Abstract
While medulloblastoma, a pediatric tumor of the cerebellum, is characterized by aberrations in developmental pathways, the majority of genetic determinants remain unknown. An unbiased Sleeping Beauty transposon screen revealed MyoD as a putative medulloblastoma tumor suppressor. This was unexpected, as MyoD is a muscle differentiation factor and not previously known to be expressed in cerebellum or medulloblastoma. In response to deletion of one allele of MyoD, two other Sonic hedgehog-driven mouse medulloblastoma models showed accelerated tumor formation and death, confirming MyoD as a tumor suppressor in these models. In normal cerebellum, MyoD was expressed in the proliferating granule neuron progenitors that are thought to be precursors to medulloblastoma. Similar to some other tumor suppressors that are induced in cancer, MyoD was expressed in proliferating medulloblastoma cells in three mouse models and in human medulloblastoma cases. This suggests that although expression of MyoD in a proliferating tumor is insufficient to prevent tumor progression, its expression in the cerebellum hinders medulloblastoma genesis.
Keywords: tumor suppressor, medulloblastoma, cerebellum, bHLH transcription factor, gene regulation
Introduction
Brain tumors are among the leading cause of childhood cancer-related deaths, and medulloblastoma is the most common pediatric brain malignancy with largely undetermined molecular pathogenesis. Based on molecular signatures, medulloblastomas are broadly categorized into four main subgroups – SHH-driven, WNT-driven, and the poorly characterized Group 3 and Group 4 variants (1). However, the vast majority of genetic drivers of this highly heterogeneous cancer remain unknown.
In rare instances, medulloblastomas can show evidence of differentiation along non-neuronal lineages as evidenced by melanin production or expression of muscle markers (2, 3). The rare variant of medulloblastoma that shows some microscopic features resembling muscle are classified by the World Health Organization as “medullomyoblastoma” (4). Diagnosis of these tumors is typically made by immunohistochemical staining for myogenic markers like fast myosin, desmin, myoglobin (5). Transcription factors that drive myogenic differentiation in medulloblastoma have not previously been reported.
MyoD, a basic-helix-loop transcription factor, is a critical lineage-restricted master regulator of skeletal muscle development (6, 7). Exogenous expression of MyoD is sufficient to drive non-muscle cells (e.g. fibroblasts, chondroblasts and others) into the skeletal muscle lineage (8, 9). MyoD function or expression during normal cerebellar development or tumorigenesis, however, remains unknown.
Findings in a variety of cancers implicate MyoD as a possible tumor suppressor. MyoD is epigenetically silenced in solid tumors, including prostate and colon cancer (10, 11) as well as during immortalization of cell lines (12, 13), yet the functional significance of this has not been elucidated. MyoD has not previously been considered as a possible tumor suppressor in medulloblastoma.
Here we identify MyoD as a candidate medulloblastoma tumor suppressor in an unbiased Sleeping Beauty transposon-based in vivo screen. We confirm that single allele loss of MyoD is sufficient to accelerate Sonic hedgehog (Shh)-driven medulloblastoma genesis and that the chromosomal region that expresses MyoD is deleted in some human medulloblastomas. We show that MyoD is expressed in normal cerebellar development, in the cells that are thought to be precursors of Shh-driven medulloblastomas. Our study of MyoD as a novel tumor suppressor in medulloblastoma adds a new dimension to the functional versatility of this lineage-restricted muscle determinant, while providing a unique insight into the critical regulation of gene expression in medulloblastoma.
Materials and Methods
Smo Transgenic and MyoD +/− Mouse Lines
The ND2:SmoA1 (SmoA1), ND2:SmoA2 (SmoA2) transgenic mouse lines, PtchF/F Math1-Cre conditional knock out and MyoD+/− mice and genotyping protocols have been previously described (14–17).
Human Tissue Samples
Collection and use of human tissue samples were approved by the Institutional Review Boards of each institution.
Histopathology, Immunohistochemistry and Immunofluorescence
Mice were euthanized using CO2 inhalation and tissue snap-frozen for RNA studies or fixed in 10% formalin for histological examination. Formalin-fixed paraffin-embedded tissues were cut into 4-μm sections. For IHC, anti-MyoD (5.8A, BD Biosciences 1:200) followed by anti-mouse Fab frag-ME kit/CSA detection kit, and anti-Ki67 (Novocastra, 1:200) were used. Data were confirmed using additional MyoD antibodies (rat Active motif 1:75; rabbit Santa Cruz Biotechnology M-318). Slides were developed using DAB Plus reagent followed by DAKO Hematoxylin counterstain (DAKO). For human MYOD detection, anti-MYOD (Novocastra, 1:80) and (Epitomics, 1:500) were used. For immunofluorescence assays, the following antibodies and protocols were used: MyoD - rat anti-MyoD (Active Motif 1:75) followed by goat anti-Rat HRP (1:500) and CSAII Amplification Reagent with FITC. This was confirmed by mouse anti-MyoD (5.8A, BD Biosciences 1:200), followed by anti-mouse Fab-fragment ME-kit, secondary CSA-SA Alexa 350 polymer (Molecular probes, Invitrogen). Antigen-retrieval was performed using the Biocare Rodent Decloaker system. Ki67 - rabbit anti-Ki67 (Novocastra, 1:100) followed by goat anti-rabbit Alexa 647 (Molecular Probes, Invitrogen) pseudocolored red in image. Math1: rabbit anti-Math1 (LS Bio, 1:50) followed by goat anti-rabbit Alexa 647 (Molecular Probes, Invitrogen) pseudocolored red in image. NeuN: mouse anti-NeuN (Millipore/Chemicon, 1:75) followed by unconjugated Rabbit anti-mouse with ME kit and goat anti-rabbit Alexa 647 (Molecular Probes, Invitrogen) pseudocolored red in image. DAPI was used as nuclear counterstain.
Image Acquisition
Images were acquired using the following methods: (1) Transmitted light color images of stained tissue sections were acquired on a Nikon E800 microscope fitted with a Nikon 10/0.45 or 20/0.75 Plan Apo objective and Photometrics Coolsnap cf color CCD camera; (2) 3-D stacks of optical sections were acquired on an Applied Precision Deltavision microscope fitted with an Olympus 100/1.35 UPlan Apo oil immersion objective, and a Photometrics Coolsnap HQ CCD camera. The image stacks were deconvolved using the manufacturer’s SoftWorx software; (3) tissue sections were imaged with a Aperio ScanScope FL slide scanner or 3D Histech’s Panoramic 250 Flash whole slide scanner using a 20× objective. Minimal Image adjustments in accordance with journal regulations were made using Image J or Adobe Photoshop CS5.
Image processing method
Regions of interest (ROIs) (1024 × 1024) were manually extracted using ImageScope software v11.1.2.752 from scans acquired with an Aperio ScanScope FL slide scanner using a 20× objective. Cell abundance and protein expression measurements of all DAPI positive cells co-stained with anti-Ki67 and anti-MyoD were quantified at the single cell level using a classifier rule set generated with CellProfiler and CellProfiler Analyst (version r11710) (18, 19). Cumulative distribution functions of the classification rule versus fractional relative cell frequency were generated in Graphpad Prism 6.02 to assess protein expression level changes.
Microarray Analysis
RNA was extracted using Qiagen RNeasy mini kit from whole tumor lysates (n=3 per group) and the Agilent 2100 Bioanalyzer Expert was used for quality assessment. Samples were processed at the FHCRC Genomics Shared Resource according to Illumina standard protocols. Array analysis was carried out using the Illumina MouseWG-6 v2.0 Expression BeadChip Kit. Microarray data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE51219. Data were processed using the Bioconductor package ‘limma’ (20) and quantile normalized using the lumi bioconductor package (21–24). A probe was determined to being significantly differentially expressed if: |logFC| > .585 & adj.P.Val < 0.05.
qRT-PCR
RNA was isolated using miRNAeasy Kit (Invitrogen), DNase (Qiagen) treated and converted to cDNA using High Capacity Reverse Transcription kit (Applied Biosystems). Reactions were set up using ABI SYBR green or Taqman Master Mix and run on an ABI 7900HT Fast Real-Time PCR System. Taqman Gene Expression Assays were used for mouse MyoD, b2m, Human MYOD, PPIA. For SYBR (Invitrogen) assays, primers (Document S1) were designed using Primer3 software (25). Data were analyzed using SDS 2.3 software. All conditions were run in triplicates and normalized to b2m or Ppia/PPIA endogenous controls.
Western Blot Analysis
Protein lysates were prepared using RIPA Buffer (Millipore) with Halt Protease Inhibitor Cocktail (Pierce), Phosphatase Inhibitor Cocktails (Calbiochem/Sigma). 25ug protein from each sample were subject to SDS-PAGE using NuPAGE Novex Bis-Tris gels, transferred to nitrocellulose membranes using X-Cell SureLock Mini cell (Invitrogen), probed with primary and corresponding secondary antibodies (Document S1). Proteins were detected using ECL chemiluminiscent substrate (Pierce).
Molecular Classification of Human Medulloblastomas
The molecular classification of medulloblastoma tumors using a nanoString-based assay was described previously (26). Briefly, the RNA expressions of markers were measured using a nanoString assay. The expression values were log-transformed, batch-corrected, normalized to endogenous controls, and used as features for class prediction using the Prediction Analysis for Microarrays (PAM) (27) algorithm, as implemented in the pamr package (v 1.51). The class predictions were then filtered using pre-defined confidence score thresholds for bona fide predictions.
Statistical Analysis
For the analysis of qRT-PCR data, statistical significances of differences between means from two groups were tested using two-tailed Student t-test. Survival curves were plotted using Kaplan-Meier method(28) and compared using two-sided log-rank test(29). Statistical analyses were performed in R statistical systems (http://www.r-project.org). Survival analyses used animal death times as events and mice that were still alive at the time of analysis were censored. A nonparametric Kolmogorov-Smirnov statistical test was performed to determine if differences in MyoD single cell expression measurements from each genotype (cumulative distribution functions) are statistically significant. The level of significance for all tests was 0.05 (alpha)
Results
Genomic loss of MYOD is observed in medulloblastoma
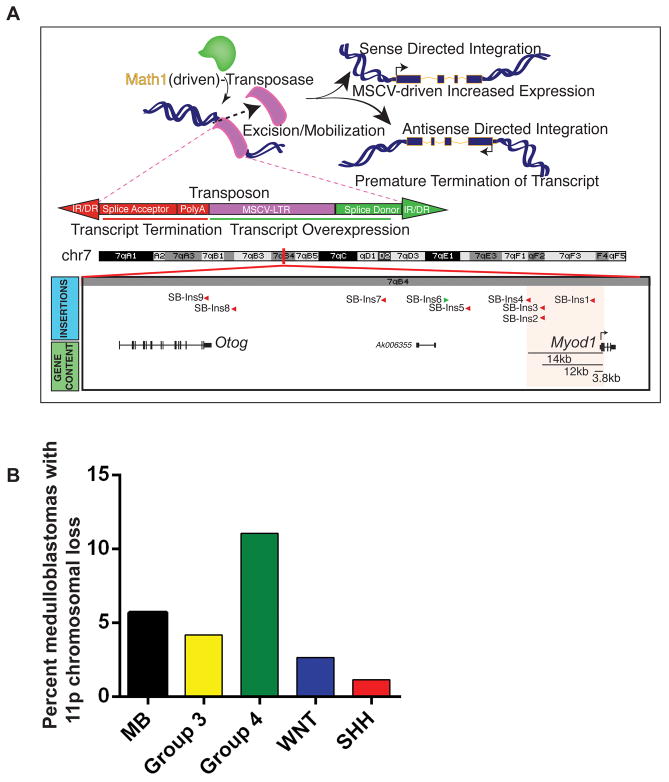
The Sleeping Beauty (SB) Transposon system is an unbiased, in vivo genetic tool allowing identification of oncogenes and tumor suppressor genes through random integration and clonal expansion in a model of medulloblastoma (30). Using this system, MyoD was identified as a gene-centric common insertion site (gCIS) (Figure 1A). The targeting of MyoD by loss-of-function insertions suggested a selective pressure to reduce MyoD expression.
Figure 1. Genomic loss of MyoD in medulloblastoma.
A, MyoD locus is a gene-centric common insertion site in the Sleeping Beauty transposon-driven primary mouse medulloblastoma on a Ptch +/− background (30). Sleeping Beauty Transposon integration map shows location of transposons in the MyoD loci 3–14 kb upstream of the MyoD translational start site in mouse chromosome 7. The transposons are integrated in a direction antisense to MyoD thereby disrupting transcription from the MyoD loci. B, Copy number analysis of primary medulloblastomas show hemizygous deletion of the MYOD genomic loci (11p15.1) in 6% (47/827) of medulloblastomas, being significantly enriched in enriched in Group 4 tumors (35/317) followed by Group 3 (7/168), WNT (2/76) and SHH (3/266) subgroup tumors (p < 1.37E-02 based on Fisher’s Exact Test).
Further to this finding, we investigated whether a similar phenomenon occurred in human medulloblastomas. While no mutations were observed in MYOD across a cohort of previously sequenced tumors (0/310) (31–34), copy number analysis revealed hemizygous deletion of the 11p arm encompassing the MYOD genomic loci (11p15.1) in 6% (47/827) of medulloblastomas (Figure 1B). This cytogenetic event was observed in 2/76 WNT tumors, 3/266 SHH tumors, 7/168 Group 3 tumors and more enriched in the highly aggressive Group 4 tumors (35/317).
Loss of MyoD accelerates tumorigenesis in mouse models of medulloblastoma
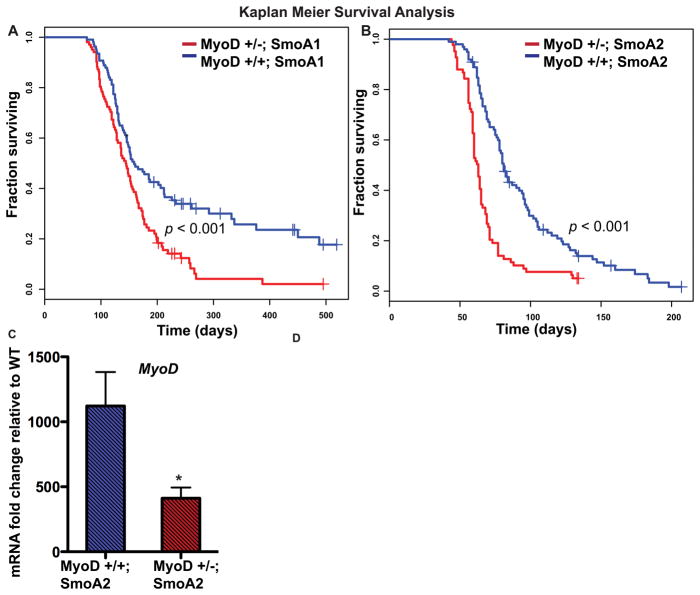
Our lab previously generated and characterized two mouse models of medulloblastoma (14, 15, 35). To directly assess whether MyoD reduction functionally contributed to medulloblastoma genesis in vivo, we crossed these two different medulloblastoma mouse models, SmoA1 or SmoA2 mice, to MyoD +/− mice (17) to obtain mice with reduced MyoD. Interestingly, SmoA1 or SmoA2 mice homozygous null for MyoD were born in sub-mendelian ratios with compromised general health and the majority died within a few weeks of postnatal life. The cause of death is unknown but unrelated to tumorigenesis (Figure S1). Heterozygous reduction in MyoD expression led to significant acceleration of tumor formation in both MyoD+/−; SmoA1 and MyoD+/−; SmoA2 mice as compared to MyoD+/+; SmoA1 and MyoD+/+; SmoA2 mice, respectively (Figure 2A, B). In stage-matched tumors, we observed a trend towards a higher proliferative index in the faster onset MyoD+/−; SmoA2 tumors compared to MyoD+/+; SmoA2 (Figure S2). We validated the reduction of MyoD in the SmoA2 MyoD+/− mice at the mRNA and protein level, confirming our genetic model (n=5, p<0.05) (Figure 2C, S2). IHC analysis suggests that the reduction in MyoD in the MyoD+/−; SmoA2 tumors compared to the MyoD+/+;SmoA2 group, stems from possibly both an overall reduction in the number of MyoD+ cells as well as the level of expression at a cellular level (Figure S2). These functional mouse genetic experiments, together with the Sleeping Beauty screen, confirmed MyoD to be a tumor suppressor in medulloblastoma genesis.
Figure 2. MyoD deficiency leads to accelerated tumorigenesis in medulloblastoma models.
A, B, Kaplan-Meier analyses comparing MyoD+/+; SmoA1 (n=108) with MyoD+/−; SmoA1 (n=102) mice and MyoD+/+; SmoA2 (n=99) with MyoD+/−; SmoA2 (n=83) mice, show accelerated tumorigenesis in SmoA1 (p = 1.44E-04) and SmoA2 mice (p =3.27E-08) lacking one allele of MyoD. C, MyoD levels in MyoD+/−;SmoA2 tumors are reduced compared to MyoD+/+; SmoA2 tumors (n=5 per genotype, *p < 0.05 by two-tailed Student t-test) as determined by qRT-PCR. β2m was used for data normalization.
MyoD is expressed in the developing mouse cerebellum and hyperproliferative cells of SmoA2 mice
The finding that the MyoD locus was a gene-centric common insertion site in the Sleeping Beauty screen and that reduced MyoD accelerated medulloblastomas in both the SmoA1 and SmoA2 models clearly demonstrated a functional role of MyoD as a medulloblastoma tumor suppressor in vivo. Because MyoD expression has not previously been reported in the developing cerebellum or in medulloblastoma, we sought to carefully characterize MyoD expression in both normal cerebella and in mouse medulloblastomas.
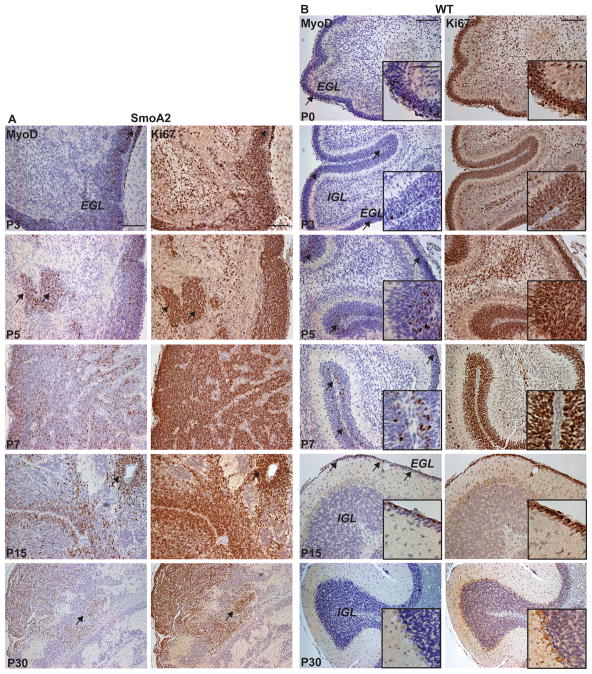
We first assessed MyoD expression in the brains of the SmoA2 mouse medulloblastoma model. Immunohistochemical (IHC) analysis revealed MyoD expression in the expanded external granule layer (EGL) and hyperproliferative lesions within the interior of the SmoA2 developing cerebellum (Figure 3A). MyoD is expressed in the normal developing cerebellum as well, restricted to the Ki67+ outermost, undifferentiated EGL from postnatal day (P) 0 to P15 in mice (Figure 1B). A comparison of WT versus the SmoA2 cerebella revealed pronounced differences in MyoD expression (Figure 3A, B). Interestingly, MyoD expression is fully silenced in the mature normal cerebellum (P30) yet persists in the undifferentiated proliferating cells of the SmoA2 cerebellum through adulthood. While our focus was on the role of MyoD in the context of medulloblastoma genesis, its expression in normal development prompted us to evaluate MyoD −/− cerebella at P5, which did not show any apparent abnormalities.
Figure 3. MyoD is expressed during cerebellar development.
A–B, MyoD and Ki67 IHC analysis during cerebellar development from P0–P30 in (A) SmoA2 and (B) WT mouse cerebella show abundant MyoD+ cells in the proliferative regions of the dysplastic SmoA2 cerebella and in a subset of cells in the Ki67+ WT outer EGL. MyoD expression is undetectable in the P30 WT cerebellum but continues to be expressed in SmoA2. Arrows indicate overlapping regions of Ki67 and MyoD positivity. Scale bar: 100μm; inset 50 μm.
MyoD is expressed in proliferating medulloblastoma cells in three SHH medulloblastoma mouse models
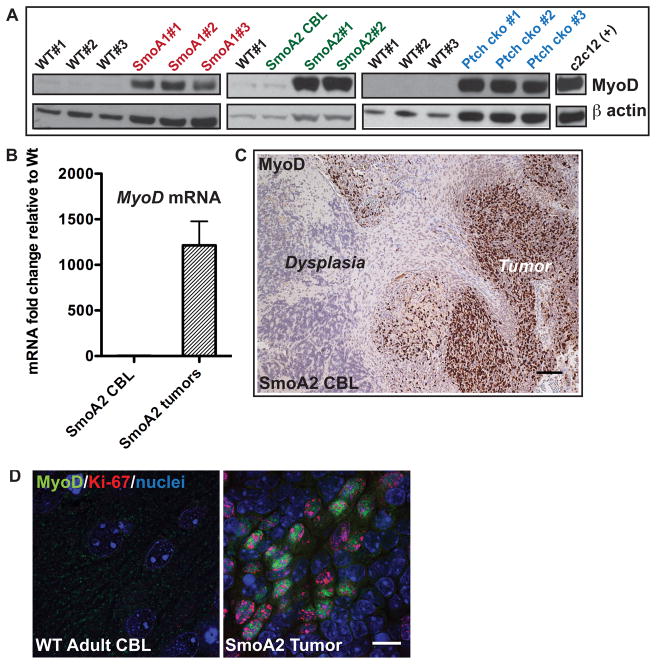
Whereas some tumor suppressors are absent or diminished in cancer, others are upregulated in proliferating cancer cells in an unsuccessful attempt to regulate proliferation or differentiation. In our mouse medulloblastoma models, the latter seems to be the case. We found high levels of MyoD in three independent SHH-subgroup mouse models - SmoA1 (14, 15), SmoA2 (35) and PtchF/F Math1-Cre conditional knockout (Ptch cko) tumors (16) (Figure 4A). Importantly, MyoD mRNA and protein were not expressed in regions of cerebellar dysplasia, that commonly occur in the adult SmoA2 mice, but rather only in neoplastic tissue (Figure 4B, C). The lack of expression of MyoD in SmoA2-expressing non-tumor cells suggests that MyoD is not a direct target of the conditionally active Smoothened protein, but its expression is maintained as a consequence of the cellular transformation process. To confirm our hypothesis, we examined the relationship between proliferation (via Ki67+) and MyoD expression using immunofluorescence. MyoD expression was observed exclusively in Ki67+ cells in tumors derived from SmoA1, SmoA2 and Ptch cko mice (Figure 4D, S3). The proportion of MyoD+ cells appeared to be highest in the outer region of the tumors known to develop from the hyperproliferative EGL, as compared to the inner core (Figure S4). No proliferating (Ki67+) or MyoD+ cells were detected in the WT adult cerebellum. Thus, like P53, TP73, pRb2/p130, ARF and p16INK4a in selected types of cancer (36–41), MyoD appears to be a tumor suppressor that is expressed in response to oncogenic signaling, but insufficient as a single protein to prevent medulloblastoma progression.
Figure 4. MyoD is expressed in proliferating medulloblastoma cells.
A, MyoD protein is expressed in cerebellar tumors from three mouse models of medulloblastoma – SmoA1, SmoA2 and PtchF/F Math1-Cre conditional knockout (Ptch cko) tumors at levels comparable to c2c12 myoblast cells as determined by Western blot analysis. MyoD remains undetectable in adult WT or adult non-tumor SmoA2 CBL. β actin was used as the loading control. B, SmoA2 tumors express high levels of MyoD relative to WT adult cerebellum (CBL) (n=5 per genotype) while expression in non-tumor adult SmoA2 CBL is similar to WT, as determined by qRT-PCR. β2m was used for data normalization. All data represent mean +/− SEM. C, IHC analysis shows MyoD is expressed only in tumor cells but not in cells in the contiguous region of dysplasia in representative SmoA2 CBL (n=5). Scale Bar: 100um D, MyoD (green) is localized only in Ki67+ (red) tumor cells as determined by immunofluorescence analysis (n=8 tumors). Scale Bar: 10μm.
MyoD co-localizes with Math1 in granule neuronal precursors
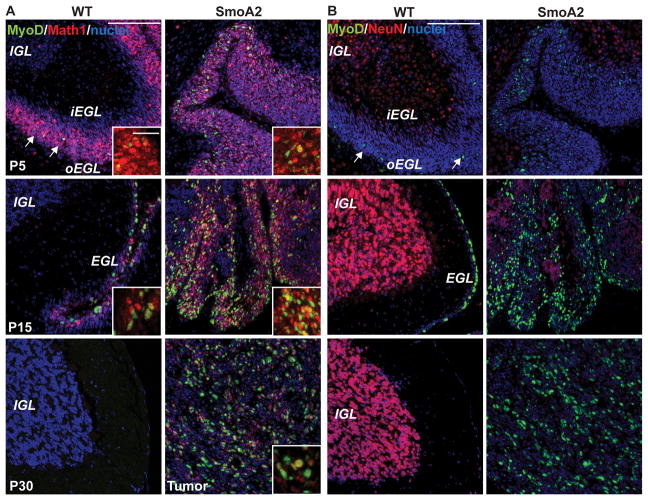
Granule neuron precursors (GNPs) are a transiently proliferative population of cells that form the EGL and are considered to be the cell-of-origin for Shh medulloblastomas (42). To characterize the relationship of MyoD to normal cerebellar development and Shh-driven medulloblastoma genesis, we carried out dual immunofluorescence for MyoD with Math1, a GNP marker and NeuN, a marker of differentiated granule neurons. Our results demonstrate that MyoD is observed exclusively in the Math1 compartment and mutually exclusive with NeuN (Figure 5A,B). Thus it appears that MyoD is expressed in proliferating normal GNPs during the final stage prior to cell cycle arrest and differentiation. It is possible, but unproven that Shh induction of MyoD in these normal cells and in proliferating Shh-induced medulloblastoma cells is part of an onco-fetal development program that is aberrantly activated in medulloblastoma.
Figure 5. MyoD co-localizes with Math1 in granule neuron progenitors and is not expressed in NeuN-expressing differentiated granule neurons.
A, MyoD (green) co-localizes with Math1 (red) in P5 and P15 outer EGL of WT and SmoA2 developing cerebella (n=3 per group). Both MyoD and Math1 are silenced in the WT adult cerebellum at P30, but continue to be expressed in the SmoA2 tumors. B, MyoD (green) remains exclusive with NeuN (red) which is expressed in differentiated granule neurons in the IGL at P5 and P15. SmoA2 tumor cells are predominantly negative for NeuN. Scale Bar: 100μm, inset 25 μm
The tumor suppressor function of MyoD is not executed through the canonical myogenic differentiation program
To determine whether the canonical myogenic differentiation program was involved in increased tumorigenicity following loss of MyoD, we performed gene expression analysis comparing MyoD+/+; SmoA2 with MyoD+/−;SmoA2 tumors. Surprisingly, no single gene candidate passed statistical significance between the two genotypes. To identify subtle transcriptional differences possibly beyond the detection limit of an array-based approach, we carried out quantitative reverse transcription (qRT)-PCR analyses on canonical genes involved in the MyoD-mediated skeletal muscle differentiation program. Myf5, Myog, Desmin, Cdh15 showed no difference between the two groups. The only gene involved in myogenic differentiation that differed between the two genotypes was Id3, which was reduced by approximately two-fold (Figure S5). Notably Id3 is involved in many differentiation programs, including hematopoeisis and neurogenesis. These tumors did not show any histological evidence of muscle differentiation and were negative for myogenic markers, desmin and myoglobin by IHC as well. Taken together, these data demonstrate that MyoD does not utilize the canonical myogenic differentiation program as the basis of tumor suppression.
MyoD is expressed in a subset of human medulloblastomas
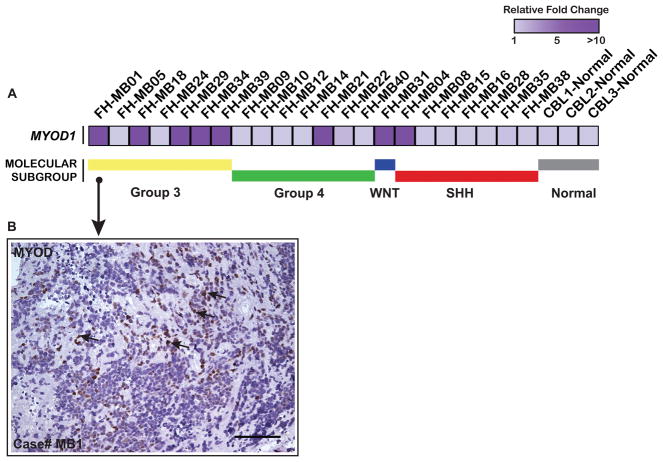
Medulloblastomas with myogenic differentiation (medullomyoblastomas) have been described based on histopathological criteria. To determine whether MyoD was expressed in human medulloblastomas and learn whether expression correlated with molecular subtype, we performed qRT-PCR analysis of MYOD expression in a cohort of primary medulloblastomas (n=22) after establishing molecular subgroup affiliations. MYOD expression was detected in 36% (8/22) of medulloblastomas across the four molecular subgroups, most prominent in Group 3 medulloblastomas. FH-MB01, the tumor with the highest level of MYOD expression (Figure 6A), was confirmed to express nuclear MYOD protein by IHC analysis albeit sparse and heterogeneous throughout the tumor section (Figure 6B). There was no histological evidence of differentiated skeletal muscle cells, consistent with the patient’s pathology report and our findings that MyoD did not appear to execute a canonical muscle differentiation program in brain or brain tumors. Subgroup-specific expression patterns of MYOD were confirmed across an independent and non-overlapping validation series (n=103) (Figure S6).
Figure 6. MYOD is expressed in a subset of human medulloblastomas.
A, Heatmap analysis of MYOD mRNA expression data obtained by qRT-PCR analysis, in human medulloblastoma specimens shows 36% (8/22) human medulloblastomas to express MYOD. PPIA was used for data normalization. B, Case # FH-MB01 with highest MYOD mRNA in (A), shows heterogeneous nuclear MYOD expression (arrows) as determined by IHC analysis. Scale Bar: 100um.
Discussion
Medulloblastoma is a pediatric malignancy characterized by aberrant developmental pathways. Here we present the first report of MyoD expression in GNPs during normal cerebellar development restricted to the outermost proliferative and undifferentiated EGL in mice. While quiescent in the normal adult cerebella (i.e. beyond P15 as the GNPs in the EGL exit the cell cycle and migrate inwards to form the differentiated internal granular layer (IGL)), MyoD expression is retained in the neoplastic expansion of the EGL in mouse models of medulloblastoma. We demonstrate through genetic studies with multiple lines of mice that loss of one allele of MyoD significantly accelerates tumorigenesis in vivo establishing its role as a tumor suppressor gene in medulloblastoma.
The absence of detectable MyoD expression in non-tumor cells from SmoA1 or SmoA2 mice, that differentiate into mature neurons, suggests that MyoD is not a direct target of conditionally activated Smoothened protein; rather the developmental expression of MyoD occurs as a result of neoplastic transformation.
MyoD expression in Ki67+ tumor cells is consistent with other examples of anti-proliferative networks being activated by oncogenic signals such as p53 induction by oncogenes E1A/Myc/E2F or ARF activation by Ras/TSC/mTORC1 signaling as responses to counter abnormal hyperproliferative cues (36, 39). In cancer, overexpression of tumor suppressors have been described as inherent protective responses – similar examples include TP73 overexpression in medulloblastoma associated with improved survival outcomes (37); pRb2/p130 in hepatocellular carcinoma (38); p16INK4a induction in HPV-induced tumors as an attempt to arrest proliferation (40); wildtype p53 overexpression in human glioma (41). The normal developmental expression of MyoD in proliferative GNPs only during a period of intense growth suggests an important role in regulation of cell proliferation prior to neuronal maturation.
Ki67 is expressed in all active phases of the cell cycle (absent in G0) (43) whereas MyoD has been shown to undergo a biphasic expression pattern in proliferating myoblasts, peaking in the middle of G1 and at the end of G2 (44). Our results therefore suggest that the Ki67+/MyoD+ medulloblastoma cells may be in G1 or G2. Since cell cycle exit and induction of differentiation can occur in G1 or in late G2/mitosis, a strong expression of MyoD may define a temporal window in which differentiation can occur (44). Moreover, MyoD has also been shown to induce cell cycle arrest independent of differentiation in normal as well as transformed cells (45, 46). Therefore, functions of MyoD in cell cycle regulation (47–49) provide important leads for further understanding the mechanism of action of MyoD as a tumor suppressor.
Pro-proliferative molecules like Id3 are known to antagonize the function of MyoD in the myogenic program (6) as well as play key neurodevelopmental functions such as inhibition of differentiation during neurogenesis (50). Our observation of elevated Id3 following the loss of MyoD together with the known contributions of Id genes to tumor growth, metastasis and vascularization in cancer (50, 51) suggest a possible connection of Id3 to the MyoD-regulated tumor suppressor network in medulloblastoma. A recent study by Bai et al. shows oncogene Otx2 to repress the MyoD enhancer in medulloblastoma cells whereby the loss of Otx2 causes activation of the myogenic differentiation program in vitro (4). These results directly support our finding of a tumor suppressor role of MyoD in medulloblastoma in vivo. As MyoD is a functionally versatile molecule that binds thousands of genes to exert its complex genetic and epigenetic regulatory functions, further investigation of the downstream targets in the MyoD-regulated tumor suppressor network remains an important future step.
The role of MyoD in tumor suppression and in mammalian brain development is novel and fascinating, with a highly complex underlying biology. Based on our current findings, we propose the following model for MyoD activity in cerebellar development and medulloblastoma genesis: During development, MyoD is transiently expressed in the proliferating GNPs potentially playing a key role in the maturation of the normal cerebellum through mechanisms unrelated to its myogenic targets. While MyoD is normally silenced as GNPs differentiate into mature neurons in the normal cerebellum, medulloblastoma tumor cells either retain MyoD or re-induce expression of MyoD under the influence of oncogenes such as the constitutively active Shh pathway. While aberrant expression of MyoD in proliferating medulloblastoma cells is insufficient to prevent tumor formation, reduction of endogenous MyoD expression in developing cerebellum is sufficient to accelerate tumor formation. This is reminiscent of multiple other tumor suppressors that happen to be activated by oncogenic pathways, yet fail to fully impede tumor progression (36–41).
Supplementary Material
Acknowledgments
Human tissue was obtained from the Brain Tumor Tissue Bank, London, Ontario funded by The Brain Tumor Foundation of Canada, NICHD Brain Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD and Seattle Children’s Hospital Cancer Biorepository, Seattle, WA, with appropriate approval from Institutional Review Boards. We wish to thank Dr. Julie-Randolph Habecker, Kimberly Melton and the FHCRC Experimental Histopathology Shared Resource, Ying-tzang Tien at University of Washington (UW) Histopathology Lab, Dr. Paul Swanson and Farinaz Shokri at UW Medicine Anatomic Pathology Lab, Dr. Lauren Snider, Barbara Pullar for colony management and genotyping, Dr. Martine Roussel, Dr. Daisuke Kawaichi for reagents, Dr. Julio Vazquez and David L. McDonald at FHCRC Scientific Imaging, Dr. Jeff Delrow and Ryan Bosom at FHCRC Genomics Shared Resource, Betty Lu and Marc Remke for Nanostring analysis, Drs. Richard Klinghoffer, Michelle Cook Sangar, Michelle Lee, Sally Ditzler, Ilona Tretyak for guidance and technical help.
Grant Support
This work was supported by NIH grants 5R01CA11456705 and 5R01CA112350; NIH NIAMS R01AR045113; The Pediatric Brain Tumor Foundation (MDT). JD was supported by Pre-doctoral Developmental Biology Training Grant 5T32HD007183 from the National Institute of Child Health and Human Development (NICHD), AMD was supported by Canadian Institutes of Health Research (CIHR) Vanier Scholarship, BM was supported by U54CA149237 from the Integrative Cancer Biology Program of the National Cancer Institute.
Footnotes
Conflict of Interest Statement: The authors declare no conflict of interest
Author Contributions
Conception and Design: J.D, J.M.O, S.J.T, A.M.D, M.D.T; Experimental work: J.D, A.M.D, X.W., P.A.N; Technical support: K.D.P, Data analysis: J.D., A.M.D, D.T., P.A.N, X.W., D.S., B.M., M.L.B, manuscript writing and review: J.D., A.M.D., J.M.O., M.D.T., Study supervision: J.M.O., S.J.T., M.D.T
References
- 1.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2011 doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neuro Oncol; Abstracts of the 15th International Symposium on Pediatric Neuro-Oncology; June 24–27 2012; Toronto, Ontario, Canada. 2012. pp. i1–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheema ZF, Cannon TC, Leech R, Brennan J, Adesina A, Brumback RA. Medullomyoblastoma: case report. J Child Neurol. 2001;16:598–9. doi: 10.1177/088307380101600812. [DOI] [PubMed] [Google Scholar]
- 4.Bai RY, Staedtke V, Lidov HG, Eberhart CG, Riggins GJ. OTX2 Represses Myogenic and Neuronal Differentiation in Medulloblastoma Cells. Cancer research. 2012;72:5988–6001. doi: 10.1158/0008-5472.CAN-12-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eberhart CG. Molecular diagnostics in embryonal brain tumors. Brain Pathol. 2011;21:96–104. doi: 10.1111/j.1750-3639.2010.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–95. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 7.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–95. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:5434–8. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:7988–92. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer research. 1998;58:5489–94. [PubMed] [Google Scholar]
- 11.Mishra DK, Chen Z, Wu Y, Sarkissyan M, Koeffler HP, Vadgama JV. Global methylation pattern of genes in androgen-sensitive and androgen-independent prostate cancer cells. Mol Cancer Ther. 2010;9:33–45. doi: 10.1158/1535-7163.MCT-09-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones PA, Wolkowicz MJ, Rideout WM, 3rd, Gonzales FA, Marziasz CM, Coetzee GA, et al. De novo methylation of the MyoD1 CpG island during the establishment of immortal cell lines. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:6117–21. doi: 10.1073/pnas.87.16.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rideout WM, 3rd, Eversole-Cire P, Spruck CH, 3rd, Hustad CM, Coetzee GA, Gonzales FA, et al. Progressive increases in the methylation status and heterochromatinization of the myoD CpG island during oncogenic transformation. Mol Cell Biol. 1994;14:6143–52. doi: 10.1128/mcb.14.9.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 15.Hatton BA, Villavicencio EH, Tsuchiya KD, Pritchard JI, Ditzler S, Pullar B, et al. The Smo/Smo model: hedgehog-induced medulloblastoma with 90% incidence and leptomeningeal spread. Cancer Res. 2008;68:1768–76. doi: 10.1158/0008-5472.CAN-07-5092. [DOI] [PubMed] [Google Scholar]
- 16.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–45. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–90. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 18.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1826–31. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyth GK. Limma: linear models for microarray data. In: Gentleman RVCS, Dudoit RI, Huber W, editors. ‘Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 21.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 22.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du P, Kibbe WA, Lin SM. nuID: a universal naming scheme of oligonucleotides for illumina, affymetrix, and other microarrays. Biol Direct. 2007;2:16. doi: 10.1186/1745-6150-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 26.Northcott PA, Shih DJ, Remke M, Cho YJ, Kool M, Hawkins C, et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta neuropathologica. 2012;123:615–26. doi: 10.1007/s00401-011-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–72. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Journal of American Statistical Association. 1958;53:457–81. [Google Scholar]
- 29.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemotherapy Reports. 1966;50:163–70. [PubMed] [Google Scholar]
- 30.Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–33. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–5. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–10. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–8. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dey J, Ditzler S, Knoblaugh SE, Hatton BA, Schelter JM, Cleary MA, et al. A distinct smoothened mutation causes severe cerebellar developmental defects and medulloblastoma in a novel transgenic mouse model. Molecular and cellular biology. 2012;32:4104–15. doi: 10.1128/MCB.00862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–15. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 37.Castellino RC, De Bortoli M, Lin LL, Skapura DG, Rajan JA, Adesina AM, et al. Overexpressed TP73 induces apoptosis in medulloblastoma. BMC Cancer. 2007;7:127. doi: 10.1186/1471-2407-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huynh H. Overexpression of tumour suppressor retinoblastoma 2 protein (pRb2/p130) in hepatocellular carcinoma. Carcinogenesis. 2004;25:1485–94. doi: 10.1093/carcin/bgh154. [DOI] [PubMed] [Google Scholar]
- 39.Miceli AP, Saporita AJ, Weber JD. Hypergrowth mTORC1 signals translationally activate the ARF tumor suppressor checkpoint. Molecular and cellular biology. 2012;32:348–64. doi: 10.1128/MCB.06030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reuschenbach M, Waterboer T, Wallin KL, Einenkel J, Dillner J, Hamsikova E, et al. Characterization of humoral immune responses against p16, p53, HPV16 E6 and HPV16 E7 in patients with HPV-associated cancers. Int J Cancer. 2008;123:2626–31. doi: 10.1002/ijc.23837. [DOI] [PubMed] [Google Scholar]
- 41.Alderson LM, Castleberg RL, Harsh GRt, Louis DN, Henson JW. Human gliomas with wild-type p53 express bcl-2. Cancer research. 1995;55:999–1001. [PubMed] [Google Scholar]
- 42.Kho AT, Zhao Q, Cai Z, Butte AJ, Kim JY, Pomeroy SL, et al. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes & development. 2004;18:629–40. doi: 10.1101/gad.1182504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–22. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 44.Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb NJ, Fernandez A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J Cell Biol. 1998;142:1447–59. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crescenzi M, Fleming TP, Lassar AB, Weintraub H, Aaronson SA. MyoD induces growth arrest independent of differentiation in normal and transformed cells. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8442–6. doi: 10.1073/pnas.87.21.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorrentino V, Pepperkok R, Davis RL, Ansorge W, Philipson L. Cell proliferation inhibited by MyoD1 independently of myogenic differentiation. Nature. 1990;345:813–5. doi: 10.1038/345813a0. [DOI] [PubMed] [Google Scholar]
- 47.de la Serna IL, Roy K, Carlson KA, Imbalzano AN. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J Biol Chem. 2001;276:41486–91. doi: 10.1074/jbc.M107281200. [DOI] [PubMed] [Google Scholar]
- 48.Otten AD, Firpo EJ, Gerber AN, Brody LL, Roberts JM, Tapscott SJ. Inactivation of MyoD-mediated expression of p21 in tumor cell lines. Cell Growth Differ. 1997;8:1151–60. [PubMed] [Google Scholar]
- 49.Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, et al. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. Embo J. 1997;16:369–83. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O’Reilly R, et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–7. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 51.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–14. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.