Key Points
Mice that express a mutation in STAT3 phenocopy patients with HIES.
Bone marrow transplantation does not fully correct the susceptibility of these animals to bacterial infection.
Abstract
Mutations of STAT3 underlie the autosomal dominant form of hyperimmunoglobulin E syndrome (HIES). STAT3 has critical roles in immune cells and thus, hematopoietic stem cell transplantation (HSCT), might be a reasonable therapeutic strategy in this disease. However, STAT3 also has critical functions in nonhematopoietic cells and dissecting the protean roles of STAT3 is limited by the lethality associated with germline deletion of Stat3. Thus, predicting the efficacy of HSCT for HIES is difficult. To begin to dissect the importance of STAT3 in hematopoietic and nonhematopoietic cells as it relates to HIES, we generated a mouse model of this disease. We found that these transgenic mice recapitulate multiple aspects of HIES, including elevated serum IgE and failure to generate Th17 cells. We found that these mice were susceptible to bacterial infection that was partially corrected by HSCT using wild-type bone marrow, emphasizing the role played by the epithelium in the pathophysiology of HIES.
Introduction
The autosomal dominant (AD) hyperimmunoglobulin E syndrome (HIES) is a multisystem disorder characterized by recurrent and severe cutaneous and sinopulmonary bacterial infections, chronic dermatitis, elevated serum IgE, and connective tissue abnormalities.1 In contrast to other disorders associated with elevations in IgE, AD-HIES is the result of dominant-negative mutations of STAT3.2,3
STAT3 belongs to a family of genes that encode latent cytosolic transcription factors. Each of the 7 members of this family has diverse functions in transmitting cytokine signals. Some STAT family members, including STAT4 and STAT6, have relatively discrete actions.4 By contrast, STAT3 and STAT5 are activated by diverse cytokines and have broad functions in multiple tissues.4 For example, germline deletion of Stat3 in mice is embryonically lethal due to the role played by STAT3 downstream of leukemia inhibitory factor signaling and placental integrity.5 Within immune and hematopoietic cells, the roles of STAT3 are both pleotropic and sometimes contradictory. For example, Stat3 deficiency in myeloid cells results in defective dendritic cell maturation and altered neutrophil homeostasis.6,7 It is also associated with a fatal autoimmunity that stems from an inability of cells to respond to IL-10 stimulation,8-10 leading to a deficiency in myeloid suppressor cell function.11 Stat3-deficient T cells have defects in helper cell differentiation, but with the opposite outcome to that seen in myeloid cells. Stat3−/− CD4+ T cells are unable to express the inflammatory cytokine IL-17,12-14 and mice with Stat3-deficient T cells are resistant to a number of mouse models of autoimmune and alloimmune disease.15-17 In addition to the classical functions of STAT3 as a transcription factor, new roles for STAT3 in regulation of metabolism and mitochondrial function have been discovered.18
Given the finding that Stat3-deficient mice die in utero and the complex phenotypes associated with tissue-specific deletion of Stat3, the discovery that STAT3 mutations underlie AD-HIES was unexpected. This suggests that the presence of the mutant allele results in reduced, but not absent STAT3 function. The susceptibility to infection is explained in part by the failure of CD4+ T cells from HIES patients to produce IL-17, a cytokine important for host defense against Staphylococcus aureus and fungi, infections to which these patients are susceptible.19-21 If the failure of immune cells to produce IL-17 and the functionally related IL-22 is the major cause of morbidity and mortality in these patients, then it would seem logical that hematopoietic stem cell transplantation (HSCT) would be an effective treatment of this disorder. Contradicting this assumption: mice that lack Stat3 within the gut epithelia demonstrate impaired recovery after exposure to dextran sodium sulfate (DSS) drinking water.22 IL-22 is critical for protection against DSS injury,23 and it is dependent on STAT3 both for its expression and its action on epithelial cells.22 Taken together, these data would argue that if STAT3 function in epithelial cells is most critical, then HSCT would be of limited utility.
To try to resolve these issues and to obtain a clearer picture of STAT3's function as it relates to HIES, we generated a murine model of this disease. We found that mice expressing a patient-derived Stat3 allele display both impaired STAT3 DNA-binding activity and expression of IL-17. The mice recapitulated additional aspects of the human disease, including elevated serum IgE and a reduced ability to clear bacterial infection. In addition, challenge of these mice with lipopolysaccharide (LPS) was associated with heightened expression of inflammatory cytokines. To explore the relative contributions of impaired STAT3 signaling within cells of the immune system and cells of the epithelia, we reconstituted HIES mice with wild-type (WT) bone marrow (BM) and vice versa. We found that impaired STAT3 signaling in both compartments contributes to impaired host defense and abnormal inflammatory responses in response to infection with Citrobacter rodentium. That is, reconstitution of mice bearing the mutant Stat3 allele with normal BM, partially, but not completely, reversed host defense defects. These results will need to be borne in mind in consideration of HSCT as a therapy for HIES.
Methods
Generation of mut-S3 transgenic mice
The BAC transgene was constructed by modifying a 185-kb mouse BAC containing the mouse Stat3 gene (RP24-236G5). A 1-kb construct carrying 2 arms of homology (∼500 each) was ligated into the pSV1-RecA shuttle vector, which was transformed into DH10B-competent cells expressing the RP24-236G5 BAC. Proper insertion of the deletion and final bacterial selection on chloramphenicol/fusaric acid plates were monitored by polymerase chain reaction (PCR) using primers internal and external to the homologous construct. The deletion consisted of a 1,163 bp DNA fragment starting with the last codon of Stat3 exon 15 and ending at the 5ʹ of exon 16; the construct also included silent mutations in the last 7 remaining codons of exon 15 to allow specific amplification of WT and mutant transcripts by PCR.
Immunizations
Extracts from Schistosoma mansoni eggs (SEA) were prepared as described.24 The water-soluble fraction was injected intraperitoneally (50 μg) 2 times per week for 2 weeks into mut-Stat3 or control mice. After an additional 3-week rest period sera were collected. Other immunizations were in hind foot pads with 100 μg NP25-CGG precipitated in Alum (Thermo Fisher Scientific). Draining lymph nodes were harvested on day 7.
LPS shock
For the assessment of in vivo responsiveness to LPS, mice were injected intraperitoneally with 250 μg of LPS from Salmonella enterica serovar Minnesota (Sigma-Aldrich). Blood was collected from the tail vein 90 minutes after injection with LPS. Animals were monitored for viability every 4 hours.
BM transplantation
WT and mut-Stat3 mice were irradiated with 900 cGy in a single fraction followed by an injection of 10 million BM cells obtained from either WT or mut-Stat3 mice.
C rodentium infection
C rodentium was prepared by incubation with shaking at 37°C for 6 hours in Luria Broth. After 6 hours, the bacterial density was assessed by absorbance at an optical density of 600 nm and confirmed by plating of serial dilutions. Inoculation of mice was by oral gavage with 5 × 109 colony-forming units (CFU). Animals were monitored for weight loss and after 14 days all animals were euthanized. Sections of spleen, liver, and colon were analyzed for the presence of viable bacteria. Serum samples were taken for measurement of anti-C rodentium specific immunoglobulin. Sections of colon were removed for histology, analysis of messenger RNA (mRNA) expression and lamina propria lymphocytes were isolated and analyzed for cytokine expression by intracellular staining.
Cell isolation and culture
CD4+ T cells were obtained from spleens and lymph nodes using positive selection with anti-CD4 microbeads (Miltenyi Biotec); B cells were isolated from spleens by negative selection using CD43 MACS beads (Miltenyi Biotec). CD11c+ splenocytes were isolated using positive selection with anti-CD11c MACS beads (Miltenyi Biotec). To isolate lamina propria lymphocytes, large intestines were removed, cut into pieces and washed in Hank’s balanced-salt solution. Epithelial layer cells were removed and the remaining tissue was digested at 37°C with collagenase and DNAse I (both Roche). Lamina propria lymphocytes were recovered from the supernatant and pelleted in a solution of 40% Percol.
All cells were cultured in mouse media (RPMI 1640, 10% FCS, 1× antibiotic-antimycotic, 1% glutamine, 1× nonessential amino acids, 1% sodium pyruvate, 14.2 M 2-β-mercaptoethanol, and 10 mM HEPES). T cells were activated with plate-bound anti-CD3/anti-CD28 (5 μg/mL each; eBiosciences). Th17 conditions contained 10 ng/mL IL-6 (Invitrogen), 5 ng/mL transforming growth factor (TGF)-β1 (R&D Systems), 10 μg/mL anti–IL-4, 10 μg/mL anti–interferon (IFN)-γ (BioXCell). To induce class switch recombination/immunoglobulin isotype switching ex vivo, B cells were activated for 72 or 96 hours in the presence of 25 μg/mL LPS (Escherichia coli, 0111:B4; Sigma-Aldrich) and 5 ng/mL IL-4 to induce class switch recombination (CSR) to IgG1/IgE, or LPS and 2 ng/mL IFN-γ (PeproTech) to induce IgG2a CSR. Cell proliferation was monitored by carboxyfluorescein succinimidyl ester staining, according to the manufacturer’s instructions (Invitrogen).
Quantitative real-time PCR
To detect WT Stat3, mutant Stat3, and germline transcripts, B cells from mut-Stat3 or control mice were cultured in the presence of LPS, plus IL-4 for 48 hours. RNA was extracted with RNAqueous-Micro (Applied Biosystems/ABI), treated twice with DNaseI, and reverse transcribed using random hexamers (ABI). Transcript levels were monitored by real-time PCR using SYBR-green Master Mix (ABI) and normalized to glyceraldehyde-3-phosphate dehydrogenase. Quantitative PCR was performed with an ABI 7500 Fast Real-Time PCR System using Taqman site-specific primers and probes (ABI).
Microarray
WT and mut-Stat3 CD11c+ splenocytes were stimulated with either LPS or LPS and IL-10 for 24 hours. RNA was extracted and gene expression was analyzed by microarray using an Affymetrix chip. These data have been deposited in the National Center for Biotechnology Information’s (NCBI’s) Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE55607 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55607).
Histological assessment of colitis
Phosphate-buffered saline/0.1% bovine serum albumin washed sections of distal colon and cecum were fixed in buffered 10% formalin. After 2 weeks, sections (4.0 mm) were prepared from fixed tissue samples using a microtome and stained with hematoxylin and eosin. Inflammation was graded according to the following scoring system: each sample was graded semiquantitatively from 0 to 3 for 5 criteria: (1) degree of epithelial hyperplasia and goblet cell depletion; (2) leukocyte infiltration in the lamina propria; (3) submucosal edema and inflammation, (4) area of tissue affected; and (5) the presence of markers of severe inflammation, such as crypt abscesses and ulcers. Scores for each criterion were added to give an overall inflammation score for each sample of 0 to 15. Scoring was performed by 2 researchers in a blind fashion according to the criteria described as follows. Cecum and distal colon sections were calculated independently.
Western blot analysis
Purified CD4+ T cells were activated with plate-bound anti-CD3/CD28 and expanded with IL-2. After a 2 hour rest in fresh media, cells were re-stimulated for 5 to 45 minutes with 100 ng/mL IL-21. B cells were purified and stimulated with LPS and IL-4 for 48 hours. Cells were lysed in Triton detergent buffer containing Complete Mini Protease Inhibitors Cocktail (Roche). Total protein was separated by polyacrylamide gel electrophoresis, transferred to nitrocellulose and blotted with antibodies recognizing actin, STAT3, pTyr-STAT3 (Cell Signaling), and Id2 (sc489, Santa Cruz Biotechnology).
STAT3 DNA binding-enzyme-linked immunosorbent assay (ELISA)
CD4+ T cells were activated as previously described and were re-stimulated for 15 minutes with 100 ng/mL IL-21. STAT3 binding to a candidate oligonucleotide was assayed using the TransAm STAT3 assay Kit (Active Motif), according to the manufacturer’s instructions. Whole cell lysate (2 mg) from stimulated or unstimulated cells was used for each assay.
Flow cytometry
Unless otherwise stated, all antibodies were from BD Biosciences: B220-PercP-Cy5.5, B220-FITC, CD19-PercP-Cy5.5, IgM-APC (Jackson ImmunoResearch Laboratories), IgG1-PE, IgG2a-FITC, IgE-FITC, CD95-PE, GL7-FITC, CD24-FITC, CD43-PE, CD127-PE, CD138-PE, CD4-PercP-Cy5.5, CD8-APC, IFN-γ-APC, IL-4-PE, IL-17A-PE, and FoxP3-FITC (eBiosciences). To-Pro-3 (Invitrogen) or 4′,6 diamidino-2-phenylindole (DAPI; Sigma-Aldrich) were used to exclude dead cells. The cells were purified using the MoFlo Cell Sorter (Dako Cytomation), acquired on the FACSCaliber (Becton Dickinson) or Cyan (Dako Cytomation) flow cytometers, and analyzed with FlowJo software (Tree Star Inc.). Intracellular staining for cytokine expression was performed on cells stimulated for 2 hours with phorbol 12-myristate 13-acetate, ionomycin, and brefeldin A, followed by fixation using a BD Fix Perm Kit (Becton Dickinson).
ELISA
Serum immunoglobulin levels were measured from 8- to 12-week-old mice by ELISA (Bethyl Laboratories), using 3,3ʹ,5,5ʹ-tetramethylbenzidine (Pierce) as a developing substrate. Levels of SEA-specific antibody were determined by ELISA as previously described.25
Statistics
Statistical analysis comparing 2 groups was performed with a Mann Whitney U test. Where 3 or more groups were analyzed (Figure 7), a one-way analysis of variance test was performed. A single asterisk denotes a P value of <.05, a double asterisk denotes P < .01, a triple asterisk denotes P < .001. Statistical analysis was determined using GraphPad Prism (version 5) software. Two-dimensional cluster analysis (hierarchical, Pearson uncentered, average linkage, unsupervised) was performed on a list of IL-10 responsive genes using Genespring software.
Figure 7.
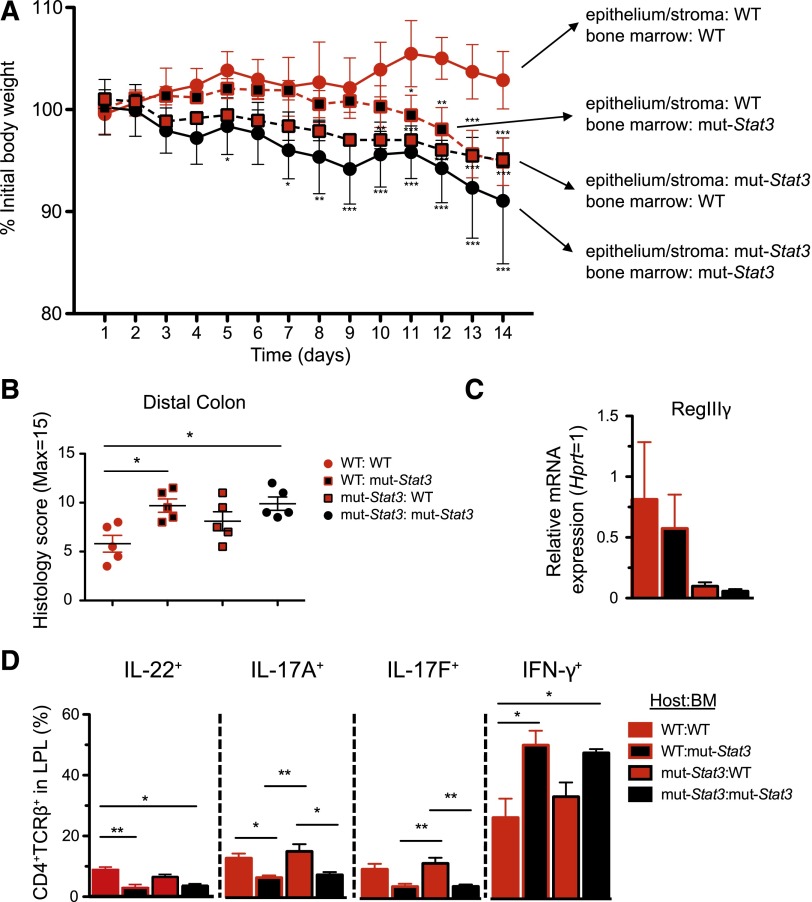
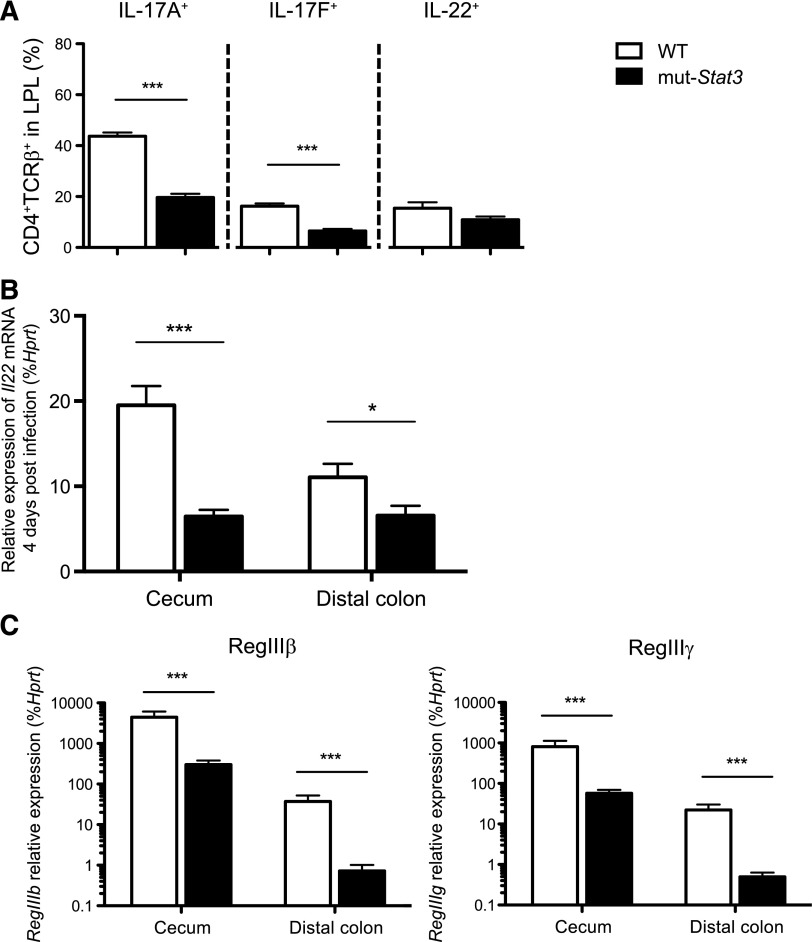
Hematopoetic and nonhematopoetic compartments contribute to increased susceptibility of mut-Stat3 mice to C rodentium. BM chimeras were generated by lethal irradiation of WT and mut-Stat3 mice followed by intravenous injection of 107 WT or mut-Stat3 BM cells. Mice rested for 8 weeks and were then orally challenged with 5 × 109 CFU C rodentium. Mice were assessed for weight loss (A). After 14 days animals were sacrificed and gut histology (B), relative expression of antimicrobial peptide mRNA (C), and CD4+ colonic lamina propria T-cell cytokine expression (D) was measured. Histogram columns represent mean values + standard error of the mean. Data are representative of 2 independent experiments (n = 5 per group, except for IL-22 staining where n = 4 per group). *P < .05; **P < .01.
Results
Mut-Stat3 mice recapitulate biochemical and functional features of human HIES
To create a mouse model of HIES, we generated transgenic mice that expressed a 185 kb bacterial artificial chromosome (BAC) that encoded a mutated mouse Stat3 gene. We chose a common mutation identified in multiple patient cohorts, namely deletion of V463 in STAT3ʹs DNA-binding domain,2,3,26 and used BAC recombineering27 to generate transgenic mice, which expressed this mutant allele (denoted mut-Stat3) (Figure 1A). From 6 transgenic lines generated, 1 was used for all subsequent experiments as it expressed 2 copies of the transgene (Figure 1B), providing the appropriate ratio of WT to mutant alleles seen in HIES patients.
Figure 1.
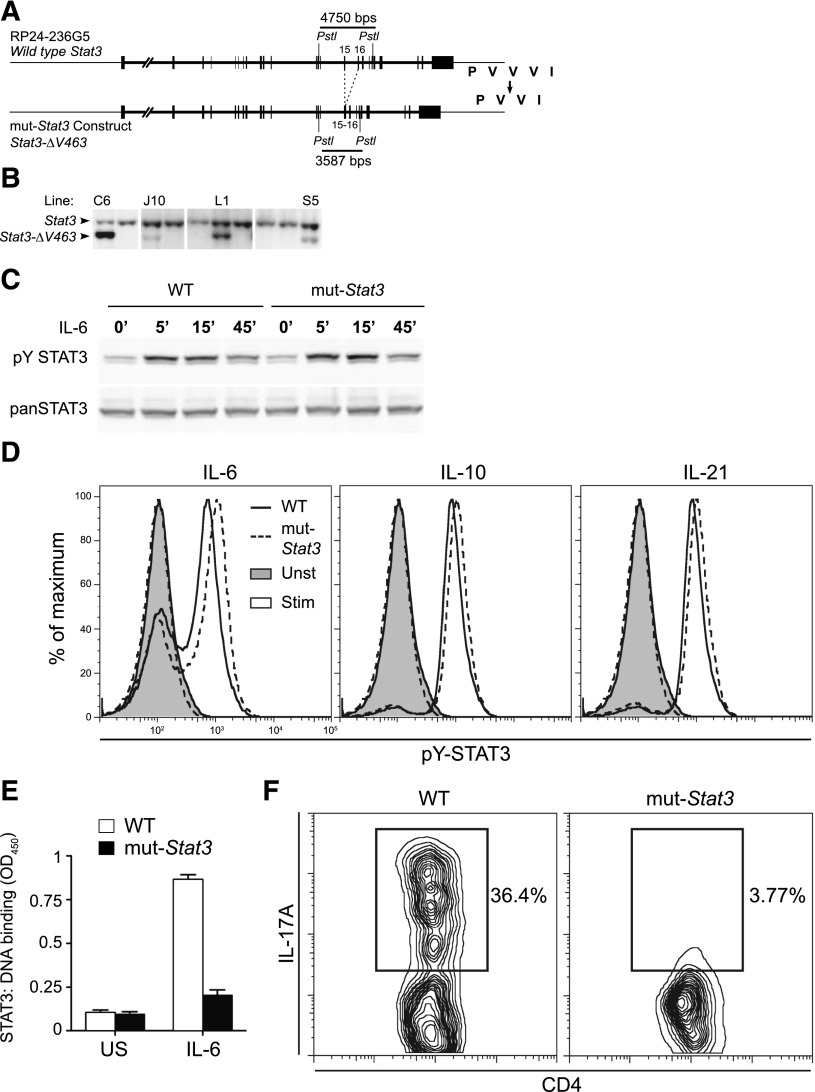
Generation of a mouse model that recapitulates key aspects of human AD-HIES. Depiction of mouse Stat3 gene within RP24-236G5 BAC. Exons 15 and 16, which encode part of STAT3 DNA binding domain, are highlighted. Based on mutations observed in HIES patients, the BAC was modified to delete valine 463 from the binding domain (from PVVVI in WT to PVVI in mutant) (A). Throughout the text, the Stat3-ΔV463 allele is referred to as mut-Stat3. The intron separating exons 15 and 16 was also deleted to permit estimation of transgene copy number (relative to WT) by Southern blotting after tail DNA digestion with PstI, as shown in panel (B). Transgenic line L1, which contains 2 transgene copies, was used in all subsequent experiments. Western blot analysis of STAT3 and Y705 phosphorylated STAT3 levels from CD4+ T cells unstimulated or stimulated with 100 ng/mL IL-21 for 5 to 15 minutes. Cells were isolated from 6- to 8-week-old WT and mut-Stat3 mice, expanded in IL-2 and rested for 2 hours prior to stimulation. Data are representative of 3 independent experiments (C). Flow cytometry analysis of Y705 phosphorylated STAT3 levels in freshly isolated splenic B cells left unstimulated (shaded) or stimulated with 50 ng/mL IL-6, IL-10, or IL-21 for 30 minutes (unshaded). Cells were isolated from 6- to 12-week-old WT (solid line) or mut-Stat3 (dotted-line) mice. Data are representative of 6 mice in 2 independent experiments (D). CD4+ T cells were prepared as in (C) and stimulated for 15 minutes with 100 ng/mL IL-21. STAT3 binding to a candidate oligonucleotide was assayed using the TransAm STAT3 Assay Kit (Active Motif) according to the manufacturer’s instructions (E). Flow cytometry analysis of IL-17A expression in CD4+ T cells stimulated with αCD3/αCD28, TGF-β-1, and IL-6 for 70 hours. Cells were isolated from 6-week-old WT and mut-Stat3 mice. Data are representative of 8 independent experiments (F).
In contrast to Stat3-deficient mice, we found that mut-Stat3 mice were viable and fertile and the disease-associated allele was transmitted in the expected Mendelian frequency. Previous work has demonstrated that AD-HIES-associated DNA-binding domain mutations of STAT3 do not impair cytokine-dependent ligand-dependent phosphorylation.19 Accordingly, we found that stimulation of cells from WT and mut-Stat3 mice with STAT3-activating cytokines resulted in comparable STAT3 Y705 phosphorylation, as measured by immunoblotting and intracellular staining (Figure 1C-D). Despite normal tyrosine phosphorylation, dominant-acting mutants in HIES patient cells interfere with STAT3 DNA binding and activation of key STAT3 target genes.19 Consistent with this, we found that cells from mice also exhibited impaired STAT3 DNA binding to a canonical STAT3 target gene (Figure 1E).
Patients with AD-HIES have normal numbers of CD4 and CD8 T cells.1 Again, mut-Stat3 mice also exhibited normal thymic development and compared with control mice, had normal proportions and absolute numbers of CD4+ and CD8+ T cells in the thymus, spleen, and lymph nodes (supplemental Figure 1A, available on the Blood Web site). STAT3 is also critical for CD4 T-cell production of IL-17,12-14,17,28 and HIES patients have severely reduced numbers of IL-17-producing T cells.19-21,29,30 We found impaired IL-17 expression in naive CD4+ T cells from mut-Stat3 mice compared with control cells when stimulated in the presence of TGF-β and IL-6 (Figure 1F). By contrast, in vitro T helper (Th)1 and Th2 cell polarization of naïve mut-Stat3 Th cells was unchanged compared with WT cells (supplemental Figure 1B). Next, we measured the number of IL-17-, IFN-γ-, or FoxP3- expressing CD4+ T cells in the lamina propria of WT and mut-Stat3 mice. Again, we found that mut-Stat3 mice had fewer IL-17+ cells and similar numbers of IFN-γ+ Th cells compared with WT animals (supplemental Figure 1C).
Mut-Stat3 mice exhibit dysregulation of B-cell function and recapitulate overproduction of IgE
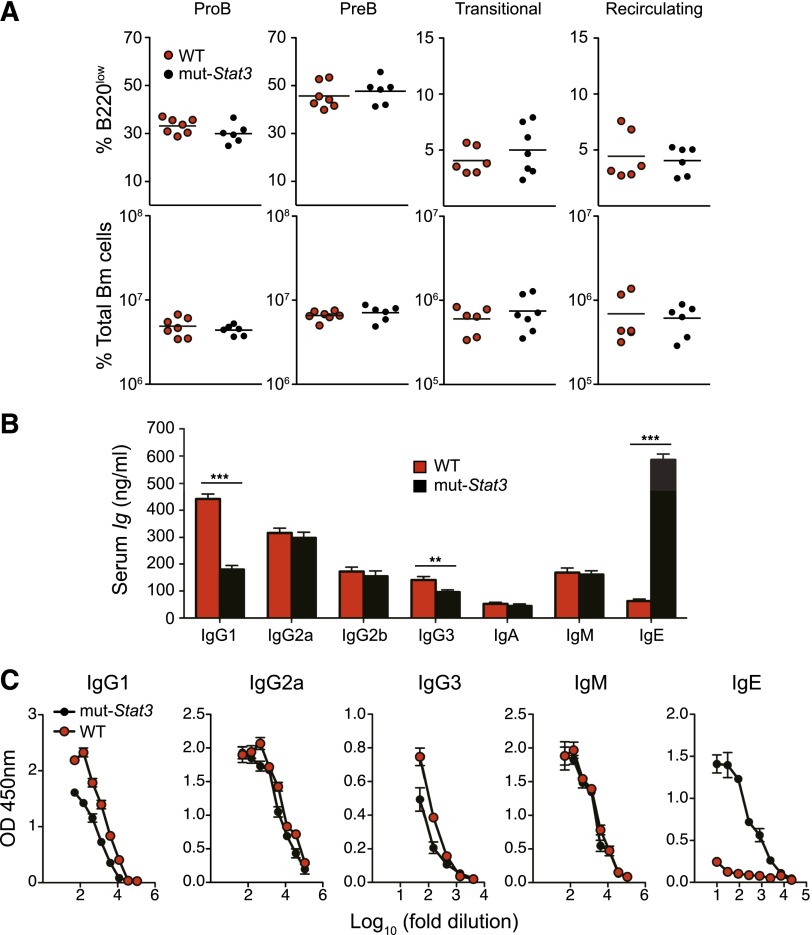
A defining feature of HIES is overproduction of IgE indicative of a role for STAT3 in the function of mature B cells. As mut-Stat3 B cells were present in normal numbers and proportions (Figure 2A), we next turned to the question of whether our model would mimic AD-HIES in terms of dysregulation of IgE production. We found that serum IgE levels in mut-Stat3 mice were sixfold higher than in WT littermate controls (Figure 2B). In addition, serum IgG1, and to a lesser extent IgG3, were significantly reduced in mut-Stat3 mice (P = .003), whereas the concentration of other isotypes was not different from controls. To assess whether this abnormality would also extend to the setting of neoantigen stimulation, we immunized mice with a soluble antigen isolated from SEA, which induces a potent Th2-driven immune response.31 As shown in Figure 2C, SEA-specific IgE production was also greatly enhanced in mut-Stat3 mice, whereas IgG1 production was reduced relative to controls. Notably, in addition to dysregulation of IgE, impaired IgG responses postimmunization have been reported in HIES patients32-34 and in IL-21-deficient mice35 These data suggest that the mut-Stat3 mice are able to reproduce another key aspect of the immune dysregulation seen in HIES.
Figure 2.
Normal B-cell development, but elevated serum IgE in mut-Stat3 mice. (A) Flow cytometry analysis of isolated BM cells from mut-Stat3 and WT mice. Populations were defined as: pro-B [B220lowCD25−IgM−], pre-B [B220lowCD25+IgM−], immature (Imm.) [B220lowCD25−IgM+], and recirculating (Rec.) [B220hiIgM+]. Populations are depicted as either percentage of B200 low cells (upper row), or absolute number of cells per BM (lower row). N = 7, not significant (ns): P > .05. Dead cells were excluded from analysis by DAPI staining. (B) ELISA measurement of serum immunoglobulin levels from naïve WT and mut-Stat3 mice aged 8 to 15 weeks (N = 23). (C) Serum Schistosoma mansoni soluble egg antigen–specific immunoglobulin levels were measured by ELISA 3 weeks after peritoneal injections of Serum Schistosoma mansoni soluble egg antigen in WT and mut-Stat3 mice. Shown is a representative result of 3 independent experiments (N = 4 for each experiment).
To better define the consequences of interfering with STAT3 function in B cells, we compared the transcriptomes of B cells from WT and mut-Stat3 mice after stimulation with LPS and IL-4, a standard protocol known for both inducing autocrine expression of IL-6 and triggering antibody class switching.36,37 mRNA-sequence analysis revealed some 120 genes that displayed greater than a twofold difference between WT and mut-Stat3 B cells (supplemental Figure 2A). Curiously, because the number of genes with elevated expression in the B cells derived from HIES mice was twice that of genes with elevated expression in WT B cells (supplemental Figure 2B), it is suggested that Stat3 functions more as a negative than a positive regulator of gene expression in activated B cells. Known STAT3 targets, such as the Socs3 gene, were significantly different between WT and mutant cells.
Marked susceptibility of mut-Stat3 mice to intestinal Citrobacter infection
Next, we turned to the question as to whether mut-Stat3 mice had impaired defense against bacterial infection. For this, we orally infected animals with C rodentium, a robust well-examined mouse model of bacterial infection, susceptibility to which is highly dependent on IL-17 and IL-22.38,39 Whereas, normal mice were resistant to Citrobacter infection (Figure 3A-B); mut-Stat3 mice lost significantly more weight and began dying at 11 to 14 days.
Figure 3.
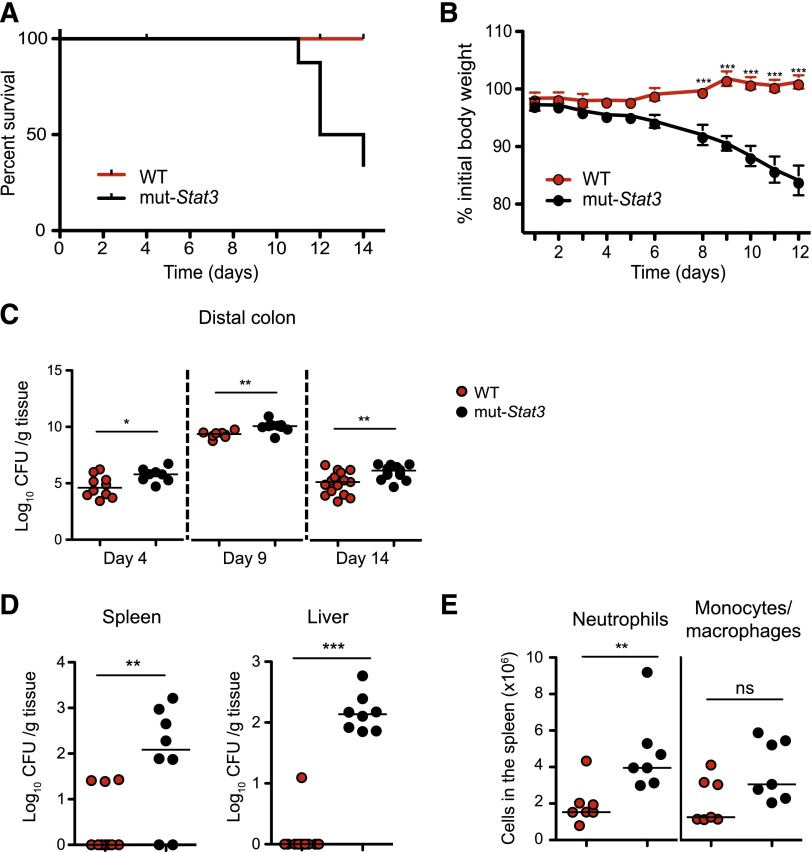
The mut-Stat3 mice show increased susceptibility to C rodentium infection. Survival (A) and weight (B) plots of WT and mut-Stat3 mice after receiving an oral dose of 5 × 109 CFU C rodentium. Bacterial load present in the distal colon was determined at 4, 9, and 14 days postinfection, (C) and in the spleen and liver 14 days postinfection (D). Splenic neutrophil and monocyte/ macrophage cell numbers were determined 14 days postinfection (E). Data represents the cumulative results of 3 independent experiments (n = 32 for WT and n = 28 for mut-Stat3).
Because of the diverse roles of STAT3, the mortality seen with C rodentium infection in mut-Stat3 mice might have been due to defects in host defense, exaggerated inflammatory responses, or both. To begin to understand the basis of susceptibility of mut-Stat3 to this infection, next, we compared the ability of WT and mut-Stat3 animals to clear these bacteria. Two weeks after infection, there were significantly higher numbers of bacteria in the distal colons of mut-Stat3 mice (Figure 3C). Furthermore, mut-Stat3 mice were unable to contain systemic infection with elevated numbers of bacteria cultured in their spleens and livers compared with infected WT animals (Figure 3D). This was associated with increased numbers of neutrophils in the spleen (Figure 3E).
Given the importance of IL-17 and IL-22 in host defense against Citrobacter, we investigated the production of these next, and other cytokines in colonic lamina propria (Figure 4A). CD4+ T-cell production of IL-17A and IL-17F in cells from infected mut-Stat3 mice was significantly reduced compared with controls when measured by intracellular staining 2 weeks after infection. There was a decline in IL-22 production that was significant when measured by quantitative PCR (Figure 4B). IL-22 provides antibacterial defense by activating STAT3 in epithelia, which, in turn, induces the expression of a number of antimicrobial proteins including Reg family peptides. In addition to reduction in IL-22 production in mut-Stat3, we anticipated that IL-22 signaling would likely to be impaired in the epithelia of mut-Stat3 mice. Therefore, we measured expression of these factors in infected mut-Stat3 animals and found that their production was significantly impaired (Figure 4C). As both mut-Stat3 mice and patients with HIES have dysregulated immunoglobulin production, we considered that this may also be a contributing factor in the susceptibility of these mice. However, we found no significant differences (supplemental Figure 3).
Figure 4.
The mut-Stat3 mouse lamina propria lymphocytes have altered cytokine expression in response to C rodentium infection. Mean values of cytokine expression (A) in viable CD4+ colonic lamina propria T cells isolated from WT or mut-Stat3 mice 14 days after oral infection with 5 × 109 CFU C rodentium. The mRNA expression of Il22 (B) and its associated antimicrobial peptides (C) were determined by quantitative PCR in the cecums and distal colons from WT or mut-Stat3 mice 14 days after oral infection with 5 × 109 CFU C rodentium. Histograms represent mean values + SEM, data are representative of 2 independent experiments (n = 8 per group). *P < .05; ***P < .001.
The presence of IL-22 and its protective actions on the gut epithelium have been shown to be essential for recovery from colitis induced byDSS.23 To confirm our findings in Citrobacter-infected mice, we measured the effect of treating WT and mut-Stat3 mice with DSS drinking water, again mut-Stat3 lost significantly more weight (supplemental Figure 4).
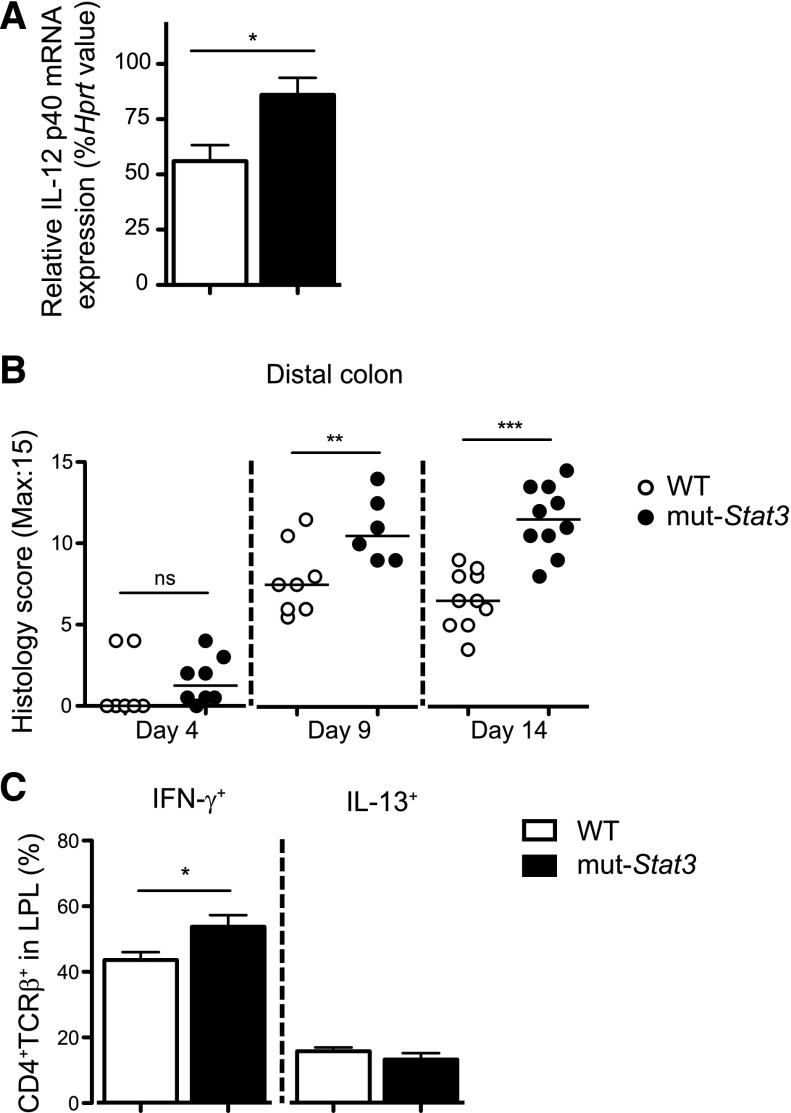
In contrast with its role in lymphocytes, STAT3 has been implicated as an anti-inflammatory factor in myeloid cells. We measured colonic IL-12 p40 mRNA by quantitative PCR 4 days after infection and found elevated expression in the mut-Stat3 mice compared with controls (Figure 5A). Next, we investigated the degree of inflammation within the infected colons of WT and mut-Stat3 mice by histology. The large bowels of mut-Stat3 mice developed significantly more inflammation compared with WT inflammation compared with WT animals after 9 days (Figure 5B). This was associated with elevated numbers of IFN-γ expressing CD4+ T cells within the colonic lamina propria (Figure 5C).
Figure 5.
The mut-Stat3 mice have a heightened inflammatory response after infection with C rodentium. Relative expression of IL-12 p40 subunit mRNA in the colons of WT and mut-Stat3 mice 4 days after infection with C rodentium (A). Histology analysis of inflammation present in the distal colon was determined at days 4, 9, and 14 postoral infection with 5 × 109 CFU C rodentium (B). IFN-γ and IL-13 expression in the colonic lamina propria CD4+ T cells at day 14 postinfection was determined by intracellular staining (C). Histograms represent mean values + standard error of the mean, data are representative of 2 independent experiments (n = 8 per group). ns = not significant. *P < .05; **P < .01; ***P < .001.
Increased mortality in mut-Stat3 mice treated with LPS
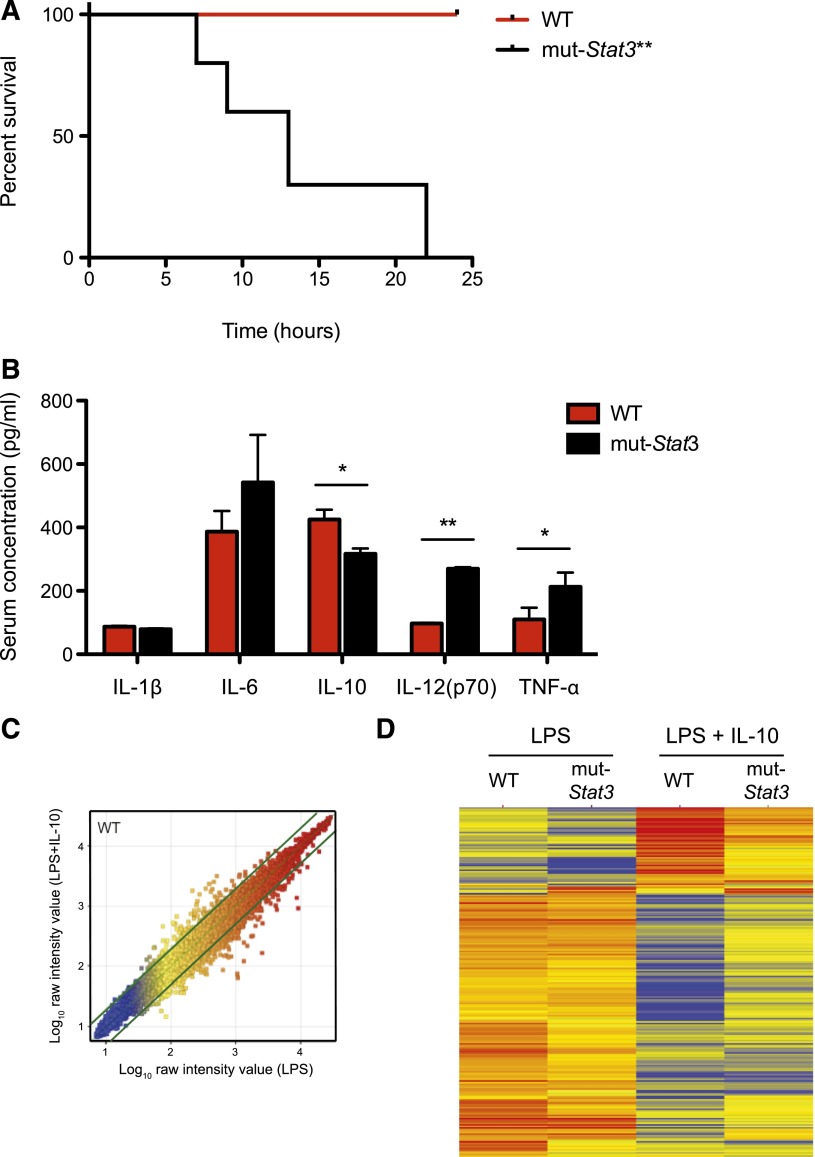
Collectively, our data suggested that mut-Stat3 mice clearly have impaired host defense against Citrobacteria together with an exaggerated inflammatory response in response to the bacterial infection. To explore this further, we compared the effect of a systemic dose of LPS on WT and mut-Stat3 mice. We found that mut-Stat3 were highly susceptible to LPS-induced shock (Figure 6A). The changes in survival were associated with a significant elevation in serum IL-12 and tumor necrosis factor-α in mut-Stat3 mice compared with WT animals together with a significant drop in serum IL-10 concentration (Figure 6B).
Figure 6.
The mut-Stat3 mice were susceptible to LPS-induced septic shock. WT and mut-Stat3 mice were intraperitonally injected with 250 µg LPS, animals were monitored for survival over the following 24 hours. **P = .0015. (A) Serum cytokine measurements by Bead array and ELISA were made 90 minutes after LPS injection (B). Histograms represent mean values + standard error of the mean data were pooled from 2 experiments (n = 10 per group). CD11c+ splenocytes from WT and mut-Stat3 mice were stimulated with either LPS or LPS and IL-10 for 24 hours. Gene expression by Affymetrix microarray comparing LPS and LPS+IL-10 treated WT CD11c+ cells is shown as a scatter plot (C). Genes that had a greater than twofold change in gene expression between conditions were defined as IL-10 responsive genes (indicated by the thick green lines). Two dimensional cluster analysis of the IL-10 responsive gene list is presented (D). Each row corresponds to a transcript and each column to the mean expression values of a sample, pooled from 3 independent microarrays. The color denotes Log10 raw intensity value (blue: 1–red: 4). *P < .05; **P < .01.
STAT3 is a key-signaling factor downstream of the anti-inflammatory cytokine, IL-10.40 Next, we investigated the effect of IL-10 on LPS stimulated splenic (CD11c+) dendritic cells from WT and mut-Stat3 animals and analyzed gene expression by differential array. A list of IL-10 responsive genes was determined by comparing WT dendritic cells stimulated in the presence or absence of IL-10 (supplemental Table 1 and Figure 6C). Analyzing this list, in the absence of IL-10, there was little difference between WT and mut-Stat3 gene expression, but in the presence of IL-10, substantial differences appeared between the 2 as a consequence of a diminished effect of IL-10 in cells from mut-Stat3 mice (Figure 6D).
Taken together, our data indicated that there were multiple explanations for the susceptibility of mut-Stat3 mice to bacterial infection. There were clear host defects related to impaired production of key cytokines like IL-17A, IL-17F, and IL-22. There was impaired responsiveness of epithelial cells to IL-22 and consequent failure to produce antimicrobial peptides. Compounding these was an exaggerated inflammatory response, part of which can be explained by failure of IL-10 to exert its anti-inflammatory actions.
Partial Utility of HSCT in mut-Stat3 mice
Our findings indicated important roles for STAT3 in preserving host defense based on its functions in hematopoietic cells and in epithelial cells, as well as limiting inflammatory responses.40 To determine whether HSCT may be an effective therapy for humans with HIES, we investigated the effect of transplantation on the susceptibility of mut-Stat3 animals to C rodentium. WT mice transplanted with WT BM were resistant to C rodentium infection. In contrast, mut-Stat3 mice transplanted with mut-Stat3 BM were susceptible to this infection, losing significantly more weight. To assess the contribution of STAT3 in the hematopoietic compartment and to mimic a therapeutic scenario, mut-Stat3 mice were transplanted with BM from WT mice (Figure 7A). The data show that these mice were more resistant to C rodentium infection than mut-Stat3 mice that received BM from mice with impaired STAT3 function. However, the effect was only partial (Figure 7A-B). Consistent with this and arguing for a key role of STAT3 in nonhematopoietic cells, WT mice transplanted with BM from mut-Stat3 mice also lost less weight than mice in which STAT3 signaling was impaired in both compartments. Accordingly, expression of RegIII-γ was dependent on intact STAT3 signaling in the nonhematopoietic cells (Figure 7C). Similarly, transplantation of mut-Stat3 mice with normal BM restored IL-17A, IL-17F, and IL-22 production (Figure 7D); and this was associated with a reduction in IFN-γ secretion consistent with the hyperinflammatory phenotype associated with STAT3 deficiency in myeloid cells.
Discussion
Mouse embryos that have a complete ablation of Stat3 die in utero; in this respect, the discovery of a group of patients with dominant-acting Stat3 mutations was unanticipated.3 The finding immediately raised the question as to whether the existence of such patients could be explained by incomplete impairment in STAT3 function or whether there were species-specific differences in the function of STAT3, as had been previously observed in another key cytokine signaling gene, Jak3.41 To address these points, we set out to create a mouse model of HIES. In the present study, we replicated the classical multisystem HIES by expressing a patient-derived mutant allele that affects STAT3 DNA binding activity in a dominant negative fashion.3 In this context, our system stands in contrast to models in which Stat3 is fully deleted in the germline.5 The disease is probably best characterized as resulting from impaired, but not abrogated STAT3 function. The mut-Stat3 mice displayed many features characteristic of patients with HIES, namely, impaired Th17 differentiation and the dysregulation of IgE in the steady state.
To investigate the animal’s susceptibility to bacterial infections, we infected WT and mut-Stat3 mice with C rodentium, and we found that mut-Stat3 mice demonstrated an impaired ability to control the bacteria within the distal colon and prevent the bacteria from spreading systemically. This was associated with a heightened inflammatory response, which may be explained by the finding that mut-Stat3 antigen-presenting cells stimulated with LPS demonstrated a heightened inflammatory response consistent with the findings seen by other groups,42 and mut-Stat3 mice were more susceptible to LPS-induced toxic shock.
Occasionally, patients with HIES have been treated with allogeneic BM transplantation, either as an attempt to treat their underlying immune-deficiency or to treat lymphoma, which is more common in this patient group compared with the general population. The effectiveness of this treatment is controversial with different transplantation groups coming to different conclusions.43-45 To explore this, we rescued lethally irradiated WT animals with BM from mut-Stat3 mice and vice versa. We found that WT mice with mut-Stat3 BM and mut-Stat3 mice with WT BM showed a similar susceptibility to C rodentium, and both groups lost significantly more weight than WT animals transplanted with WT BM. Despite a similar loss of weight, the different groups presented with different immunological changes: there was a significantly reduced inflammatory infiltrate seen in animals transplanted with WT compared with HIES BM, irrespective of the genotype of the host animal. Conversely the induction of anti-bacterial peptides in response to C rodentium infection was more dependent on the genotype of the host animal rather than the type of BM that was used to rescue the animal. Thus, we conclude that many factors contribute to the susceptibility of mut-Stat3 mice to bacterial infection, some of which cannot be corrected by HSCT. If a similar partial response occurs in HIES patients, it may explain the variable responses documented by different transplant centers. Recently, 2 HIES patients with somatic mosaicism have been reported, both of these had preserved populations of Th17 cells derived from normal T cells, but regions of epithelia that expressed mutated STAT3. The authors noted that both patients had an intermediate phenotype consistent with our findings.46 As with our model, such patients argue for the importance of STAT3 in host defense in cells outside the hematopoietic system.
In conclusion, we show an example by which the new mouse model should prove crucial in dissecting out the role of STAT3 in other features of the disease, hopefully leading to better treatments for not only HIES patients, but also others suffering from disorders in which STAT3 activation plays a role.
Supplementary Material
Acknowledgments
S.M.S.-T. is a member of the National Institutes of Health-Oxford/Cambridge Scholars Program. A.K. and S.M.S.-T. were funded by the Howard Hughes Medical Institute-National Institutes of Health research scholar program.
This work was supported in part by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health.
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.M.S.-T., A.L., R.C., Y.K., A.V.V., S.K., J.J.O.S., S.M.H., J.D.M., and F.P. designed research; S.M.S.-T., A.L., R.C., Y.K., A.V.V., S.K., A.K., E.A.W., G.S., K.H., K.J., and L.F. performed research; G.V., H.W.S., and W.R. analyzed data; and S.M.S.-T., A.L., R.C., and J.J.O.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Arian Laurence, Molecular Immunology and Inflammation Branch, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, 10 Center Dr, MSC 1930, Building 10, Room 13C120-11B, Bethesda, MD 20892-1930; e-mail: laurencea@mail.nih.gov.
References
- 1.Heimall J, Freeman A, Holland SM. Pathogenesis of hyper IgE syndrome. Clin Rev Allergy Immunol. 2010;38(1):32–38. doi: 10.1007/s12016-009-8134-1. [DOI] [PubMed] [Google Scholar]
- 2.Holland SM, DeLeo FR, Elloumi HZ, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357(16):1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 3.Minegishi Y, Saito M, Tsuchiya S, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448(7157):1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 4.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 5.Takeda K, Noguchi K, Shi W, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94(8):3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Nguyen-Jackson H, Panopoulos AD, Li HS, Murray PJ, Watowich SS. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood. 2010;116(14):2462–2471. doi: 10.1182/blood-2009-12-259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamassia N, Castellucci M, Rossato M, et al. Uncovering an IL-10-dependent NF-kappaB recruitment to the IL-1ra promoter that is impaired in STAT3 functionally defective patients. FASEB J. 2010;24(5):1365–1375. doi: 10.1096/fj.09-145573. [DOI] [PubMed] [Google Scholar]
- 8.Takeda K, Clausen BE, Kaisho T, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10(1):39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 9.Welte T, Zhang SS, Wang T, et al. STAT3 deletion during hematopoiesis causes Crohn’s disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci U S A. 2003;100(4):1879–1884. doi: 10.1073/pnas.0237137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CK, Raz R, Gimeno R, et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17(1):63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- 11.Vasquez-Dunddel D, Pan F, Zeng Q, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest. 2013;123(4):1580–1589. doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurence A, Tato CM, Davidson TS, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26(3):371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Mathur AN, Chang HC, Zisoulis DG, et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178(8):4901–4907. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- 14.Yang XO, Panopoulos AD, Nurieva R, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282(13):9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 15.Durant L, Watford WT, Ramos HL, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32(5):605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurence A, Amarnath S, Mariotti J, et al. STAT3 transcription factor promotes instability of nTreg cells and limits generation of iTreg cells during acute murine graft-versus-host disease. Immunity. 2012;37(2):209–222. doi: 10.1016/j.immuni.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris TJ, Grosso JF, Yen HR, et al. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol. 2007;179(7):4313–4317. doi: 10.4049/jimmunol.179.7.4313. [DOI] [PubMed] [Google Scholar]
- 18.Wegrzyn J, Potla R, Chwae YJ, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323(5915):793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milner JD, Brenchley JM, Laurence A, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452(7188):773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma CS, Chew GY, Simpson N, et al. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205(7):1551–1557. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minegishi Y, Saito M, Nagasawa M, et al. Molecular explanation for the contradiction between systemic Th17 defect and localized bacterial infection in hyper-IgE syndrome. J Exp Med. 2009;206(6):1291–1301. doi: 10.1084/jem.20082767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickert G, Neufert C, Leppkes M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206(7):1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29(6):947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jankovic D, Kullberg MC, Caspar P, Sher A. Parasite-induced Th2 polarization is associated with down-regulated dendritic cell responsiveness to Th1 stimuli and a transient delay in T lymphocyte cycling. J Immunol. 2004;173(4):2419–2427. doi: 10.4049/jimmunol.173.4.2419. [DOI] [PubMed] [Google Scholar]
- 25.Jankovic D, Kullberg MC, Noben-Trauth N, et al. Schistosome-infected IL-4 receptor knockout (KO) mice, in contrast to IL-4 KO mice, fail to develop granulomatous pathology while maintaining the same lymphokine expression profile. J Immunol. 1999;163(1):337–342. [PubMed] [Google Scholar]
- 26.Jiao H, Tóth B, Erdos M, et al. Novel and recurrent STAT3 mutations in hyper-IgE syndrome patients from different ethnic groups. Mol Immunol. 2008;46(1):202–206. doi: 10.1016/j.molimm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Crouch EE, Li Z, Takizawa M, et al. Regulation of AID expression in the immune response. J Exp Med. 2007;204(5):1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z, Laurence A, Kanno Y, et al. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci U S A. 2006;103(21):8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renner ED, Rylaarsdam S, Anover-Sombke S, et al. Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced T(H)17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J Allergy Clin Immunol. 2008;122(1):181–187. doi: 10.1016/j.jaci.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Beaucoudrey L, Puel A, Filipe-Santos O, et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J Exp Med. 2008;205(7):1543–1550. doi: 10.1084/jem.20080321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jankovic D, Steinfelder S, Kullberg MC, Sher A. Mechanisms underlying helminth- induced Th2 polarization: default, negative or positive pathways? Chem Immunol Allergy. 2006;90:65–81. doi: 10.1159/000088881. [DOI] [PubMed] [Google Scholar]
- 32.Dreskin SC, Goldsmith PK, Gallin JI. Immunoglobulins in the hyperimmunoglobulin E and recurrent infection (Job’s) syndrome. Deficiency of anti-Staphylococcus aureus immunoglobulin A. J Clin Invest. 1985;75(1):26–34. doi: 10.1172/JCI111683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheerin KA, Buckley RH. Antibody responses to protein, polysaccharide, and phi X174 antigens in the hyperimmunoglobulinemia E (hyper-IgE) syndrome. J Allergy Clin Immunol. 1991;87(4):803–811. doi: 10.1016/0091-6749(91)90126-9. [DOI] [PubMed] [Google Scholar]
- 34.Buckley RH, Wray BB, Belmaker EZ. Extreme hyperimmunoglobulinemia E and undue susceptibility to infection. Pediatrics. 1972;49(1):59–70. [PubMed] [Google Scholar]
- 35.Shang XZ, Ma KY, Radewonuk J, et al. IgE isotype switch and IgE production are enhanced in IL-21-deficient but not IFN-gamma-deficient mice in a Th2-biased response. Cell Immunol. 2006;241(2):66–74. doi: 10.1016/j.cellimm.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Savelkoul HF, Termeulen J, Coffman RL, Van der Linde-Preesman RA. Frequency analysis of functional Ig C epsilon gene expression in the presence and absence of interleukin 4 in lipopolysaccharide-reactive murine B cells from high and low IgE responder strains. Eur J Immunol. 1988;18(8):1209–1215. doi: 10.1002/eji.1830180810. [DOI] [PubMed] [Google Scholar]
- 37.Barr TA, Shen P, Brown S, et al. B cell depletion therapy ameliorates autoimmune disease through ablation of IL-6-producing B cells. J Exp Med. 2012;209(5):1001–1010. doi: 10.1084/jem.20111675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangan PR, Harrington LE, O’Quinn DB, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441(7090):231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 39.Zheng Y, Valdez PA, Danilenko DM, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14(3):282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 40.Murray PJ. Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr Opin Pharmacol. 2006;6(4):379–386. doi: 10.1016/j.coph.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 41.O’Shea JJ, Husa M, Li D, et al. Jak3 and the pathogenesis of severe combined immunodeficiency. Mol Immunol. 2004;41(6-7):727–737. doi: 10.1016/j.molimm.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Saito M, Nagasawa M, Takada H, et al. Defective IL-10 signaling in hyper-IgE syndrome results in impaired generation of tolerogenic dendritic cells and induced regulatory T cells. J Exp Med. 2011;208(2):235–249. doi: 10.1084/jem.20100799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nester TA, Wagnon AH, Reilly WF, Spitzer G, Kjeldsberg CR, Hill HR. Effects of allogeneic peripheral stem cell transplantation in a patient with job syndrome of hyperimmunoglobulinemia E and recurrent infections. Am J Med. 1998;105(2):162–164. doi: 10.1016/s0002-9343(98)00200-9. [DOI] [PubMed] [Google Scholar]
- 44.Gennery AR, Flood TJ, Abinun M, Cant AJ. Bone marrow transplantation does not correct the hyper IgE syndrome. Bone Marrow Transplant. 2000;25(12):1303–1305. doi: 10.1038/sj.bmt.1702446. [DOI] [PubMed] [Google Scholar]
- 45.Goussetis E, Peristeri I, Kitra V, et al. Successful long-term immunologic reconstitution by allogeneic hematopoietic stem cell transplantation cures patients with autosomal dominant hyper-IgE syndrome. J Allergy Clin Immunol. 2010;126(2):392–394. doi: 10.1016/j.jaci.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Hsu AP, Sowerwine KJ, Lawrence MG, et al. Intermediate phenotypes in patients with autosomal dominant hyper-IgE syndrome caused by somatic mosaicism. J Allergy Clin Immunol. 2013;131(6):1586–1593. doi: 10.1016/j.jaci.2013.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.