Abstract
The objective of the present study was to delineate the mechanisms of GC-A/natriuretic peptide receptor-A (GC-A/NPRA) gene (Npr1) expression in vivo. We used all-trans retinoic acid (ATRA) and histone deacetylase (HDAC) inhibitor, sodium butyrate (NaBu) to examine the expression and function of Npr1 using gene-disrupted heterozygous (1-copy; +/−), wild-type (2-copy; +/+), and gene-duplicated heterozygous (3-copy; ++/+) mice. Npr1+/− mice exhibited increased renal HDAC and reduced histone acetyltransferase (HAT) activity; on the contrary, Npr1++/+ mice showed decreased HDAC and enhanced HAT activity compared with Npr1+/+ mice. ATRA and NaBu promoted global acetylation of histones H3-K9/14 and H4-K12, reduced methylation of H3-K9 and H3-K27, and enriched accumulation of active chromatin marks at the Npr1 promoter. A combination of ATRA-NaBu promoted recruitment of activator-complex containing E26 transformation–specific 1, retinoic acid receptor α, and HATs (p300 and p300/cAMP response element–binding protein-binding protein–associated factor) at the Npr1 promoter, and significantly increased renal NPRA expression, GC activity, and cGMP levels. Untreated 1-copy mice showed significantly increased systolic blood pressure and renal expression of α-smooth muscle actin (α-SMA) and proliferating cell nuclear antigen (PCNA) compared with 2- and 3-copy mice. Treatment with ATRA and NaBu synergistically attenuated the expression of α-SMA and PCNA and reduced systolic blood pressure in Npr1+/− mice. Our findings demonstrate that epigenetic upregulation of Npr1 gene transcription by ATRA and NaBu leads to attenuation of renal fibrotic markers and systolic blood pressure in mice with reduced Npr1 gene copy number, which will have important implications in prevention and treatment of hypertension-related renal pathophysiological conditions.
Introduction
Cardiac hormones, atrial natriuretic peptides (ANPs), and brain natriuretic peptides elicit natriuretic/diuretic, vasorelaxant, and antimitogenic responses, largely directed to the reduction of blood pressure and blood volume (de Bold, 1985; Levin et al., 1998; Pandey, 2008). GC-A/natriuretic peptide receptor-A (GC-A/NPRA) is one of the principal loci involved in the regulatory actions of ANPs and brain natriuretic peptides, which produces the intracellular second messenger cGMP in response to hormone binding (Pandey and Singh, 1990; Drewett and Garbers, 1994). The ANP/cGMP signaling through its downstream effector proteins, including cGMP-dependent protein kinases, phosphodiesterases, and cyclic nucleotide–gated ion channels, mediates the cellular and physiologic effects of NPRA (Garbers et al., 2006; Pandey, 2011). Gene targeting and functional expression studies of the Npr1 gene (coding for GC-A/NPRA) have shown the hallmark significance of this receptor protein in providing protective effects against renal and cardiac hypertrophic and fibrotic growth, extracellular matrix deposition, and cell proliferative responses (Vellaichamy et al., 2005; Li et al., 2009; Das et al., 2010). Previous studies have indicated an association of Npr1 gene variants with left ventricular mass index and septal wall thickness in human essential hypertension (Pitzalis et al., 2003; Rubattu et al., 2006). Moreover, a longer thymidine adenine repeat unit in the spontaneously hypertensive rat model has been shown to regulate Npr1 gene transcription, thus affecting diastolic blood pressure (Tremblay et al., 2003). The activity and expression of NPRA, assessed primarily through ANP-stimulated cGMP production, are mediated by agents, including autoregulation involving natriuretic peptides, osmotic sensors, and other regulatory hormones (Garg and Pandey, 2003, 2005; Chen et al., 2004; Arise and Pandey, 2006). However, the exact mechanisms of Npr1 gene expression and regulation are not yet clearly understood.
Retinoic acid (RA) is a group of derivatives of vitamin A, including all-trans RA (ATRA), 9-cis RA, and 13-cis RA, which play critical regulatory roles in various physiologic and pathophysiological conditions of numerous organ systems (Zile, 2010; Lee and Jeong, 2012). A considerable number of studies have shown that RA and its receptor agonists protect the structure and function of kidneys in numerous animal models of renal diseases, ascribing to its anti-inflammatory and antiproliferative properties (Schaier et al., 2004; Liu et al., 2008). The physiologic effects of retinoic acid are mediated by binding to retinoic acid receptors (RARs) and retinoid X receptors, which interact with a multitude of coregulators, chromatin modifiers, and transcription machinery (Wei, 2003; Bastien and Rochette-Egly, 2004). Chromatin remodeling by post-translational modifications of histone tails, including acetylation, methylation, and phosphorylation, plays critical roles in regulating gene expression and function by controlling the availability of key regulatory elements to chromatin (Latham and Dent, 2007; Lennartsson and Ekwall, 2009).
Epigenetic mechanisms including changes in histone acetylation/deacetylation have been shown to be involved in altered gene expression under various pathologic conditions (Marumo et al., 2010; Lee et al., 2012). Inhibition of histone deacetylases (HDACs) by sodium butyrate (NaBu), trichostatin A (TSA), and valproic acid has shown improved cardiovascular and renal disease conditions (Bhaumik et al., 2007; Bush and McKinsey, 2010). Recently, it has been indicated that NaBu attenuated gentamicin-induced nephrotoxicity by enhancing renal antioxidant enzyme activity and expression of prohibitin protein (Sun et al., 2013). Moreover, hyaluronan mixed esters of butyric and retinoic acid have also been shown to enhance cardiac repair in infarcted rat and pig hearts (Ventura et al., 2007; Simioniuc et al., 2011). Previously, we have reported that ATRA induces Npr1 gene transcription in cultured mouse mesangial cells (MMCs) in association with E26 transformation–specific 1 (Ets-1), specificity protein 1 (Sp1), and histone acetylation (Kumar et al., 2010). However, the cross-talk between retinoid signaling and other regulatory factors including chromatin modifiers that modulate the expression and regulation of Npr1 gene under in vivo conditions is still unknown. In the present study, we examined the effect of ATRA- and NaBu-mediated regulation of Npr1 gene transcription and expression in gene-targeted mutant mouse models. Furthermore, we have analyzed the epigenetic marks including histone H3 modifications, effected by ATRA and NaBu in controlling the Npr1 gene transcription and expression, an important player in prevention of renal pathophysiology and hypertension.
Materials and Methods
Colorimetric histone acetylation and methylation quantification kits, global dimethyl histone H3-K9 (H3-K9me2), trimethyl histone H3-K9 (H3-K9me3), trimethyl histone H3-K4 (H3-K4me3), trimethyl histone H3-K27 (H3-K27me3), acetyl histone H3-K9 (H3-K9ac), acetyl histone H4-K12 (H4-K12ac), histone acetyltransferase (HAT) activity/inhibition assay kit, histone purification kit, and antibodies for p300 and H3-K9me3 were purchased from Epigentek (Brooklyn, NY). RNeasy mini-kit for total RNA isolation, RT2 First Strand cDNA kit, and RT2 SYBR Green/ROX master mix were obtained from Qiagen (Valencia, CA). The chromatin immunoprecipitation (ChIP)-IT Express enzymatic kit, HDAC assay kit, and H3-K4me3 were purchased from Active Motif (Carlsbad, CA). All-trans retinoic acid and NaBu were purchased from Sigma-Aldrich (St. Louis, MO), and ATRA-NaBu hybrid drug was obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Generation of Npr1 Gene-Targeted Mice.
Npr1 gene-disrupted and gene-duplicated mice were produced by homologous recombination in embryonic stem cells as described previously (Oliver et al., 1997, 1998; Pandey et al., 1999). The mice were bred and maintained at the Tulane University Health Sciences Center animal facility. Animals were handled under protocols approved by the Institutional Animal Care and Use Committee. The mouse colonies were housed under 12-hour light/dark cycles at 25°C and fed regular chow (Purina Laboratory, Framingham, MA) and tap water ad libitum. All animals were littermate progeny of the C57/BL6 genetic background and were designated as Npr1 gene-disrupted heterozygous (Npr1+/−, 1-copy), wild-type (Npr1+/+, 2-copy), and gene-duplicated heterozygous (Npr1++/+, 3-copy) mice. All protocols were approved by the Institutional Animal Care and Use Committee at Tulane University Health Sciences Center.
ATRA and HDAC Inhibitor Treatment.
The stock solutions of ATRA and ATRA-NaBu hybrid drug were prepared at 20- and 10-mg/ml concentrations in dimethylsulfoxide, respectively, and stored at −70°C. On the day of injection, the drugs were thawed, diluted with olive oil to appropriate concentrations, vortexed for 2 minutes at room temperature, and administered intraperitoneally. NaBu was prepared at 20 mg/ml in phosphate-buffered saline (PBS; pH 7.4) and injected intraperitoneally. Control groups were injected with vehicle (dimethylsulfoxide and olive oil). Twenty-four-week age-matched male adult 1-copy (n = 32), 2-copy (n = 32), and 3-copy (n = 32) mice were used in the following treatment groups: group I (n = 8), vehicle-treated (control); group II (n = 8), ATRA-treated (0.5 mg/kg/day); group III (n = 8), NaBu-treated (0.5 mg/kg/day); and group IV (n = 8), ATRA-NaBu combination–treated (1 mg/kg/day). Animals were injected with the drugs intraperitoneally for 2 weeks and assays were performed.
Blood Pressure Analysis.
Systolic blood pressures were measured by a noninvasive computerized tail-cuff method using Visitech 2000 (Visitech Systems, Apex, NC) and calculated as the average of three to five sessions per day for 5 consecutive days as described previously (Shi et al., 2001). All mice were first trained for 7 days (for acclimatization) and actual blood pressure was measured on and from the 11th day till the 15th day of the treatment.
Tissue Collection.
Mice were euthanized by administration of a high concentration of CO2. Kidney tissues were dissected, frozen in liquid nitrogen, and stored at −80°C.
Cell Culture and Treatments.
MMCs were isolated and cultured in Dulbecco's modified Eagle’s medium supplemented with 10% fetal calf serum and insulin-transferrin-sodium selenite, as described previously (Pandey et al., 2000). Cells were treated with ATRA-NaBu and stilbene-based retinoid TTNPB (4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid), a RAR-specific agonist, in Dulbecco’s modified Eagle’s medium containing 0.1% bovine serum albumin (BSA) for 24 hours. All cultures were kept at 37°C under an atmosphere of 5% CO2 and 95% O2.
Histone Purification.
Total histone was extracted from frozen kidney tissues using Epigentek’s EpiQuik extraction kit following the manufacturer’s protocol. In brief, pooled kidney tissues from three animals were weighed and cut into small pieces and homogenized in 1× prelysis buffer, transferred in a 2-ml tube, and centrifuged at 10,000g for 1 minute at 4°C. The supernatant was removed; tissue pellet was resuspended in 3 volumes of lysis buffer, incubated on ice for 30 minutes, and centrifuged at 12,000g for 5 minutes at 4°C. Balance-dithiothreitol (DTT) buffer (0.3 volumes) was added to the supernatant, which was stored at −80°C. The protein concentration of the eluted histone was estimated using a Bradford protein detection kit (Bio-Rad, Hercules, CA) using BSA as a standard.
Quantification of Global Histone H3 Modifications.
Histone extracts were prepared as described earlier. Quantification of global levels of lysine-specific histone modifications was performed using an EpiQuik global H3-K9/14ac, H4-K12ac, H3-K4me3, H3-K9me2, H3-K9me3, H3-K27me3 colorimetric kit following the manufacturer’s protocol. In the assay, acetylated or methylated histones were captured with specific antibody and detected with a labeled detection antibody, followed by a color development reagent. Absorbance was read at 450 nm, and results were calculated using a standard curve following the manufacturer’s instructions.
Cytosolic and Nuclear Extract Preparation.
Cytosolic and nuclear proteins were prepared from frozen kidney tissues as described previously (Das et al., 2010). The kidney tissues were homogenized in an ice-cold 10 mM Tris-HCl buffer (pH 8.0) containing 0.32 M sucrose, 3 mM calcium chloride (CaCl2), 2 mM magnesium acetate (MgOAc), 0.1 mM EDTA, 0.5% NP40, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM orthovanadate, 30 mM sodium fluoride (NaF), and 10 µg/ml leupeptin and aprotinin. The homogenate was centrifuged at 8000g at 4°C for 20 minutes, and the supernatant was separated and stored as a cytosolic fraction. The pellet was washed in 3× wash buffer (same as described earlier except for NP40) by resuspending with a 20-gauge needle and centrifugation at 6000g. The pellet was resuspended in a low-salt buffer [20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA, 25% glycerol, 0.5 mM DTT, and 0.5 mM PMSF], incubated on ice for 5 minutes, and mixed with an equal volume of high-salt buffer containing 20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 800 mM KCl, 0.2 mM EDTA, 1% NP40, 25% glycerol, 0.5 mM DTT, 0.5 mM PMSF, 1 mM orthovanadate, 30 mM NaF, and 10 µg/ml leupeptin and aprotinin. The mixture was incubated on ice for 30 minutes and centrifuged at 14,000g for 20 minutes. The supernatant was separated and stored as a nuclear fraction at −80°C.
Western Blot Assay.
Cytoplasmic fraction (40–60 µg) or nuclear extract (30–50 μg) was mixed with sample loading buffer and separated using 10% SDS-PAGE essentially as described earlier (Das et al., 2010). Proteins were electrotransferred onto a polyvinylidene difluoride membrane, and the membrane was blocked with 1× Tris-buffered saline–Tween 20 (25 mM Tris, 500 mM NaCl, and 0.05% Tween 20, pH 7.5) containing 5% fat-free milk for 1 hour and incubated overnight in Tris-buffered saline–Tween 20 containing 3% fat-free milk at 4°C with primary antibodies (1:250 dilution). The membrane was treated with corresponding secondary antirabbit or antimouse horseradish peroxidase–conjugated antibodies (1:5000 dilutions). Protein bands were visualized by enhanced chemiluminescence plus detection system with an Alpha Innotech phosphoimager from Proteinsimple (Santa Clara, CA). The antibodies used in Western blot assay are listed in Supplemental Table 1.
Plasma Membrane Preparation and GC Activity Assay.
The plasma membranes from kidney tissues were prepared as described previously (Khurana and Pandey, 1996). In brief, kidney tissues were homogenized in 5 volumes of 10 mM sodium phosphate buffer (pH 7.4) containing 250 mM sucrose, 150 mM NaCl, 1 mM PMSF, 5 mM benzamidine, 5 mM EDTA, and 10 μg/ml each of leupeptin and aprotinin, centrifuged at 400g for 10 minutes at 4°C, and the supernatant collected was recentrifuged at 80,000g for 1 hour at 4°C. The resultant supernatant was discarded, and the pellet was resuspended in 1 ml of 50 mM HEPES buffer (pH 7.4) containing 150 mM NaCl, 1 mM PMSF, 5 mM benzamidine, 5 mM EDTA, and 10 μg/ml each of leupeptin and aprotinin, and centrifuged at 80,000g for 1 hour at 4°C. The final pellet was suspended in 200 μl of HEPES buffer (pH 7.4). GC activity was assayed as described by Leitman et al. (1988) with modifications (Khurana and Pandey, 1996). A 50-μg aliquot of plasma membrane was added to 100 μl of GC assay buffer containing 50 mM Tris-Cl buffer (pH 7.6), 4 mM MnCl2, 2 mM 3-isobutyl-1-methylxanthine, 1 mM BSA, 5 units of creatinine phosphokinase, 7.5 mM creatine phosphate, and 0.5 mM GTP. The samples were incubated in a water bath at 37°C for the indicated time. Reaction was stopped by adding 900 μl of 55 mM sodium acetate (pH 6.2), and sample tubes were placed in a boiling water bath for 5 minutes and then on ice for 15 minutes to stop the reaction. Samples were centrifuged at 13,000g for 5 minutes, supernatant was collected, and the generated cGMP was determined using a direct cGMP immunoassay kit.
cGMP Assay.
Frozen kidney tissue samples were homogenized in 10 volumes of 0.1 M HCl containing 1% Triton X-100. The homogenate was centrifuged at 10,000g at 4°C, and the supernatant was collected and used for cGMP assay. MMCs were treated with TTNPB and ATRA-NaBu for 24 hours and stimulated with ANP at 37°C for 15 minutes in the presence of 0.2 mM 3-isobutyl-1-methylxanthine, washed three times with PBS, and scraped into 0.5 N HCl. Cell suspension was subjected to five cycles of freeze and thaw. The mixture was centrifuged at 10,000g for 15 minutes. The supernatant thus collected was used for the cGMP assay. Cyclic GMP levels were determined using a direct cGMP immunoassay kit according to the manufacturer’s protocol. The results are expressed as picomoles of cGMP/milligram of protein.
HDAC and HAT Activity Assay.
Total HDAC and HAT enzyme activities were measured in drug-treated and control kidney nuclear extracts using a colorimetric enzyme-linked immunosorbent assay kit from Active motif and Epigentek (Farmingdale, NY), respectively. The HDAC activity was calculated by measuring the amount of HDAC-deacetylated product, which was directly proportional to the HDAC enzyme activity. For the HAT enzyme activity assay, the amount of acetylated product was measured. Absorbance was read at 450 nm, and results were calculated using a standard curve following the manufacturer’s instructions and expressed as nanograms/hour/milligram of protein.
Quantitative Chromatin Immunoprecipitation Assay.
Chromatin was prepared from frozen kidney tissue samples, and ChIP was performed using a ChIP-IT Express kit. Tissue samples were weighed and finely minced on ice, and 25 mg of tissue for each IP was used. Tissue was transferred to a 2-ml tube containing PBS and protease inhibitor cocktail, and 37% formaldehyde (final concentration 1.5%) was added with rocking at room temperature for 20 minutes to cross-link protein to DNA. Cross-linking was stopped by adding 100 μl of 10× glycine, and the solution was mixed for 5 minutes and centrifuged at 1500g for 5 minutes at 4°C. Pellet was washed with PBS, centrifuged at 1500g at 4°C, and resuspended in PBS containing protease inhibitors. Pellet was disaggregated using Dounce’s homogenizer, centrifuged at 1500g at 4°C, and used for nuclei preparation and chromatin digestion. The pellet was resuspended in 1 ml of digestion buffer and 50 μl of enzymatic shearing mixture and incubated at 37°C for 10 minutes. The reaction was stopped by adding 20 μl of 0.5 M EDTA followed by chilling on ice for 10 minutes. Sheared DNA was centrifuged at 13,000g at 4°C for 10 minutes, and supernatant was collected. Ten percent of supernatant was saved as input DNA and processed for further use as a positive control. Immunoprecipitation was performed using protein G magnetic beads and 5 μg of antibodies of H3-K9/14ac, H4-K12ac, H3-K4me3, H3-K9me3, p300/cAMP response element–binding protein-binding protein–associated factor (PCAF), and p300 or control IgG at 4°C with overnight rotation. Beads were pelleted and washed sequentially once with ChIP buffer 1 and twice with ChIP buffer 2. After washing the magnetic beads, bound protein was eluted by gentle rotation for 25 minutes in elution buffer at 22°C. Cross-linking of the protein/DNA complex was reversed at 65°C overnight to release DNA. Immunoprecipitated DNA was sequentially treated with RNase A and proteinase K and then purified. The Npr1 proximal promoter region containing Ets-1 and Sp1 transcription factor binding sites was polymerase chain reaction (PCR) amplified using purified DNA as a template and the forward (5′-gaggggaggattcgctgc-3′) and reverse (5′-ctaagaagagcgaggggagc-3′) primers. The antibodies used in ChIP assay are listed in Supplemental Table 1. For quantitative ChIP assay, real-time PCR was performed with RT2 real-time SYBR Green/ROX PCR Master Mix (SABiosciences, Valencia, CA).
Real-Time Reverse-Transcription PCR Assay.
Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen). Thirty milligrams of kidney tissue was used to extract RNA. First-strand cDNA was synthesized from 1 μg of total RNA in a final volume of 20 μl using the RT2 First Strand kit (SABiosciences). Primers for Npr1 and β-actin were purchased from SABiosciences. The Ets-1 gene was amplified using forward (5′-atcagctggacaggagattgc-3′) and reverse (5′-cgcgtatacgtaggcgttgc-3′) primers. Real-time reverse-transcription PCR was performed using the Mx3000P real-time PCR system, and data were analyzed with MxPro software (Stratagene, La Jolla, CA). The reaction mixture without template cDNA was used as a negative control. The Npr1 and Ets-1 expression values were normalized to β-actin. Relative expression of the Npr1 and Ets-1 gene was determined by the comparative Ct value.
Immunohistochemistry of Proliferating Cell Nuclear Antigen and α-Smooth Muscle Actin.
Immunohistochemical staining for proliferating cell nuclear antigen (PCNA) and α-smooth muscle actin (α-SMA) proteins was done on 5-µm sections of paraffin-embedded kidney tissues as previously described (Das et al., 2010). In brief, after dewaxing in xylene, the kidney sections were dehydrated by serial dilutions of alcohol (100, 90, and 70%) for 10 minutes each and washed in PBS for 5 minutes. Sections were incubated in 0.3% H2O2 prepared in methanol for 30 minutes to block the endogenous peroxidase activity. After washing in PBS for 20 minutes, antigen unmasking was done using warm 10 mM sodium citrate buffer (pH 6.0) and washed in PBS for 10 minutes. The sections were sequentially incubated at room temperature with normal blocking serum (goat serum) for 40 minutes, primary antibody (mouse monoclonal PCNA and α-SMA) diluted in PBS-Tween 20 containing 1% BSA at 1:1000 dilution overnight at 4°C, secondary antibody (biotinylated goat antimouse IgG) for 30 minutes, and avidin-biotin horseradish peroxidase complex for 45 minutes, using the ABC staining kit (Santa Cruz Biotechnology). Peroxidase activity was visualized with 0.1% 3,3′-diaminobenzidine. The slides were then washed in tap water, counterstained with hematoxylin, mounted using aqueous mounting medium, and coverslipped. Immunohistochemically stained sections were visualized, and the percentage of PCNA- and α-SMA–positive area to the total kidney was calculated using an Olympus BX51 camera and photographed with integrated Magnafire SP Digital Firewire camera software (Olympus America Inc., Melville, NY).
Statistical Analysis.
The results are expressed as the mean ± S.E. The statistical significance was evaluated by one-way analysis of variance, followed by Dunnett’s multiple comparison tests using Prism software (GraphPad Software, San Diego, CA). Student’s unpaired t test with two-tailed analysis was also performed for comparison between groups. A P value of 0.05 was considered significant.
Results
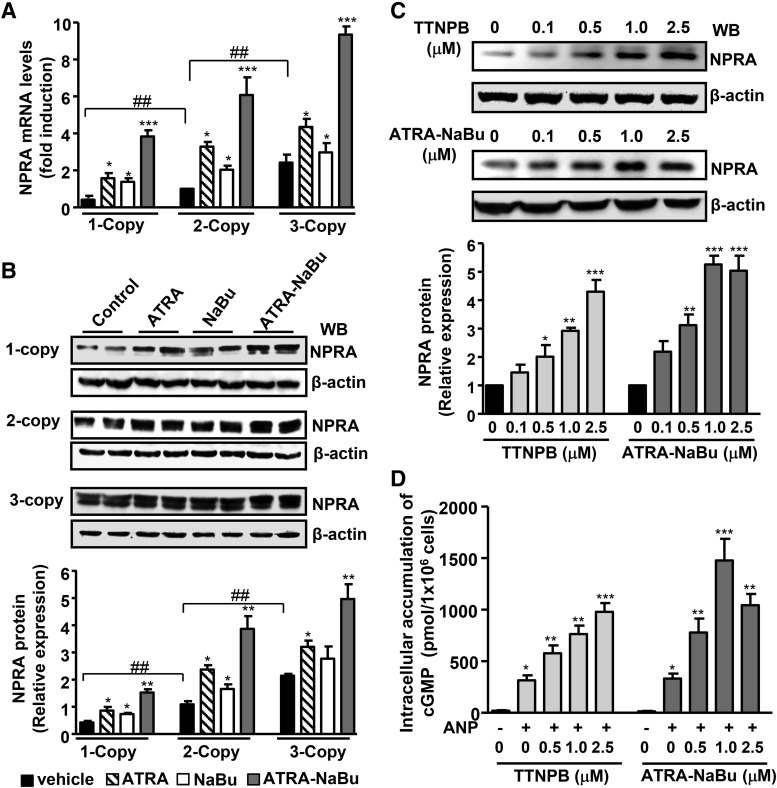
The results showed that there was a significant reduction in systolic blood pressure of Npr1 gene-disrupted heterozygous 1-copy mice treated with ATRA and NaBu (114.6 ± 2.2, P < 0.01 and 119.2 ± 1.7, P < 0.05, respectively) versus vehicle-treated (128.2 ± 2.1) 1-copy mice (Table 1). The treatment with ATRA-NaBu hybrid also considerably lowered systolic blood pressure in 1-copy mice (108.8 ± 4.3, P < 0.001) compared with 1-copy control mice. Renal GC activity and cGMP levels were significantly reduced in 1-copy mice by 63 and 65%, respectively, and were higher in gene-duplicated heterozygous 3-copy mice by 1.7- to 2-fold compared with wild-type 2-copy mice (Table 1). There was an increase in renal GC activity and cGMP levels in ATRA- and NaBu-treated Npr1 mice with different gene copy numbers; however, treatment with ATRA-NaBu combination markedly augmented renal GC activity and cGMP levels in treated groups compared with their vehicle-treated control mice. The endogenous Npr1 mRNA levels were considerably lower in 1-copy mice kidneys compared with 2-copy mice and higher in 3-copy mice (Fig. 1A). Treatment with ATRA and NaBu considerably increased the mRNA levels in 1-, 2-, and 3-copy mice compared with their vehicle-treated controls. On the other hand, ATRA-NaBu combination significantly augmented Npr1 mRNA levels in 1-copy (9.5-fold), 2-copy (6-fold), and 3-copy (3.2-fold) mice compared with vehicle-treated respective controls. The Western blot analysis showed attenuated levels of NPRA protein in 1-copy mice kidney compared with 2-copy mice and higher levels in 3-copy mice (Fig. 1B). Treatment with ATRA-NaBu combination synergistically induced renal NPRA protein levels in all treated groups compared with their vehicle-treated controls. Retinoic acid–dependent induction in NPRA signaling was further confirmed under in vitro conditions utilizing the RAR agonist TTNPB and ATRA-NaBu combination drug in MMCs. There was a dose-dependent increase in NPRA protein expression in cells treated with TTNPB and ATRA-NaBu combination drug compared with untreated controls (Fig. 1C). Treatment with TTNPB and ATRA-NaBu combination drug showed 90- and 145-fold stimulation in cGMP levels, respectively, in the presence of ANP compared with untreated controls (Fig. 1D).
TABLE 1.
Comparisons of blood pressure, kidney GC activity, and cGMP levels among Npr1 genotypes treated with ATRA and NaBu
Measurement of systolic blood pressure, renal GC activity, and cGMP levels in ATRA- and NaBu-treated mice. Blood pressure was measured by a computerized tail-cuff method in Npr1 gene-targeted mice. Kidney GC activity and cGMP levels were quantitated by enzyme-linked immunosorbent assay as described in Materials and Methods. Values represent the mean ± S.E. of three independent experiments in triplicate.
| Treatments/Parameters |
Npr1 Genotype |
||
|---|---|---|---|
| 1-Copy | 2-Copy | 3-Copy | |
| Systolic blood pressure (mm Hg) | |||
| Vehicle | 128.2 ± 2.1### | 103.4 ± 1.8 | 94.7 ± 1.1# |
| ATRA | 114.6 ± 2.2** | 100.0 ± 1.9 | 92.8 ± 0.9 |
| NaBu | 119.2 ± 1.7* | 101.9 ± 3.8 | 92.4 ± 1.6 |
| ATRA + NaBu | 108.8 ± 3.1*** | 97.9 ± 1.3 | 88.43 ± 2.1 |
| GC activity (pmol cGMP/mg protein/30 min) | |||
| Vehicle | 24.8 ± 2.1## | 68.6 ± 3.3 | 116.2 ± 4.5# |
| ATRA | 40.7 ± 7.4* | 91.4 ± 3.5** | 145.7 ± 6.3* |
| NaBu | 31.8 ± 1.8* | 79.9 ± 8.8* | 133.3 ± 11.4* |
| ATRA + NaBu | 58.5 ± 2.5** | 106.1 ± 5.6** | 195.2 ± 7.5** |
| cGMP (pmol/mg protein) | |||
| Vehicle | 11.8 ± 0.9# | 34.3 ± 1.8 | 82.3 ± 6.8# |
| ATRA | 21.9 ± 1.3* | 57.4 ± 5.1** | 121.7 ± 10.1* |
| NaBu | 19.4 ± 0.7* | 46.5 ± 5.9* | 101.5 ± 5.3* |
| ATRA + NaBu | 36.3 ± 1.8** | 84.4 ± 7.6** | 160.2 ± 12.5** |
P < 0.05; **P < 0.01; ***P < 0.001 (vehicle-treated control versus drug-treated same gene copy number); #P < 0.05; ##P < 0.01; ###P < 0.001 (vehicle-treated 1- and 3-copy versus vehicle-treated 2-copy); n = 6 in each group.
Fig. 1.
Effect of ATRA, NaBu, and TTNPB on renal Npr1 gene expression in gene-targeted mice and MMCs. (A) Relative mRNA expression of renal Npr1 in drug-treated and control mice as determined by real-time reverse-transcription PCR, normalized to β-actin mRNA. (B) Western blot and densitometry analyses of renal NPRA protein expression in drug- or vehicle-treated Npr1 gene-targeted mice. β-Actin was used as a loading control. (C) Western blot analysis of NPRA protein expression in cells stimulated with increasing concentrations of TTNPB and ATRA-NaBu, and β-actin expression is shown as a loading control. (D) Intracellular accumulation of cGMP in cells treated with increasing concentrations of TTNPB and ATRA-NaBu and induced with or without 100 nM ANP. Bar represents mean ± S.E. of three independent experiments. WB, Western blot. *P < 0.05; **P < 0.01; *** P < 0.001 (vehicle-treated versus drug-treated same group); ##P < 0.01; ###P < 0.001 (1-copy or 3-copy versus 2-copy); n = 8 mice per group.
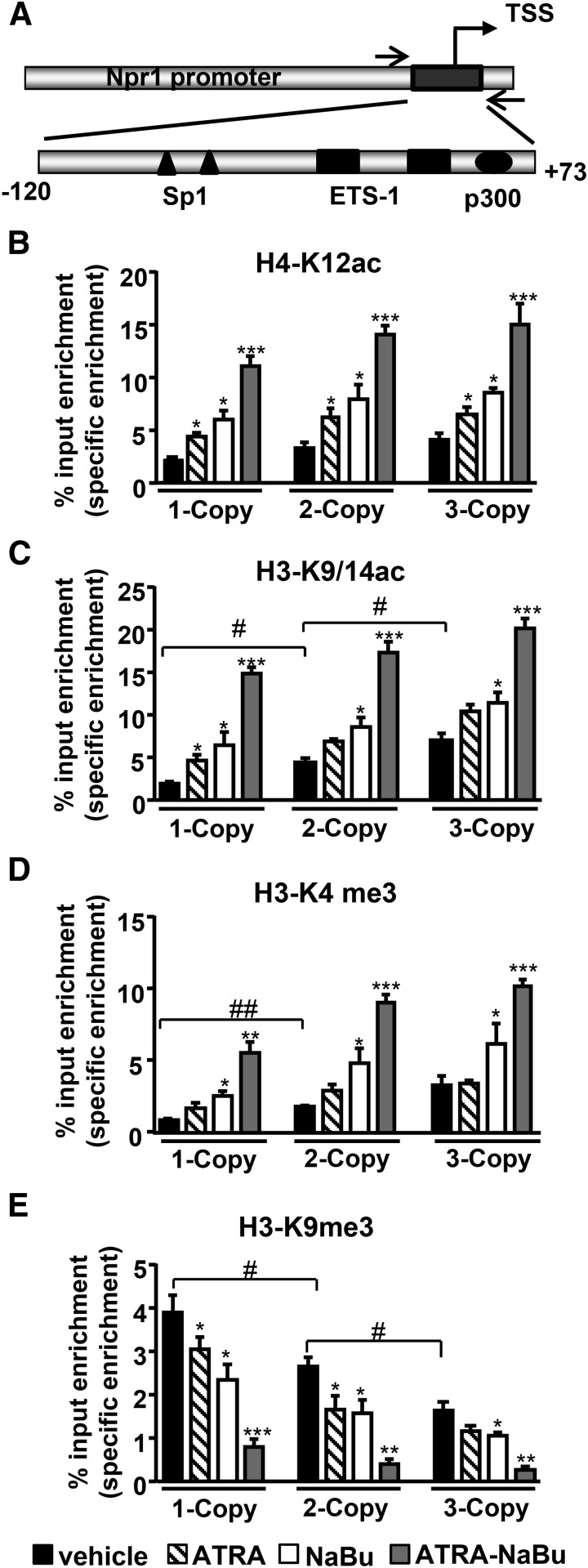
In vivo quantitative ChIP assay was performed in kidney tissues to examine ATRA- and NaBu-mediated accumulation of acetylated and methylated histone marks at the Npr1 proximal promoter region −120 to +73 (Fig. 2A). One-copy mice exhibited attenuated binding of active histone marks H4-K12ac and H3-K9/14ac with Npr1 promoter, whereas 3-copy mice showed higher binding of acetylated histone H3 and H4 compared with 2-copy mice (Fig. 2, B and C). There was a synergistic increase in H4-K12ac and H3-K9/14ac occupancy at the Npr1 promoter in ATRA-NaBu combination–treated mice compared with their controls. ATRA and NaBu treatment significantly enhanced binding of H3-K4me3 at Npr1 promoter in treated mice compared with controls (Fig. 2D). There was augmented binding of repressive histone code H3-K9me3 at Npr1 promoter in 1-copy mice and attenuated binding in 3-copy mice compared with 2-copy mice (Fig. 2E). However, promoter occupancy of H3-K9me3 was markedly reduced in ATRA-NaBu combination–treated mice compared with their respective controls. Representative gel images have been generated from ChIP assays as analyzed by conventional PCR in drug-treated Npr1 gene-targeted mice (Supplemental Fig. 1).
Fig. 2.
Effect of ATRA and NaBu on binding of acetylated and methylated histones at the proximal promoter of Npr1 gene. (A) Schematic representation of Npr1 proximal promoter region (−120 to +73) amplified for ChIP assay having Ets-1, Sp1, and p300 transcription factor binding sites. Recruitment of H4-K12ac (B), H3-K9ac (C), H3-K4me3 (D), and H3-K9me3 (E) to the Npr1 promoter in kidneys of ATRA- and Nabu-treated 1-, 2-, and 3-copy mice. Values are expressed as the mean ± S.E. of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (vehicle-treated versus drug-treated same group); #P < 0.05; ##P < 0.01 (1- or 3-copy versus 2-copy); n = 7 mice per group. TSS, transcription start site.
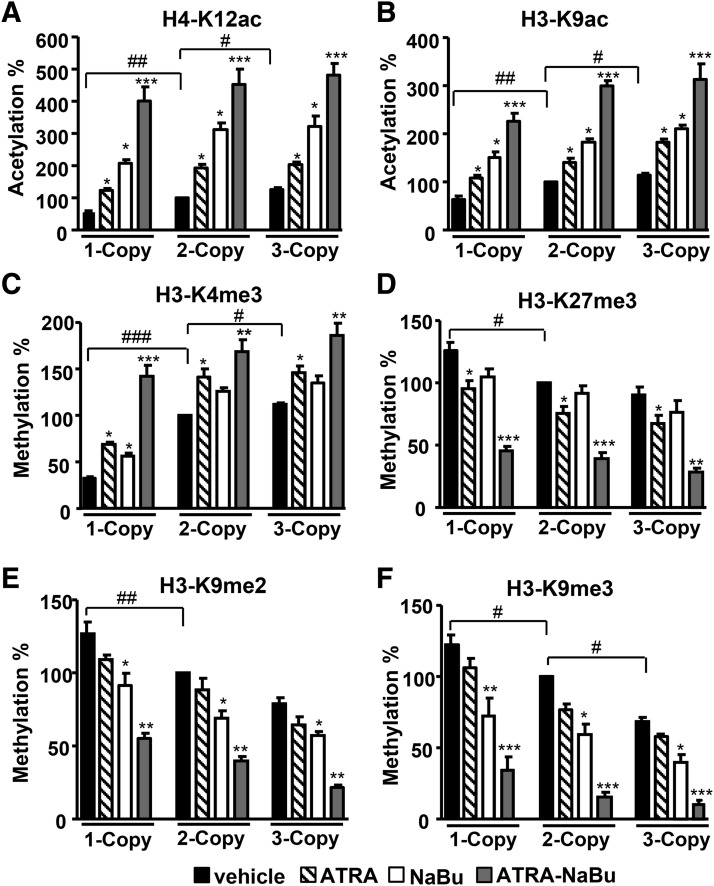
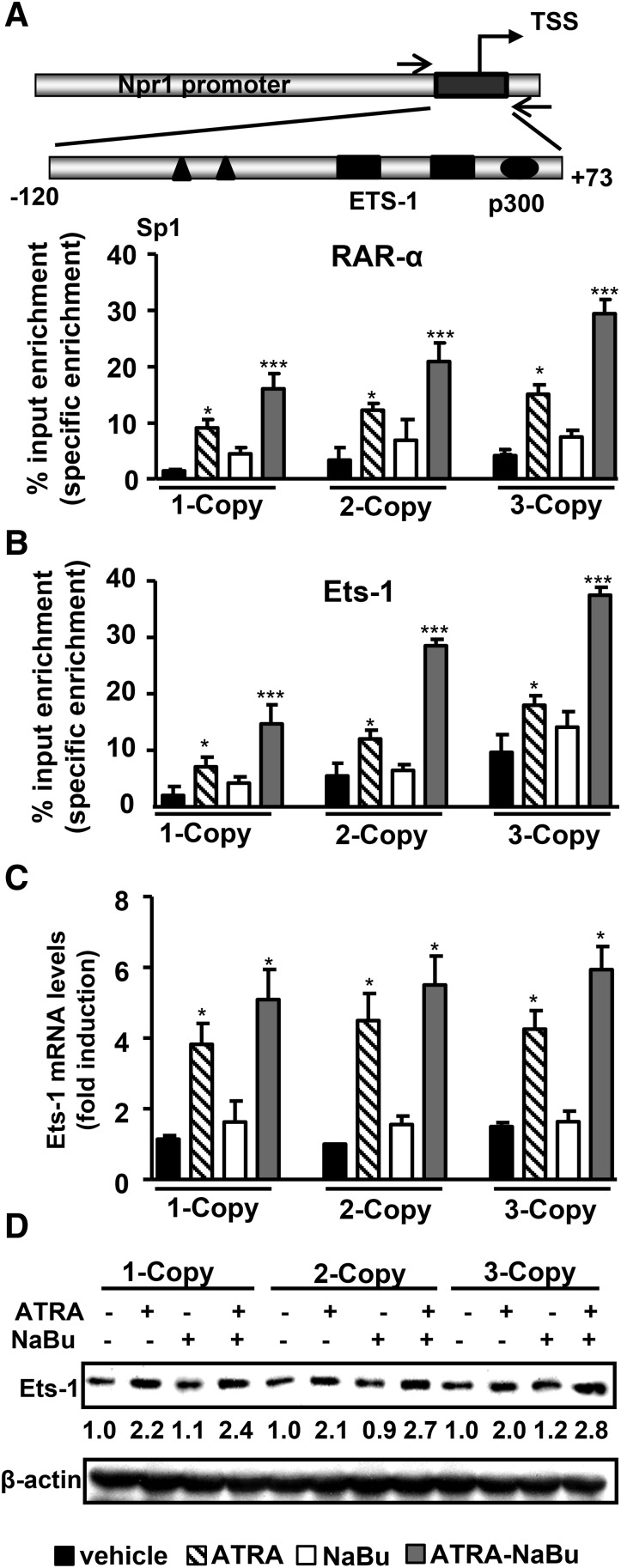
The quantitative analyses of renal histone modifications showed lower levels of active histone marks H4-K12ac, H3-K9ac, and H3-K4me3 in 1-copy mice compared with 2- and 3-copy mice (Fig. 3, A–C). Treatment with ATRA-NaBu combination synergistically increased protein levels of H4-K12ac, H3-K9ac, and H3-K4me3 in treated mice compared with their controls. One-copy mice exhibited increased levels of repressive histone codes H3-K27me3, H3-K9me2, and H3-K9me3 compared with 2- and 3-copy mice (Fig. 3, D–F). Treatment with ATRA-NaBu combination significantly attenuated H3-K27me3, H3-K9me2, and H3-K9me3 expression in treated mice compared with vehicle-treated controls. Western blot analysis further confirmed the global increase in expression levels of H4-K12ac, H3-K9/14ac, and H3-K4me3 histone marks and a decrease in H3-K9me3 in renal tissues of ATRA- and NaBu-treated mice compared with their controls (Supplemental Fig. 2, A–E). Quantitative ChIP assay demonstrated higher enrichment of RARα to Npr1 promoter in ATRA- and ATRA-NaBu combination–treated mice compared with their vehicle-treated controls (Fig. 4A). There was marked accumulation of Ets-1 protein at Npr1 promoter in ATRA and ATRA-NaBu combination–treated 1-copy mice compared with vehicle-treated control mice (Fig. 4B). Furthermore, treatment with ATRA and ATRA-NaBu combination markedly induced renal Ets-1 mRNA and protein levels in treated mice compared with vehicle-treated controls (Fig. 4, C and D).
Fig. 3.
Quantitative analysis of renal H3 modifications in ATRA- and NaBu-treated Npr1 gene-targeted mice. Renal protein expression of H4 and H3 modifications H4-K12ac (A), H3-K9ac (B), H3-K4me3 (C), H3-K27me3 (D), H3-K9me2 (E), and H3-K9me3 (F) at specific lysines was done by the EpiQuik quantification kit from EpiGentek. Bar represents the mean ± S.E. of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (vehicle-treated versus drug-treated same group); #P < 0.05; ##P < 0.01; ###P < 0.001 (1- or 3-copy versus 2-copy); n = 7 mice per group.
Fig. 4.
Effect of ATRA and NaBu on RARα and Ets-1 binding at the proximal promoter region of Npr1 gene. (A) Schematic representation of Npr1 proximal promoter region (−120 to +73) amplified for ChIP assay having Ets-1, Sp1, and p300 transcription factor binding sites. Recruitment of RARα to the Npr1 promoter in kidneys of drug-treated and control mice. (B) Ets-1 recruitment to the Npr1 promoter in response to ATRA and NaBu treatment. (C) Relative mRNA expression of renal Ets-1 in drug-treated and control mice as determined by real-time reverse-transcription PCR, normalized to β-actin mRNA. (D) Western blot analysis of renal Ets-1 protein expression in drug- or vehicle-treated Npr1 gene-targeted mice. β-Actin was used as a loading control. Pooled samples from 6 mice in each group were used for experiments. Values are expressed as the mean ± S.E. of three independent experiments. *P < 0.05; ***P < 0.001 (vehicle-treated versus drug-treated same group). TSS, transcription start site.
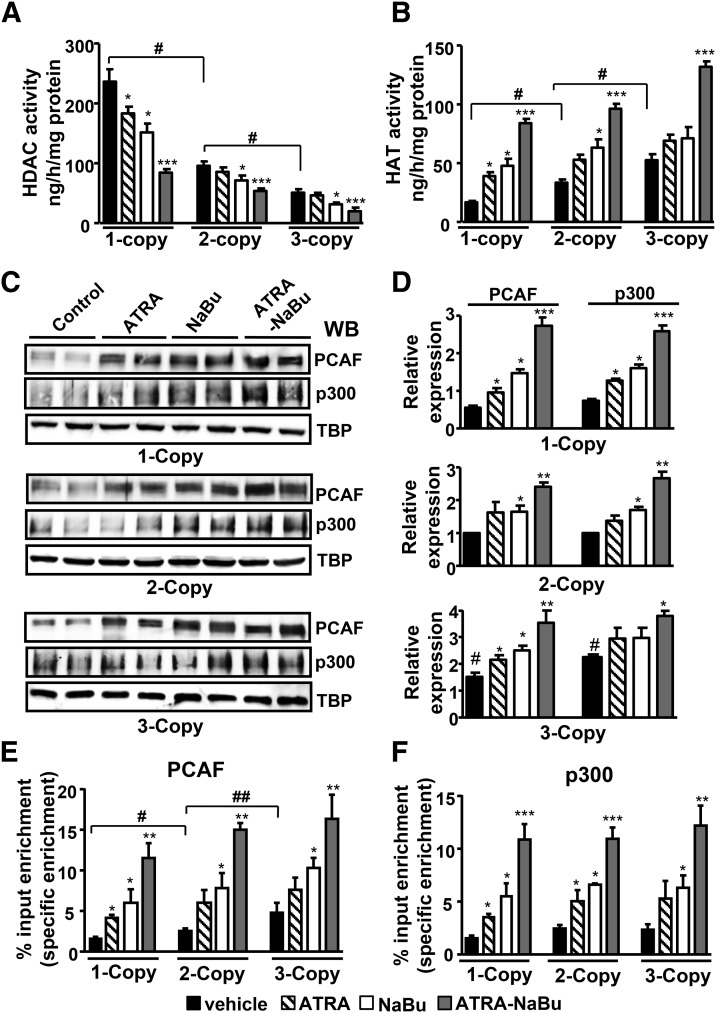
Renal HDAC activity was distinctly higher in 1-copy mice (2.5-fold) compared with 2-copy mice, but lower in 3-copy mice (Fig. 5A). However, the HDAC activity was reduced in 1-copy mice by 37% after treatment with NaBu and 66% in ATRA-NaBu combination–treated mice compared with vehicle-treated controls. Similarly, treatment with ATRA-NaBu combination noticeably attenuated HDAC activity in 2- and 3-copy mice compared with controls. On the other hand, HAT activity was significantly lower in 1-copy mice (50%) and higher in 3-copy mice (37%) compared with 2-copy mice (Fig. 5B). There was marked induction of HAT activity in 1-copy mice treated with ATRA and NaBu alone, and a 7.8-fold increase was observed in ATRA-NaBu combination–treated mice compared with controls. Likewise, ATRA-NaBu combination markedly increased HAT activity in 2- and 3-copy mice compared with controls. Western blot and densitometric analysis of HATs—namely, PCAF and p300—showed significant increases in PCAF and p300 protein levels in ATRA- and NaBu-treated 1-copy mice compared with vehicle-treated controls (Fig. 5, C and D). In vivo quantitative ChIP assay showed increased recruitment of PCAF at the Npr1 promoter in ATRA- and NaBu-treated 1-copy mice, whereas ATRA-NaBu combination showed a synergistic effect compared with control mice (Fig. 5E). There was a higher enrichment of p300 protein at Npr1 promoter in ATRA-NaBu combination–treated experimental mice with pronounced effects observed in 1-copy mice compared with their respective control groups (Fig. 5F).
Fig. 5.
Modulation of renal HDAC and HAT activity and expression of PCAF and p300 by ATRA and NaBu. HDAC (A) and HAT (B) activity were measured in nuclear extracts of drug-treated and vehicle-treated mice kidney tissues. (C and D) Western blot and densitometry analysis of renal protein levels of PCAF and p300 in Npr1 1-, 2-, and 3-copy mice treated with ATRA and NaBu. TATA-binding protein (TBP) was used as a loading control. Quantitative ChIP assay demonstrating in vivo recruitment of PCAF (E) and p300 (F) to the Npr1 promoter in ATRA- and NaBu-treated and vehicle-treated mice. Bar represents the mean ± S.E. WB, Western blot. *P < 0.05; **P < 0.01; ***P < 0.001 (vehicle-treated versus drug-treated same group); #P < 0.05; ##P < 0.01(1- or 3-copy versus 2-copy); n = 6 mice per group.
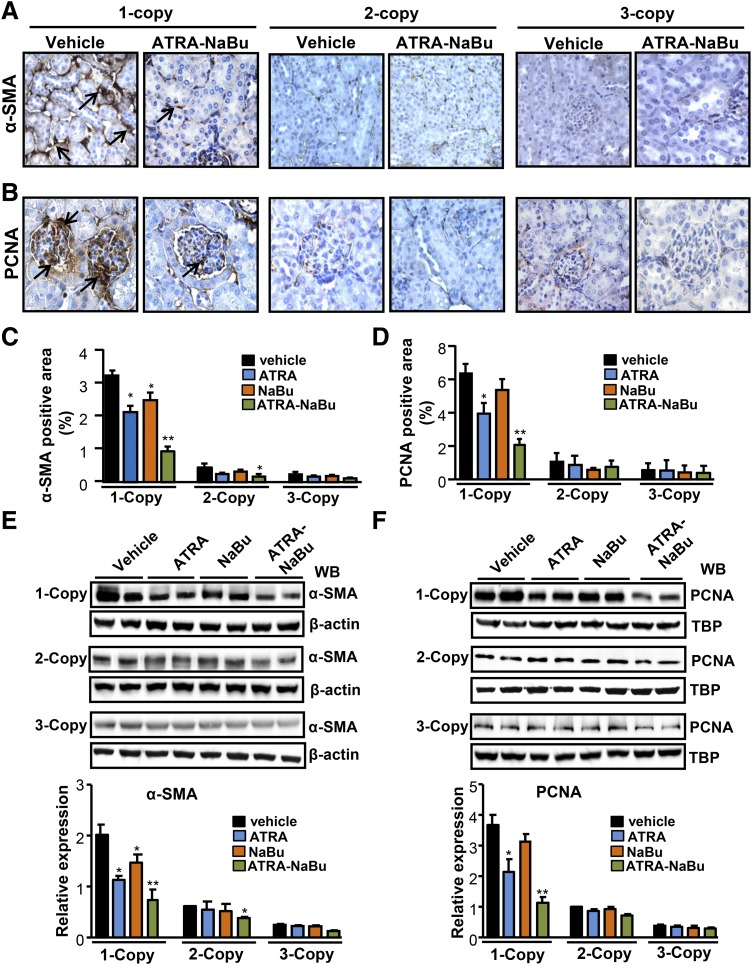
Immunohistochemical analyses for α-SMA showed significantly increased expression in tubules, arterioles, and interstitium of 1-copy mice compared with 2- and 3-copy mice (Fig. 6A). The α-SMA–positive stained cells were 6-fold higher in 1-copy mice compared with 2- and 3-copy mice (Fig. 6C). There was a significant attenuation in α-SMA expression in 1-copy mice treated with ATRA (42%, P < 0.001), NaBu (25%, P < 0.001), and ATRA-NaBu combination (70%, P < 0.001) compared with 1-copy control mice (Fig. 6A; Supplemental Fig. 3A). The immunostaining of kidney sections for PCNA showed a marked increase in PCNA-positive cells in the glomeruli of 1-copy mice compared with 2- and 3-copy mice (Fig. 6B). A substantial reduction in PCNA immunoexpression was observed in 1-copy mice treated with ATRA (36%, P < 0.001), NaBu (16%, P < 0.001), and ATRA-NaBu hybrid (68%, P < 0.001) (Fig. 6D; Supplemental Fig. 3B). Western blot and densitometric analyses showed that treatment with ATRA and NaBu alone markedly reduced renal α-SMA and PCNA protein levels, whereas ATRA-NaBu hybrid synergistically decreased α-SMA and PCNA protein expression in 1-copy treated mice compared with 1-copy control mice (Fig. 6, E and F).
Fig. 6.
Renal immunoexpression of α-SMA and PCNA in drug-treated and vehicle-treated Npr1 gene-targeted mice. Immunohistochemistry showing renal expression of α-SMA (A) and PCNA (B) in drug-treated and vehicle-treated 1-, 2-, and 3-copy mice. (C and D) Quantitative analysis of α-SMA and PCNA in drug-treated and vehicle-treated mice groups. Western blot and densitometric analyses of α-SMA (E) and PCNA (F) in drug-treated and vehicle-treated mice. β-Actin was used as a loading control. Values are expressed as the mean ± S.E. of three independent experiments. TBP, TATA-binding protein; WB, Western blot. *P < 0.05; **P < 0.01 (vehicle-treated versus drug-treated same group). In each group, five mice were used.
Discussion
The present results demonstrate that retinoic acid signaling and pharmacological inhibition of HDAC activity by NaBu markedly stimulated Npr1 gene expression in Npr1 gene-disrupted 1-copy mice. The increased Npr1 gene expression was accompanied by modulation of HDAC and HAT activity resulting in global histone modifications, including increased levels of H3-K9 and H4-K12 acetylation and decreased levels of H3-K9me2, H3-K9me3, and H3-K27me3. Previously, it has been reported that histone H3-K9/14 acetylation and H3-K4, K9, and K27 methylation are closely related to transcriptional status of the gene and are modulated by HDAC inhibitors such as TSA and NaBu to open the chromatin structure and enhance gene transcription (Bhaumik et al., 2007; Nightingale et al., 2007; Huang et al., 2011). Recently, it has been shown that treatment with valsartan in spontaneously hypertensive rats decreased local angiotensin-converting enzyme 1 mRNA and protein expressions, which were accompanied by higher levels of H3-K9me2, as well as lower levels of H3ac and H3-K4me3 (Lee et al., 2012). Furthermore, in vitro studies have shown that ATRA and HDAC inhibitors increase the binding of active histone codes (H3-K9ac and H3-K14ac) at the promoters of human pyruvate dehydrogenase kinase 4 and neurogenin1 genes and attenuate the recruitment of repressive histone code H3-K27me3 (Kwon et al., 2006; Wu et al., 2009). Findings from our present study demonstrate that ATRA and NaBu significantly enhanced binding of H3-K9/14ac, H4-K12, and H3-K4me3 at the Npr1 promoter and reduced binding of H3-K9me3, thereby modulating Npr1 gene transcription.
Evidence suggests that ATRA treatment induces epigenetic modifications at the promoter level and as a consequence of histone acetylation and opened chromatin structure; transcription factors and the basal transcriptional machinery are recruited to the region, which activate gene transcription (Kwon et al., 2006; Kashyap and Gudas, 2010). Our results show that ATRA and ATRA-NaBu combination significantly enhanced recruitment of RARα and Ets-1 at the Npr1 proximal promoter in 1-copy Npr1 mice kidneys in vivo. Earlier studies have suggested that Ets-1 is essential for normal development of mammalian kidneys and maintenance of glomerular integrity (Cederberg et al., 1999; Gomez and Norwood, 1999). The present data show that increase in histone acetylation by ATRA and NaBu facilitates recruitment of transcription factors Ets-1 and RARα at the Npr1 proximal promoter, thereby mediating ATRA-induced Npr1 gene transcription in intact animals in vivo.
The balance between HAT and HDAC is an essential regulatory switch of gene expression, and an increase in histone acetylation can be achieved through the inhibition of its deacetylation process (Gaub et al., 2010; Peserico and Simone, 2011). Several classes of HDAC inhibitors, including NaBu, TSA, vorinostat, and hydroxamic acid, have been shown to inhibit HDAC activity and shift the overall balance in favor of enhanced HAT activity (Yang and Seto, 2007; Pascual et al., 2012). Consistent with those previous findings, the present data show that treatment with ATRA and NaBu significantly reduced HDAC activity and increased HAT activity in Npr1 1-copy mice. ATRA and NaBu significantly increased protein levels and accumulation of p300 and PCAF at the Npr1 promoter in the mouse kidney. Earlier studies have shown that ATRA treatment was able to enhance p300 and PCAF protein expression and their binding to gene promoters (Dietze et al., 2002; Gaub et al., 2010; Kim et al., 2010). In the absence of ligand, the retinoic acid receptors RAR/retinoid X receptor heterodimers have been shown to bind gene promoters and interact directly with nuclear corepressor proteins, which recruit HDAC complexes and deacetylate histone tails and establish a “closed” chromatin tail (Kashyap and Gudas, 2010). Those previous findings indicated that ATRA binding to the promoters releases the corepressors and leads to the recruitment of coactivator proteins such as p300/cAMP response element–binding protein-binding protein– and PCAF complex, which mediate gene transcription (Wei, 2003; Kashyap and Gudas, 2010).
The present data suggest that ATRA and NaBu treatment elicited significant reduction in the percentage of glomeruli with PCNA-positive cells and attenuation in α-SMA immunoexpression (a marker for epithelial mesenchymal transition) in 1-copy mice. Previously, it has been shown that ATRA exerts a beneficial effect in antiglomerular basement membrane glomerulonephritis in rats and the unilateral ureteral obstruction mouse model via downregulation of PCNA, α-SMA, and collagen I expression, and has been suggested as a therapeutic strategy for the treatment of progressive renal fibrosis in diseased kidneys (Oseto et al., 2003; Kishimoto et al., 2011). On the other hand, inhibition of HDAC activity attenuates renal fibroblast activation and interstitial fibrosis in obstructive nephropathy via downregulation of α-SMA and fibronectin and modulates proinflammatory and fibrotic changes in tubulointerstitial injury, suggesting that HDACs can be a potential target for the treatment of fibrotic disorders (Pang et al., 2009; Marumo et al., 2010). Our previous studies have demonstrated that Npr1 1-copy mice exhibited increased levels of renal proinflammatory cytokines, fibrosis, and remodeling (Das et al., 2010; Zhao et al., 2013). We have also observed that a decrease in Npr1 gene copy number increases renal PCNA and α-SMA protein expression leading to fibrosis and hypertrophic growth of the kidneys in Npr1 1-copy mice compared with 2-copy mice (Das et al., 2010). It has been suggested that different cross-talk mechanisms exist between histone acetylation and methylation networks, which constitute a complex framework for epigenetic control of gene transcription during biologic or pathologic developments (Baylin and Ohm, 2006; Latham and Dent, 2007).
The treatment of Npr1 gene-disrupted 1-copy mice with ATRA-NaBu combination showed a synergistic increase in renal NPRA protein expression, GC activity, and cGMP levels via modulation of HDAC and HAT activity, histone modifications, and recruitment of transcription factors to enhance Npr1 gene transcription. The activation of NPRA/cGMP signaling in Npr1 1-copy mice inhibited renal fibrosis as evidenced by reduction in expression of renal fibrotic markers such as α-SMA and PCNA. A schematic diagram of the mechanistic pathways of the ATRA/NaBu-dependent inhibition of HDAC and activation of HAT activities leading to enhanced Npr1 gene transcription and signaling is depicted in Fig. 7. An earlier study has also shown that the activation of an endogenous NPRA system attenuates renal fibrosis in the unilateral ureteral obstruction mouse model, and ANP pretreatment significantly improved morphologic changes with increase of renal cGMP levels (Nishikimi et al., 2009). Glucocorticoids have also been shown to improve renal responsiveness to ANP by upregulating NPRA expression accompanied by remarkable increase in renal cGMP levels in decompensated heart failure (Liu et al., 2011). Recently, it has been suggested that NaBu attenuated gentamicin-induced nephrotoxicity by enhancing renal antioxidant enzyme activity and expression of prohibitin protein (Sun et al., 2013). Our present data provide direct evidence that activation of NPRA/cGMP signaling by ATRA and NaBu via epigenetic mechanisms inhibits various renal fibrotic pathways and attenuates systolic blood pressure with decreasing Npr1 gene copy number.
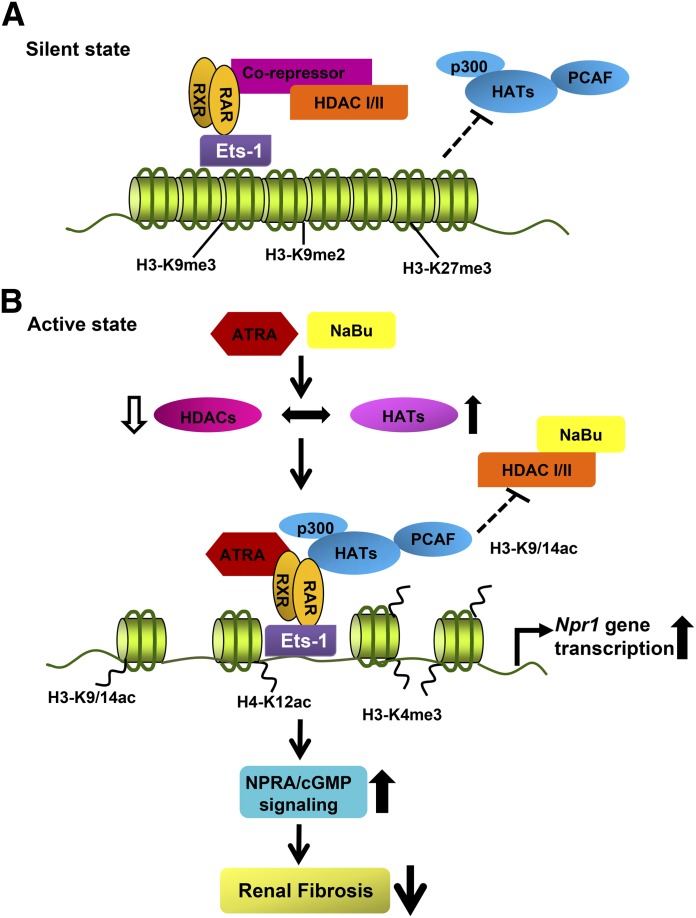
Fig. 7.
Schematic presentation of proposed epigenetic signaling during ATRA- and NaBu-mediated Npr1 gene regulation in vivo. The proposed diagram indicates that, under nonpermissive chromatin state, the repressive histone marks (H3-K9me3, H3-K9me2, and H3-K27me3) are associated with Npr1 gene promoter. HDACs, class I and II, form a corepressor complex, associate with retinoic acid receptors, and ensure that H3 remains hypoacetylated. Treatment with ATRA and NaBu inhibits HDAC activity and induces HAT activity with increased expression and recruitment of HATs (p300 and PCAF) leading to hyperacetylated H3-K9/14 and H4-K12 in the proximity of Npr1 promoter. Increased promoter occupancy of active histone marks (H3-K9/14ac, H4-K12ac, and H3-K4me3) leads to localized unwinding of DNA and allows transcription factors (Ets-1 and RARα) to bind in the promoter region and enhance Npr1 gene expression. Enhanced NPRA/cGMP signaling reduces expression of renal fibrotic markers α-SMA and PCNA in 1-copy mice. The open downward arrow indicates a decrease in HDAC activity and fibrosis, whereas the bold upward arrows indicate an increase in HAT activity and Npr1 gene transcription and expression.
In summary, the present results provide strong evidence that ATRA and NaBu inhibit HDAC activity and induce Npr1 gene transcription and expression in the kidneys of 1-copy mice via histone acetylation and enhanced recruitment of transcription factors and HAT proteins to Npr1 promoter. Our observations demonstrate that ATRA- and NaBu-mediated chromatin remodeling is involved in transcriptional regulation of Npr1 gene, which leads to an increased renal NPRA/cGMP signaling and attenuation of renal fibrosis in Npr1 gene-targeted 1-copy mice. Overall, the present study provides new insights into the epigenetic regulatory mechanisms of Npr1 gene transcription, expression, and function, which are critical for the biologic activity of NPRA for possible therapeutic targets in the diagnosis and treatment of renal disorders under reduced NPRA signaling.
Supplementary Material
Acknowledgments
The authors thank Vickie Nguyen for technical assistance and Kamala Pandey for assistance during the preparation of this manuscript. The authors sincerely thank Dr. Giri Raghavaraju for assistance during the initial stages of the experiments. The authors are indebted to Dr. Oliver Smithies (University of North Carolina, Chapel Hill) for providing early breeding pairs of Npr1 gene-targeted mice colonies.
Abbreviations
- ANP
atrial natriuretic peptide
- ATRA
all-trans retinoic acid
- BSA
bovine serum albumin
- ChIP
chromatin immunoprecipitation assay
- DTT
dithiothreitol
- Ets-1
E26 transformation–specific 1
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- H3-K9ac
acetyl histone H3-K9
- H4-K12ac
acetyl histone H4-K12
- H3-K9me2
dimethyl histone H3-K9
- H3-K9me3
trimethyl histone H3-K9
- H3-K4me3
trimethyl histone H3-K4
- H3-K27me3
trimethyl histone H3-K27
- MMC
mouse mesangial cell
- NaBu
sodium butyrate
- NPRA
natriuretic peptide receptor-A
- PBS
phosphate-buffered saline
- PCAF
p300/cAMP response element–binding protein-binding protein–associated factor
- PCNA
proliferating cell nuclear antigen
- PCR
polymerase chain reaction
- PMSF
phenylmethylsulfonyl fluoride
- RA
retinoic acid
- RAR
retinoic acid receptor
- α-SMA
α-smooth muscle actin
- Sp1
specificity protein 1
- TSA
trichostatin A
- TTNPB
4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid
Authorship Contributions
Participated in research design: Kumar, Das, Pandey.
Conducted experiments: Kumar, Periyasamy, Das, Neerukonda, Mani, Pandey.
Contributed new reagents or analytic tools: Kumar, Periyasamy, Das, Mani, Pandey.
Performed data analysis: Kumar, Periyasamy, Pandey.
Wrote or contributed to the writing of the manuscript: Kumar, Pandey.
Footnotes
This work was supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grants HL57531 and HL62147]; and received partial support from the Institutional Development Award (IDeA) Program.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Arise KK, Pandey KN. (2006) Inhibition and down-regulation of gene transcription and guanylyl cyclase activity of NPRA by angiotensin II involving protein kinase C. Biochem Biophys Res Commun 349:131–135 [DOI] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. (2004) Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328:1–16 [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. (2006) Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6:107–116 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. (2007) Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 14:1008–1016 [DOI] [PubMed] [Google Scholar]
- Bush EW, McKinsey TA. (2010) Protein acetylation in the cardiorenal axis: the promise of histone deacetylase inhibitors. Circ Res 106:272–284 [DOI] [PubMed] [Google Scholar]
- Cederberg A, Hulander M, Carlsson P, Enerbäck S. (1999) The kidney-expressed winged helix transcription factor FREAC-4 is regulated by Ets-1. A possible role in kidney development. J Biol Chem 274:165–169 [DOI] [PubMed] [Google Scholar]
- Chen S, McCormick JA, Prabaker K, Wang J, Pearce D, Gardner DG. (2004) Sgk1 mediates osmotic induction of NPR-A gene in rat inner medullary collecting duct cells. Hypertension 43:866–871 [DOI] [PubMed] [Google Scholar]
- Das S, Au E, Krazit ST, Pandey KN. (2010) Targeted disruption of guanylyl cyclase-A/natriuretic peptide receptor-A gene provokes renal fibrosis and remodeling in null mutant mice: role of proinflammatory cytokines. Endocrinology 151:5841–5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bold AJ. (1985) Atrial natriuretic factor: a hormone produced by the heart. Science 230:767–770 [DOI] [PubMed] [Google Scholar]
- Dietze EC, Caldwell LE, Marcom K, Collins SJ, Yee L, Swisshelm K, Hobbs KB, Bean GR, Seewaldt VL. (2002) Retinoids and retinoic acid receptors regulate growth arrest and apoptosis in human mammary epithelial cells and modulate expression of CBP/p300. Microsc Res Tech 59:23–40 [DOI] [PubMed] [Google Scholar]
- Drewett JG, Garbers DL. (1994) The family of guanylyl cyclase receptors and their ligands. Endocr Rev 15:135–162 [DOI] [PubMed] [Google Scholar]
- Garbers DL, Chrisman TD, Wiegn P, Katafuchi T, Albanesi JP, Bielinski V, Barylko B, Redfield MM, Burnett JC., Jr (2006) Membrane guanylyl cyclase receptors: an update. Trends Endocrinol Metab 17:251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Pandey KN. (2003) Angiotensin II-mediated negative regulation of Npr1 promoter activity and gene transcription. Hypertension 41:730–736 [DOI] [PubMed] [Google Scholar]
- Garg R, Pandey KN. (2005) Regulation of guanylyl cyclase/natriuretic peptide receptor-A gene expression. Peptides 26:1009–1023 [DOI] [PubMed] [Google Scholar]
- Gaub P, Tedeschi A, Puttagunta R, Nguyen T, Schmandke A, Di Giovanni S. (2010) HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation. Cell Death Differ 17:1392–1408 [DOI] [PubMed] [Google Scholar]
- Gomez RA, Norwood VF. (1999) Recent advances in renal development. Curr Opin Pediatr 11:135–140 [DOI] [PubMed] [Google Scholar]
- Huang PH, Chen CH, Chou CC, Sargeant AM, Kulp SK, Teng CM, Byrd JC, Chen CS. (2011) Histone deacetylase inhibitors stimulate histone H3 lysine 4 methylation in part via transcriptional repression of histone H3 lysine 4 demethylases. Mol Pharmacol 79:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap V, Gudas LJ. (2010) Epigenetic regulatory mechanisms distinguish retinoic acid-mediated transcriptional responses in stem cells and fibroblasts. J Biol Chem 285:14534–14548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana ML, Pandey KN. (1996) Atrial natriuretic peptide inhibits the phosphoinositide hydrolysis in murine Leydig tumor cells. Mol Cell Biochem 158:97–105 [DOI] [PubMed] [Google Scholar]
- Kim SH, Kang HJ, Na H, Lee MO. (2010) Trichostatin A enhances acetylation as well as protein stability of ERalpha through induction of p300 protein. Breast Cancer Res 12:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K, Kinoshita K, Hino S, Yano T, Nagare Y, Shimazu H, Nozaki Y, Sugiyama M, Ikoma S, Funauchi M. (2011) Therapeutic effect of retinoic acid on unilateral ureteral obstruction model. Nephron, Exp Nephrol 118:e69–e78 [DOI] [PubMed] [Google Scholar]
- Kumar P, Garg R, Bolden G, Pandey KN. (2010) Interactive roles of Ets-1, Sp1, and acetylated histones in the retinoic acid-dependent activation of guanylyl cyclase/atrial natriuretic peptide receptor-A gene transcription. J Biol Chem 285:37521–37530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HS, Huang B, Ho Jeoung N, Wu P, Steussy CN, Harris RA. (2006) Retinoic acids and trichostatin A (TSA), a histone deacetylase inhibitor, induce human pyruvate dehydrogenase kinase 4 (PDK4) gene expression. Biochim Biophys Acta 1759:141–151 [DOI] [PubMed] [Google Scholar]
- Latham JA, Dent SY. (2007) Cross-regulation of histone modifications. Nat Struct Mol Biol 14:1017–1024 [DOI] [PubMed] [Google Scholar]
- Lee HA, Cho HM, Lee DY, Kim KC, Han HS, Kim IK. (2012) Tissue-specific upregulation of angiotensin-converting enzyme 1 in spontaneously hypertensive rats through histone code modifications. Hypertension 59:621–626 [DOI] [PubMed] [Google Scholar]
- Lee YS, Jeong WI. (2012) Retinoic acids and hepatic stellate cells in liver disease. J Gastroenterol Hepatol 27 (Suppl 2):75–79 [DOI] [PubMed] [Google Scholar]
- Leitman DC, Andresen JW, Catalano RM, Waldman SA, Tuan JJ, Murad F. (1988) Atrial natriuretic peptide binding, cross-linking, and stimulation of cyclic GMP accumulation and particulate guanylate cyclase activity in cultured cells. J Biol Chem 263:3720–3728 [PubMed] [Google Scholar]
- Lennartsson A, Ekwall K. (2009) Histone modification patterns and epigenetic codes. Biochim Biophys Acta 1790:863–868 [DOI] [PubMed] [Google Scholar]
- Levin ER, Gardner DG, Samson WK. (1998) Natriuretic peptides. N Engl J Med 339:321–328 [DOI] [PubMed] [Google Scholar]
- Li Y, Saito Y, Kuwahara K, Rong X, Kishimoto I, Harada M, Adachi Y, Nakanishi M, Kinoshita H, Horiuchi M, et al. (2009) Guanylyl cyclase-A inhibits angiotensin II type 2 receptor-mediated pro-hypertrophic signaling in the heart. Endocrinology 150:3759–3765 [DOI] [PubMed] [Google Scholar]
- Liu C, Chen Y, Kang Y, Ni Z, Xiu H, Guan J, Liu K. (2011) Glucocorticoids improve renal responsiveness to atrial natriuretic peptide by up-regulating natriuretic peptide receptor-A expression in the renal inner medullary collecting duct in decompensated heart failure. J Pharmacol Exp Ther 339:203–209 [DOI] [PubMed] [Google Scholar]
- Liu X, Lü L, Tao BB, Zhu YC. (2008) All-trans retinoic acid inhibits the increases in fibronectin and PAI-1 induced by TGF-beta1 and Ang II in rat mesangial cells. Acta Pharmacol Sin 29:1035–1041 [DOI] [PubMed] [Google Scholar]
- Marumo T, Hishikawa K, Yoshikawa M, Hirahashi J, Kawachi S, Fujita T. (2010) Histone deacetylase modulates the proinflammatory and -fibrotic changes in tubulointerstitial injury. Am J Physiol Renal Physiol 298:F133–F141 [DOI] [PubMed] [Google Scholar]
- Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM. (2007) Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J Biol Chem 282:4408–4416 [DOI] [PubMed] [Google Scholar]
- Nishikimi T, Inaba-Iemura C, Ishimura K, Tadokoro K, Koshikawa S, Ishikawa K, Akimoto K, Hattori Y, Kasai K, Minamino N, et al. (2009) Natriuretic peptide/natriuretic peptide receptor-A (NPR-A) system has inhibitory effects in renal fibrosis in mice. Regul Pept 154:44–53 [DOI] [PubMed] [Google Scholar]
- Oliver PM, Fox JE, Kim R, Rockman HA, Kim HS, Reddick RL, Pandey KN, Milgram SL, Smithies O, Maeda N. (1997) Hypertension, cardiac hypertrophy, and sudden death in mice lacking natriuretic peptide receptor A. Proc Natl Acad Sci USA 94:14730–14735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PM, John SW, Purdy KE, Kim R, Maeda N, Goy MF, Smithies O. (1998) Natriuretic peptide receptor 1 expression influences blood pressures of mice in a dose-dependent manner. Proc Natl Acad Sci USA 95:2547–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseto S, Moriyama T, Kawada N, Nagatoya K, Takeji M, Ando A, Yamamoto T, Imai E, Hori M. (2003) Therapeutic effect of all-trans retinoic acid on rats with anti-GBM antibody glomerulonephritis. Kidney Int 64:1241–1252 [DOI] [PubMed] [Google Scholar]
- Pandey KN. (2008) Emerging roles of natriuretic peptides and their receptors in pathophysiology of hypertension and cardiovascular regulation. J Am Soc Hypertens 2:210–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey KN. (2011) The functional genomics of guanylyl cyclase/natriuretic peptide receptor-A: perspectives and paradigms. FEBS J 278:1792–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey KN, Nguyen HT, Li M, Boyle JW. (2000) Natriuretic peptide receptor-A negatively regulates mitogen-activated protein kinase and proliferation of mesangial cells: role of cGMP-dependent protein kinase. Biochem Biophys Res Commun 271:374–379 [DOI] [PubMed] [Google Scholar]
- Pandey KN, Oliver PM, Maeda N, Smithies O. (1999) Hypertension associated with decreased testosterone levels in natriuretic peptide receptor-A gene-knockout and gene-duplicated mutant mouse models. Endocrinology 140:5112–5119 [DOI] [PubMed] [Google Scholar]
- Pandey KN, Singh S. (1990) Molecular cloning and expression of murine guanylate cyclase/atrial natriuretic factor receptor cDNA. J Biol Chem 265:12342–12348 [PubMed] [Google Scholar]
- Pang M, Kothapally J, Mao H, Tolbert E, Ponnusamy M, Chin YE, Zhuang S. (2009) Inhibition of histone deacetylase activity attenuates renal fibroblast activation and interstitial fibrosis in obstructive nephropathy. Am J Physiol Renal Physiol 297:F996–F1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Do Couto BR, Alfonso-Loeches S, Aguilar MA, Rodriguez-Arias M, Guerri C. (2012) Changes in histone acetylation in the prefrontal cortex of ethanol-exposed adolescent rats are associated with ethanol-induced place conditioning. Neuropharmacology 62:2309–2319 [DOI] [PubMed] [Google Scholar]
- Peserico A, Simone C. (2011) Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol 2011:371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzalis MV, Sarzani R, Dessì-Fulgheri P, Iacoviello M, Forleo C, Lucarelli K, Pietrucci F, Salvi F, Sorrentino S, Romito R, et al. (2003) Allelic variants of natriuretic peptide receptor genes are associated with family history of hypertension and cardiovascular phenotype. J Hypertens 21:1491–1496 [DOI] [PubMed] [Google Scholar]
- Rubattu S, Bigatti G, Evangelista A, Lanzani C, Stanzione R, Zagato L, Manunta P, Marchitti S, Venturelli V, Bianchi G, et al. (2006) Association of atrial natriuretic peptide and type a natriuretic peptide receptor gene polymorphisms with left ventricular mass in human essential hypertension. J Am Coll Cardiol 48:499–505 [DOI] [PubMed] [Google Scholar]
- Schaier M, Liebler S, Schade K, Shimizu F, Kawachi H, Grone HJ, Chandraratna R, Ritz E, Wagner J. (2004) Retinoic acid receptor alpha and retinoid X receptor specific agonists reduce renal injury in established chronic glomerulonephritis of the rat. J Mol Med (Berl) 82:116–125 [DOI] [PubMed] [Google Scholar]
- Shi SJ, Nguyen HT, Sharma GD, Navar LG, Pandey KN. (2001) Genetic disruption of atrial natriuretic peptide receptor-A alters renin and angiotensin II levels. Am J Physiol Renal Physiol 281:F665–F673 [DOI] [PubMed] [Google Scholar]
- Simioniuc A, Campan M, Lionetti V, Marinelli M, Aquaro GD, Cavallini C, Valente S, Di Silvestre D, Cantoni S, Bernini F, et al. (2011) Placental stem cells pre-treated with a hyaluronan mixed ester of butyric and retinoic acid to cure infarcted pig hearts: a multimodal study. Cardiovasc Res 90:546–556 [DOI] [PubMed] [Google Scholar]
- Sun X, Zhang B, Hong X, Zhang X, Kong X. (2013) Histone deacetylase inhibitor, sodium butyrate, attenuates gentamicin-induced nephrotoxicity by increasing prohibitin protein expression in rats. Eur J Pharmacol 707:147–154 [DOI] [PubMed] [Google Scholar]
- Tremblay J, Hum DH, Sanchez R, Dumas P, Pravenec M, Krenova D, Kren V, Kunes J, Pausova Z, Gossard F, et al. (2003) TA repeat variation, Npr1 expression, and blood pressure: impact of the Ace locus. Hypertension 41:16–24 [DOI] [PubMed] [Google Scholar]
- Vellaichamy E, Khurana ML, Fink J, Pandey KN. (2005) Involvement of the NF-kappa B/matrix metalloproteinase pathway in cardiac fibrosis of mice lacking guanylyl cyclase/natriuretic peptide receptor A. J Biol Chem 280:19230–19242 [DOI] [PubMed] [Google Scholar]
- Ventura C, Cantoni S, Bianchi F, Lionetti V, Cavallini C, Scarlata I, Foroni L, Maioli M, Bonsi L, Alviano F, et al. (2007) Hyaluronan mixed esters of butyric and retinoic Acid drive cardiac and endothelial fate in term placenta human mesenchymal stem cells and enhance cardiac repair in infarcted rat hearts. J Biol Chem 282:14243–14252 [DOI] [PubMed] [Google Scholar]
- Wei LN. (2003) Retinoid receptors and their coregulators. Annu Rev Pharmacol Toxicol 43:47–72 [DOI] [PubMed] [Google Scholar]
- Wu M, Zhang Y, Wu NH, Shen YF. (2009) Histone marks and chromatin remodelers on the regulation of neurogenin1 gene in RA induced neuronal differentiation of P19 cells. J Cell Biochem 107:264–271 [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. (2007) HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26:5310–5318 [DOI] [PubMed] [Google Scholar]
- Zhao D, Das S, Pandey KN. (2013) Interactive roles of NPR1 gene-dosage and salt diets on cardiac angiotensin II, aldosterone and pro-inflammatory cytokines levels in mutant mice. J Hypertens 31:134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zile MH. (2010) Vitamin A-not for your eyes only: requirement for heart formation begins early in embryogenesis. Nutrients 2:532–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.