pagliaro LC, Kim J, Millikan RE, Ryan C, et al. (2013) Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin Cancer Res 19:3621–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. (2012) Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44:685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. (2007) Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene 26:2048–2057 [DOI] [PubMed] [Google Scholar]
- Bindra RS, Schaffer PJ, Meng A, Woo J, Måseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, Glazer PM. (2004) Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol 24:8504–8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J, et al. (2011) Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell 19:664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristow RG, Hill RP. (2008) Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer 8:180–192 [DOI] [PubMed] [Google Scholar]
- Brown ET, Holt JT. (2009) Rad51 overexpression rescues radiation resistance in BRCA2-defective cancer cells. Mol Carcinog 48:105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N, Pires IM, Bencokova Z, Coackley C, Luoto KR, Bhogal N, Lakshman M, Gottipati P, Oliver FJ, Helleday T, et al. (2010) Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res 70:8045–8054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, et al. (2009) Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361:123–134 [DOI] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. (2012) The mutational landscape of lethal castration-resistant prostate cancer. Nature 487:239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T. (2010) Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis 31:955–960 [DOI] [PubMed] [Google Scholar]
- Kachhap SK, Rosmus N, Collis SJ, Kortenhorst MS, Wissing MD, Hedayati M, Shabbeer S, Mendonca J, Deangelis J, Marchionni L, et al. (2010) Downregulation of homologous recombination DNA repair genes by HDAC inhibition in prostate cancer is mediated through the E2F1 transcription factor. PLoS ONE 5:e11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL. (2008) The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 7:686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummar S, Chen A, Ji J, Zhang Y, Reid JM, Ames M, Jia L, Weil M, Speranza G, Murgo AJ, et al. (2011) Phase I study of PARP inhibitor ABT-888 in combination with topotecan in adults with refractory solid tumors and lymphomas. Cancer Res 71:5626–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SA, Roques C, Magwood AC, Masson JY, Baker MD. (2009) Recovery of deficient homologous recombination in Brca2-depleted mouse cells by wild-type Rad51 expression. DNA Repair (Amst) 8:170–181 [DOI] [PubMed] [Google Scholar]
- Liu JJ, Zhang J. (2013) Sequencing systemic therapies in metastatic castration-resistant prostate cancer. Cancer Control 20:181–187 [DOI] [PubMed] [Google Scholar]
- LoRusso P, Ji JJ, Li J, Heilbrun LK, Shapiro G, Sausville EA, Boerner SA, Smith DW, Pilat MJ, Zhang J, et al. (2011) Phase I study of the safety, pharmacokinetics (PK), and pharmacodynamics (PD) of the poly(ADP-ribose) polymerase (PARP) inhibitor veliparib (ABT-888; V) in combination with irinotecan (CPT-11; Ir) in patients (pts) with advanced solid tumors (Abstract). J Clin Oncol 29 (Suppl):3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. (2007) RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res 67:9658–9665 [DOI] [PubMed] [Google Scholar]
- Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, Waldman T, Lord CJ, Ashworth A. (2009) Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med 1:315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng AX, Jalali F, Cuddihy A, Chan N, Bindra RS, Glazer PM, Bristow RG. (2005) Hypoxia down-regulates DNA double strand break repair gene expression in prostate cancer cells. Radiother Oncol 76:168–176 [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11:196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Huang SY, Renaud A, Zhang Y, Ji J, Takeda S, Morris J, Teicher BA, Doroshow JH, Pommier Y. (2014) Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther 13:433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. (2012) Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res 72:5588–5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AG, Flatten KS, Schneider PA, Dai NT, McDonald JS, Poirier GG, Kaufmann SH. (2012) Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes. J Biol Chem 287:4198–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu SK, Schelman WR, Wilding G, Moreno V, Baird RD, Miranda S, Hylands L, Riisnaes R, Forster M, Omlin A, et al. (2013) The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol 14:882–892 [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. (1997) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88:265–275 [DOI] [PubMed] [Google Scholar]
- Seruga B, Ocana A, Tannock IF. (2011) Drug resistance in metastatic castration-resistant prostate cancer. Nat Rev Clin Oncol 8:12–23 [DOI] [PubMed] [Google Scholar]
- Smith LM, Willmore E, Austin CA, Curtin NJ. (2005) The novel poly(ADP-Ribose) polymerase inhibitor, AG14361, sensitizes cells to topoisomerase I poisons by increasing the persistence of DNA strand breaks. Clin Cancer Res 11:8449–8457 [DOI] [PubMed] [Google Scholar]
- Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, et al. (2010) Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376:235–244 [DOI] [PubMed] [Google Scholar]
- Yang Z, Waldman AS, Wyatt MD. (2012) Expression and regulation of RAD51 mediate cellular responses to chemotherapeutics. Biochem Pharmacol 83:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TA, Sandhu SK, Carden CP, de Bono JS. (2011) Poly(ADP-ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA Cancer J Clin 61:31–49 [DOI] [PubMed] [Google Scholar]
- Zhang J, Hu S, Schofield DE, Sorensen PH, Triche TJ. (2004) Selective usage of D-Type cyclins by Ewing’s tumors and rhabdomyosarcomas. Cancer Res 64:6026–6034 [DOI] [PubMed] [Google Scholar]
- Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. (2011) Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res 39:3607–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]