Abstract
The effect of genetic markers associated with IgA nephropathy on risk of disease in sub-phenotype and progression is uncertain. Data from 2096 Chinese patients were used to create both un-weighted (uw) and weighted (w) genetic risk score (GRS). The association between GRS with disease susceptibility and clinical parameters were assessed. All nine selected single nucleotide polymorphisms (SNPs) were associated with susceptibility to IgAN. uwGRS and wGRS showed a similar fit in disease associations. With every 1-unit increase in the uwGRS, the disease risk increased by approximately 20%; whereas every one standard deviation increase in the wGRS, disease risk increased by approximately 40% ~ 60%. Association between rs3803800 and serum IgA was replicated, and risk groups in GRSs were associated with increased IgA/IgA1 levels. uwGRS9 ≥ 16 was an independent predictor for end stage renal disease (ESRD) in IgAN, with a relative risk of 2.52 (p = 6.68 × 10−3). In conclusion, we observed that GRSs comprising nine SNPs identified in a GWAS of IgAN were strongly associated with susceptibility to IgAN. The high risk GRS9 group had a high risk of ESRD in follow-up.
IgA nephropathy (IgAN) is the most prevalent primary chronic glomerular disease worldwide. The clinical manifestation and progression of IgAN varies. The 20-year predicted survival without the need for dialysis was 96% among patients with no risk factors versus 36% among those with three factors: urinary protein excretion of more than 1 g per day, hypertension (>140/90 mm Hg), and severe histological lesions at the time of renal biopsy1,2,3. Thus, risk prediction is vital for disease prevention and refining prediction strategies remains important for targeting treatment recommendations4,5,6,7,8,9. One area of potential improvement has been the discovery of genetic markers for IgAN, as well as intermediate phenotypes, such as proteinuria and blood pressure.
Genetic factors undoubtedly influence the pathogenesis of IgAN, with an estimated heritability of 40%–50%10,11. Recent efforts using genome-wide association studies (GWASs) have identified genetic markers associated with IgAN12,13,14,15,16. In a study using a standardized seven–SNP genetic risk score (GRS), disease risk increased sharply with Eastward and Northward distance from Africa, which correlated with differences in disease prevalence among world populations. In addition, it explained 4.7% of overall IgAN risk, and one standard deviation increase in the score was associated with nearly 50% increase in the odds of disease5. Thus, it strongly suggested that use of a multi-locus genetic risk score might be promising for prediction for disease susceptibility. As genetic backgrounds are stable, their presence may act over the entire life course9. However, it remains unknown whether the cumulative effects of variants identified by GWASs could benefit prediction of disease progression and treatment decisions17,18,19.
No best GRS model was recommended in the recent GRIPS Statement (recommendations for the reporting of Genetic RIsk Prediction Studies)20 and it was widely observed that the count method (risk allele counts, the total number of risk alleles an individual carries, or unweighted GRS) showed similar discriminative accuracy, but less complication in weighting process, compared with the log odds procedure (sums of the natural logarithm of the allelic odds ratio for each risk allele within and across loci, or weighted GRS) for most diseases21,22,23. Therefore, based on data from GWASs, we constructed both weighted and unweighted genetic scores24,25,26. We aimed to firstly construct models that were easy to interpret but were valid for risk prediction. Notably, a comparison with the pre-established seven–SNP GRS (a weighted score) was also conducted. The scores were then tested to assess their predictive ability in both disease/intermediate phenotype susceptibility and disease progression, using a Chinese Northern Han population. As the strongest association observed was with a subset of alleles encoding the class II Human Leukocyte Antigens (HLA), whereas several non-HLA loci also demonstrated genetic associations, both HLA allele scores and non-HLA allele scores were constructed to evaluate their respective role in specific sub-phenotypes.
Results
Association between single SNP selected and susceptibility to IgAN
As can be seen from Table 1, all the SNPs selected for further GRS analysis were associated with susceptibility to IgAN. The top seven associated IgAN alleles were also the SNPs reported in the previous GWAS conducted in our cohort, as well as the seven SNPs selected in previous seven-SNP GRS among different populations in geospatial risk analysis5,27. The association between the two novel SNPs selected from Southern Chinese Han GWAS and IgAN could also be replicated12. Although they showed less significant p values for disease association in the current study, they conferred similar risk effect compared with a previous report from a Southern Chinese population12. Odds ratio (OR) for rs2738048C and rs3803800G were 0.81 and 0.87 in our cohort, compared with 0.79 and 0.83 respectively in the previous report. Thus, the data implied that the associations between nine SNPs and IgAN were real and our current cohort could be a representative population for further risk stratification.
Table 1. SNPs used in the GRSs and their association with IgAN.
| SNP | Chr. | Loci | Position | Protective allele/risk allele | PAF (%) Cases vs. Controls | P | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| rs6677604 | 1q32 | CFH, CFHR1, CFHR3 | 194953541 | A/G | 4.10/7.26 | 8.41 × 10−6 | 0.55 (0.42–0.72) |
| rs9275224 | 6p21 | HLA-DQB1, -DQA1, -DRB1 | 32767856 | A/G | 34.37/44.00 | 2.29 × 10−10 | 0.67 (0.59–0.76) |
| rs2856717 | 6p21 | HLA-DQB1, -DQA1, -DRB1 | 32778286 | T/C | 18.76/25.83 | 4.03 × 10−8 | 0.66 (0.57–0.77) |
| rs9275596 | 6p21 | HLA-DQB1, -DQA1, -DRB1 | 32789609 | C/T | 13.66/21.98 | 1.77 × 10−12 | 0.56 (0.48–0.66) |
| rs9357155 | 6p21 | HLA-DOB, PSMB8, PSMB9, TAP1, TAP2 | 32917826 | A/G | 14.67/20.03 | 4.69 × 10−6 | 0.69 (0.58–0.81) |
| rs1883414 | 6p21 | HLA-DPB2, -DPB1, -DPA1 | 33194426 | T/C | 18.55/23.89 | 2.51 × 10−5 | 0.73 (0.62–0.84) |
| rs2738048 | 8p23 | DEFB1, DEFA6, DEFA4, DEFA1, DEFA3, DEFA5 | 6810195 | C/T | 26.93/31.26 | 2.13 × 10−3 | 0.81 (0.71–0.93) |
| rs3803800 | 17p13 | TNFSF12, TNFSF13, MPDU1, EIF4A1, CD68, TP53, SOX15 | 7403693 | G/A | 65.09/68.15 | 3.80 × 10−2 | 0.87 (0.77–0.99) |
| rs2412971 | 22q12 | HORMAD2, MTMR3, LIF, OSM, GATSL3, SF3A1 | 28824371 | A/G | 31.41/38.80 | 6.22 × 10−7 | 0.72 (0.64–0.82) |
Chr: Chromosome; PAF: protective allele frequency; CI: confidence interval.
After quality control, genotypes from 1190 cases and 899 controls were used for the association study. The statistics were slightly different from that reported in the previous GWAS because of a slight modification of the sample size (1194 cases and 902 controls in previous GWAS). As individuals with 100% non-missing genotypes across all the scored loci were analyzed, the numbers of missing samples were not deleted intentionally.
The top seven associated IgAN alleles were also the SNPs reported in the previous GWAS conducted in our cohort, as well as the seven SNPs selected in previous GRS among different populations in geospatial risk analysis.
SNPs, including rs2738048 and rs3803800, selected from Southern Chinese Han GWAS were also associated with IgAN in our Northern Chinese Han cohort.
Linkage disequilibrium (LD) analysis indicated that HLA variants rs9275224, rs2856717 and rs9275596 were in partial LD, with r2 ranging from 0.33 to 0.75; however, they were not in LD with two other HLA variants, rs9357155 and rs1883414 (r2 < 0.1). When the nine SNPs were included in a logistic model, they all showed significant associations with susceptibility to IgAN. Concordant with previous reports5, conditional analysis indicated that all nine SNPs were independently associated SNPs. However, no gene-gene interactions were observed among the nine SNPs, including the interaction between the CFHR3/R1(rs6677604) and the HORMAD2 loci (rs2412971) (p = 0.41), reported in the previous seven-SNP genetic score.
Individual association between single SNPs and clinical parameters of IgAN
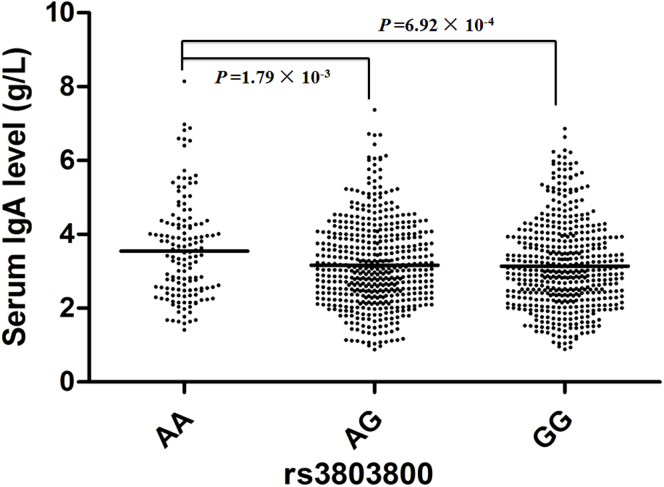
The individual association between the nine susceptibility SNPs with clinical phenotypes of IgAN were assessed in our cohort. We observed that the risk allele A of rs3803800 was associated with an increased IgA (P = 3.91 × 10−3) level in sera, which was concordant with previous reports from Southern Chinese Han GWAS12,28. The serum IgA concentrations (g/L, mean ± standard derivation) were 3.15 ± 1.21, 3.18 ± 1.19, and 3.55 ± 1.33 for rs3803800 GG, AG and AA, respectively (Figure 1). We also observed associations of rs2412971 with serum IgA and IgA1 levels, rs1883414 with gross hematuria and hypertension (Table 2). Risk genotypes seemed to be associated with higher serum IgA or IgA1 level, higher frequency of gross hematuria or higher frequency of hypertension (Table 2). However, the effect size conferred by the risk genotype was only moderate, and none of the associations survived the multiple-testing correction.
Figure 1. Associations between genotypes of rs3803800 with serum IgA level.
Table 2. Correlation of the SNPs and GRS with clinical phenotype in IgAN patients at renal biopsy.
| Genetic information | Log (Proteinuria) | Log (eGFR) | Log (IgA level) | Log (IgA1 level) | Log (gd-IgA1 level) | Gross hematuria | Hypertension | Hyperlipidemia | Hyperuricemia | CKD stage | Hass grade |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs6677604 | 0.24 | 0.77 | 0.71 | 4.22 × 10−2 (beta0.09) | 0.85 | 0.38 | 0.85 | 0.41 | 0.59 | 0.14 | 0.25 |
| rs9275224 | 0.51 | 0.62 | 0.13 | 0.45 | 0.93 | 0.75 | 0.57 | 0.32 | 0.93 | 0.66 | 0.11 |
| rs2856717 | 0.36 | 0.73 | 0.19 | 0.17 | 0.96 | 0.08 | 0.61 | 0.81 | 0.74 | 0.84 | 0.73 |
| rs9275596 | 0.40 | 0.38 | 0.15 | 0.09 | 0.90 | 0.59 | 0.57 | 0.82 | 0.82 | 0.20 | 0.97 |
| rs9357155 | 1.00 | 0.74 | 0.11 | 0.72 | 0.17 | 0.90 | 0.26 | 0.11 | 0.50 | 0.37 | 0.72 |
| rs1883414 | 0.42 | 0.13 | 0.66 | 0.97 | 0.55 | 6.76 × 10−3 (OR 1.37) | 7.32 × 10−4 (OR 1.43) | 0.23 | 0.06 | 0.94 | 0.25 |
| rs2738048 | 0.72 | 0.32 | 0.51 | 0.64 | 0.90 | 0.61 | 0.90 | 0.44 | 0.74 | 0.13 | 0.89 |
| rs3803800 | 0.40 | 0.73 | 3.91 × 10−3 (beta 0.09) | 0.13 | 0.74 | 0.35 | 0.37 | 0.91 | 0.15 | 0.93 | 0.32 |
| rs2412971 | 0.15 | 0.28 | 3.60 × 10−2 (beta 0.06) | 3.08 × 10−2 (beta0.11) | 0.53 | 0.05 | 0.09 | 0.98 | 0.23 | 0.47 | 0.37 |
| uwGRS5 | 0.61 | 0.88 | 0.30 | 0.21 | 0.62 | 0.99 | 0.14 | 0.87 | 0.33 | 0.60 | 0.13 |
| uwGRS7 | 0.26 | 0.86 | 0.08 | 2.87 × 10−2 (beta0.11) | 0.78 | 0.43 | 0.06 | 0.98 | 0.22 | 0.97 | 0.91 |
| uwGRS9 | 0.16 | 0.57 | 7.57 × 10−3 (beta 0.08) | 2.06 × 10−2 (beta0.12) | 0.70 | 0.25 | 4.08 × 10−2 (OR 0.95) | 0.84 | 0.14 | 0.74 | 0.59 |
| uwGRS4 | 0.07 | 0.20 | 8.89 × 10−4 (beta 0.10) | 1.84 × 10−2 (beta0.12) | 0.97 | 3.42 × 10−2 (OR 1.12) | 0.13 | 0.90 | 0.22 | 0.79 | 4.71 × 10−2 |
| wGRS5 | 0.48 | 0.65 | 0.26 | 0.16 | 0.70 | 0.65 | 0.34 | 1.00 | 0.50 | 0.21 | 0.06 |
| wGRS7 | 0.20 | 0.84 | 0.09 | 2.09 × 10−2 (beta0.12) | 0.81 | 0.78 | 0.19 | 0.85 | 0.39 | 1.00 | 1.00 |
| wGRS9 | 0.15 | 1.00 | 3.42 × 10−2 (beta0.06) | 1.84 × 10−2 (beta0.12) | 0.77 | 0.65 | 0.16 | 0.94 | 0.34 | 1.00 | 1.00 |
| wGRS4 | 4.48 × 10−2 (beta −0.08) | 0.27 | 5.85 × 10−3 (beta0.08) | 5.34 × 10−3 (beta0.14) | 0.86 | 2.49 × 10−2 (OR 1.56) | 0.17 | 0.86 | 0.40 | 0.96 | 0.91 |
| Standardized GRS | 0.24 | 0.93 | 0.12 | 3.70 × 10−2 (beta0.11) | 0.80 | 9.46 × 10−3 (OR 1.22) | 0.09 | 0.85 | 0.22 | 1.00 | 1.00 |
Linear regression was applied for the correlation analysis of natural log-transformed proteinuria, natural log-transformed eGFR, and natural log-transformed serum IgA level.
Binary logistic regression was carried out for the correlation analysis of history of gross hematuria, hypertension, hyperlipidemia and hyperuricemia.
Ordinal logistic regression was performed for the correlation analysis of CKD stage at the time of biopsy and Hass biopsy grade.
Effect estimates (OR/BETA) are shown only for significant associations (p < 0.05), because of space limitations.
Observed GRS in IgAN and controls
We constructed four different genetic scores involving different combinations of IgAN alleles, including GRS5 (five reported HLA alleles), GRS7 (five reported HLA alleles and two non-HLA alleles, which were the same as reported standardized GRS), GRS9 (five reported HLA alleles and four non-HLA alleles), and GRS4 (four non-HLA alleles). Every score could be weighted or un-weighted. For comparison, we also directly adopted standardized GRS as reported previously.
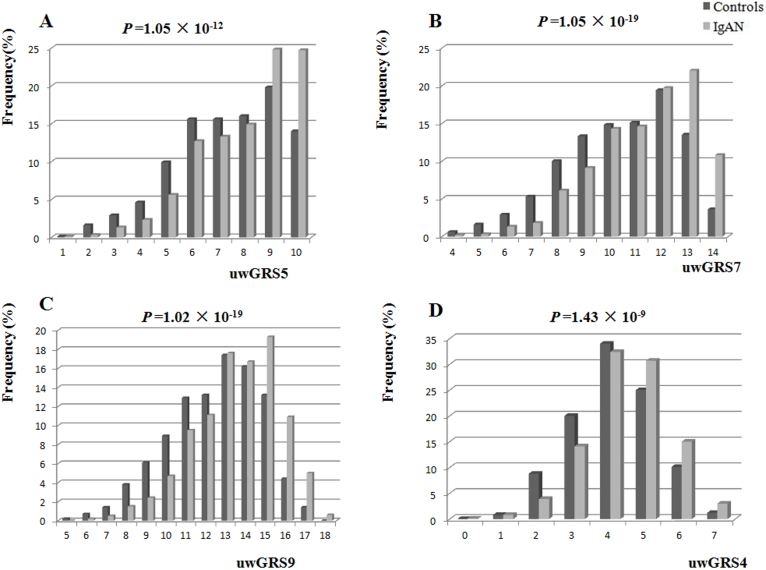
The distribution of unweighted GRSs (uwGRSs) between IgAN and controls were significantly different (Figure 2). The frequency of a higher uwGRS (more risk alleles) was higher in IgAN than in controls. With every 1-unit increase in the uwGRS or one copy increase of a risk allele, the disease risk increased by about 20% ~ 30% (Table 3). Using the difference value (differences of uwGRS between IgAN and controls, differences value = uwGRSIgAN − uwGRScontrol) as a risk function, the difference value of uwGRS5, uwGRS7, or uwGRS9 was much further from zero than that of uwGRS4 (non-HLA risk score). This might suggest that IgAN cases had one more copy of a risk allele than the controls, which was mainly from the HLA alleles.
Figure 2. Distribution of unweighted genetic risk score (uwGRS) between IgAN and controls.
(A) uwGRS5, (B) uwGRS7, (C) uwGRS9, (D) uwGRS4. The p values indicate comparison of cases and controls using a chi-squared test.
Table 3. Risk of susceptibility to IgAN based on uwGRS.
| GRS | Mean Cases vs. Controls | Difference value in mean | Median Cases vs. Controls | Difference value in median | OR (95% CI) | p | Adjusted OR (95% CI) | p |
|---|---|---|---|---|---|---|---|---|
| uwGRS5 | 8.00/7.29 | 0.71 | 8/7 | 1 | 1.22 (1.16–1.28) | 9.73 × 10−17 | 1.21 (1.15–1.27) | 2.24 × 10−14 |
| uwGRS7 | 11.29/10.36 | 0.93 | 12/11 | 1 | 1.24 (1.19–1.30) | 1.01 × 10−22 | 1.24 (1.19–1.30) | 5.19 × 10−20 |
| uwGRS9 | 13.45/12.38 | 1.07 | 14/13 | 1 | 1.24 (1.19–1.29) | 2.23 × 10−25 | 1.23 (1.19–1.29) | 6.11 × 10−22 |
| uwGRS4 | 5.45/5.09 | 0.36 | 5/5 | 0 | 1.30 (1.21–1.40) | 1.04 × 10−11 | 1.29 (1.19–1.40) | 2.28 × 10−10 |
OR in this table represents the expected change in odds of a case being associated with a 1-unit increase in the GRS, as determined by logistic regression of case/control status on GRS.
For adjusted models, the age and sex of patient were included as covariates in the logistic regression model.
Differences value = uwGRSIgAN − uwGRScontrol.
The data from the weighted GRS (wGRS) model (the risk score equations are shown in Table 4) was concordant with that from unweighted models. With one standard deviation increase in the score, disease risk increased about 40% ~ 60%. The OR for one standard deviation increase were 1.47, 1.60, 1.63, 1.42 and 1.68 for wGRS5 (OR = 1.47, 95% CI: 1.34–1.61, P = 8.83 × 10−16), wGRS7 (OR = 1.60, 95% CI: 1.45–1.76, P = 7.36 × 10−22), wGRS9 (OR = 1.63, 95% CI: 1.48–1.80, P = 5.66 × 10−24), wGRS4 (OR = 1.42, 95% CI: 1.30–1.56, P = 9.58 × 10−14) and standardized GRS (OR = 1.68, 95% CI: 1.53–1.84, P = 9.42 × 10−27), respectively. Examination of wGRS quartiles also suggested a pattern of increasing disease risk with each wGRS quartile. Using group 1 (lowest level of risk) as a reference group, quartile 4 had the highest odds of IgAN, with ORs of 2.37, 3.17, 3.34, 2.28, and 3.67 for wGRS5, wGRS7, wGRS9, wGRS4 and standardized GRS25, respectively. The trends across all categories were highly significant without restriction of the wGRS adopted (Table 5).
Table 4. Risk score equations for weighted genetic risk scores (wGRS) in the current study.
| Weighted genetic risk score | Risk score equation |
|---|---|
| wGRS5 | −0.40 × N(rs9275224:A) − 0.42 × N(rs2856717:T) − 0.58 × N(rs9275596:C) − 0.37 × N(rs9357155:A) − 0.21 × N(rs1883414:T) |
| wGRS7 | −0.60 × N(rs6677604:A) − 0.40 × N(rs9275224:A) − 0.42 × N(rs2856717:T) − 0.58 × N(rs9275596:C) − 0.37 × N(rs9357155:A) − 0.21 × N(rs1883414:T) − 0.33 × N(rs2412971:A) |
| wGRS9 | −0.60 × N(rs6677604:A) − 0.40 × N(rs9275224:A) − 0.42 × N(rs2856717:T) − 0.58 × N(rs9275596:C) − 0.37 × N(rs9357155:A) − 0.21 × N(rs1883414:T) − 0.33 × N(rs2412971:A) − 0.21 × N(rs2738048:C) − 0.14 × N(rs3803800:G) |
| wGRS4 | −0.60 × N(rs6677604:A) − 0.33 × N(rs2412971:A) − 0.21 × N(rs2738048:C) − 0.14 × N(rs3803800:G) |
| Standardized GRS (reported seven–SNP genetic risk score) | [−0.46026 × N(rs6677604:A) − 0.31127 × N(rs9275224:A) + 0.41653 × N(rs2856717:T) − 0.46857 × N(rs9275596:C) − 0.22668 × N(rs9357155:A) − 0.15722 × N(rs1883414:T) − 0.26625 × N(rs2412971:A) + 0.17821 × N(rs6677604:A) × N(rs2412971:A) − Worldwide Mean]/(Worldwide SD). |
Weights were the natural log of the effect magnitude of the allele.
N = number of reference alleles for each SNP (0, 1, or 2 per individual genotype).
Worldwide Mean = −0.8360883 = mean risk score based on the HGDP data. Worldwide SD = 0.4146805 = risk score standard deviation based on the HGDP (Human Genome Diversity Project) data. http://www.columbiamedicine.org/divisions/gharavi/calc_genetic.php).
Table 5. Risk of susceptibility to IgAN based on quartiles of wGRS.
| GRS | Group | IgAN | Controls | OR | 95% CI | P | P trend | ||
|---|---|---|---|---|---|---|---|---|---|
| Mean GRS | N (%) | Mean GRS | N (%) | ||||||
| wGRS5 | Q1 | −2.30 | 125(10.5) | −2.36 | 180(20.0) | 1 | |||
| Q2 | −1.42 | 287(24.1) | −1.44 | 261(29.0) | 1.58 | 1.19–2.10 | 1.42 × 10−3 | ||
| Q3 | −0.59 | 295(24.8) | −0.58 | 225(25.0) | 1.89 | 1.42–2.52 | 1.26 × 10−5 | ||
| Q4 | −0.11 | 383(40.6) | −0.14 | 233(25.9) | 2.37 | 1.79–3.13 | 1.16 × 10−9 | 3.13 × 10−16 | |
| wGRS7 | Q1 | −2.59 | 135(11.3) | −2.74 | 202(22.5) | 1 | |||
| Q2 | −1.69 | 270(22.7) | −1.73 | 242(26.9) | 1.67 | 1.26–2.21 | 2.97 × 10−4 | ||
| Q3 | −0.96 | 249(20.9) | −0.97 | 202(22.5) | 1.84 | 1.39–2.46 | 2.56 × 10−5 | ||
| Q4 | −0.34 | 536(45.0) | −0.41 | 253(28.1) | 3.17 | 2.43–4.13 | 2.57 × 10−18 | 9.38 × 10−19 | |
| wGRS9 | Q1 | −2.87 | 147(12.4) | −3.02 | 223(24.8) | 1 | |||
| Q2 | −1.98 | 250(21.0) | −2.00 | 226(25.1) | 1.68 | 1.27–2.21 | 2.17 × 10−4 | ||
| Q3 | −1.25 | 297(25.0) | −1.26 | 225(25.0) | 2.00 | 1.53–2.63 | 4.37 × 10−7 | ||
| Q4 | −0.59 | 496(41.7) | −0.68 | 225(25.0) | 3.34 | 2.58–4.34 | 2.51 × 10−20 | 9.81 × 10−21 | |
| wGRS4 | Q1 | −1.10 | 178(15.0) | −1.15 | 218(24.2) | 1 | |||
| Q2 | −0.75 | 229(19.2) | −0.75 | 216(24.0) | 1.30 | 0.99–1.70 | 0.06 | ||
| Q3 | −0.56 | 247(20.8) | −0.56 | 177(19.7) | 1.71 | 1.30–2.25 | 1.39 × 10−4 | ||
| Q4 | −0.28 | 536(45.0) | −0.31 | 288(32.0) | 2.28 | 1.79–2.91 | 2.53 × 10−11 | 9.04 × 10−13 | |
| Standardized GRS | Q1 | −0.75 | 133(11.2) | −0.85 | 224(24.9) | 1 | |||
| Q2 | 0.15 | 228(19.2) | 0.11 | 216(24.0) | 1.78 | 1.34–2.36 | 6.74 × 10−5 | ||
| Q3 | 0.78 | 302(25.4) | 0.75 | 217(24.1) | 2.34 | 1.78–3.09 | 1.14 × 10−9 | ||
| Q4 | 1.57 | 527(44.3) | 1.48 | 242(26.9) | 3.67 | 3.82–4.77 | 3.57 × 10−23 | 3.54 × 10−24 | |
We calculated the odds for the top group (Q4) compared with the bottom group (Q1) as the reference group.
Observed GRS and clinical parameters of IgAN
We assessed the associations between the clinical parameters of IgAN, including proteinuria, hematuria, eGFR, hypertension, hyperlipidemia, hyperuricemia, CKD stage and Hass grade at the time of renal biopsy with cumulative genetic effects of identified SNPs from GWAS (Table 2). However, no clear associations were observed, except a marginally significant association between GRS4 and gross hematuria (p < 0.05). Consistent with data from individual association between single SNPs and clinical parameters of IgAN, significant associations between IgA and IgA1 levels with GRS were observed; the sera IgA level increased with increasing uwGRS or wGRS. The associations were more prominent considering GRSs that included non-HLA alleles (GRS4, GRS7 and GRS9 instead of GRS5), suggesting that the effect was mainly driven by non-HLA alleles. However, the associations became non-significant on multiple correction.
Association between genetic information and prognosis of IgAN
rs3803800 and GRS4 were marginally associated with indicators for prognosis, including natural log-transformed time averaged mean arterial pressure and eGFR slope in linear regression (Table 6). By univariate Cox regression analysis, GRS5, GRS7 and GRS9 were associated with disease progression to end stage renal disease (ESRD), in which uwGRS showed a minimal increase of sensitivity for association. Although it seemed that the relative risks were similar, uwGRS9 showed the most significant association with progression to ESRD.
Table 6. Correlation of the SNPs and GRS with prognosis of IgAN in follow-up.
| Genetic information | Log (TA-Proteinuria) | Log (TA-MAP) | Slope | ESRD |
|---|---|---|---|---|
| rs6677604 | 0.14 | 0.49 | 0.20 | 0.80 |
| rs9275224 | 0.93 | 0.78 | 0.40 | 0.11 |
| rs2856717 | 0.62 | 0.66 | 0.23 | 0.16 |
| rs9275596 | 0.86 | 0.38 | 0.13 | 0.10 |
| rs9357155 | 0.31 | 3.31 × 10−2 (Beta 0.13) | 0.79 | 0.92 |
| rs1883414 | 0.29 | 0.21 | 0.17 | 0.14 |
| rs2738048 | 0.50 | 9.71 × 10−4 (Beta −0.19) | 0.63 | 0.20 |
| rs3803800 | 0.37 | 1.56 × 10−2 (Beta −0.15) | 4.25 × 10−2 (Beta 0.12) | 0.76 |
| rs2412971 | 0.71 | 0.79 | 0.18 | 0.29 |
| uwGRS5 | 0.64 | 0.94 | 0.11 | 4.81 × 10−2 (Beta 0.18) |
| uwGRS7 | 0.70 | 0.92 | 0.37 | 3.04 × 10−2 (Beta 0.19) |
| uwGRS9 | 0.90 | 0.11 | 0.98 | 1.67 × 10−2 (Beta 0.17) |
| uwGRS4 | 0.32 | 3.84 × 10−3 (Beta −0.16) | 1.35 × 10−2 (Beta 0.14) | 0.15 |
| wGRS5 | 0.72 | 0.89 | 0.12 | 0.05 |
| wGRS7 | 0.85 | 0.95 | 0.35 | 3.96 × 10−2 (Beta 0.44) |
| wGRS9 | 0.98 | 0.47 | 0.55 | 2.51 × 10−2 (Beta 0.46) |
| wGRS4 | 0.32 | 0.10 | 1.80 × 10−2 (Beta 0.13) | 0.20 |
| Standardized GRS | 0.65 | 0.89 | 0.89 | 2.25 × 10−2 (Beta 0.48) |
Linear regression was applied for the correlation analysis of natural log-transformed time-average proteinuria, natural log-transformed time-average mean artery pressure and eGFR slope.
Univariate Cox regression analysis was applied for the association of disease progression with ESRD.
Effect estimates (OR/BETA) are shown only for significant associations (p < 0.05).
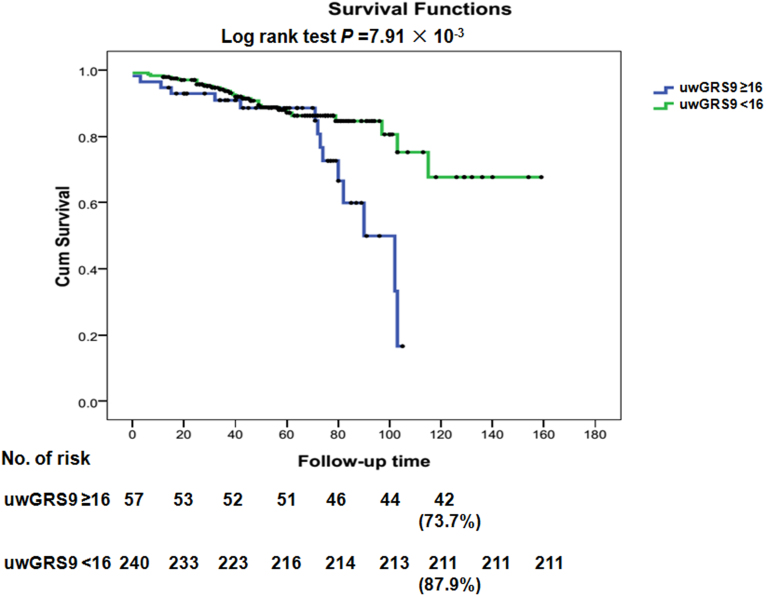
Consistent with previous reports, the statistics confirmed good discrimination between IgAN and controls regarding GRSs (AUC was about 0.6, p < 0.001)5(Table 7), in which GRS9 and standardized GRSs showed the better fit in model prediction. Using the Kaplan-Meier survival method with the optimal derived cut-off value (16 for uwGRS9 with a sensitivity 0.96 and specificity 0.93) identified by a receiver operator characteristic (ROC) curve, we observed a worse renal prognosis rate of 26.3% (Figure 3, p = 7.91 × 10−3) only in IgAN patients with uwGRS ≥ 16 at 10 years ESRD, compared with 12.1% in uwGRS < 16. When covariates of ACEI/ARB use and steroid use (yes or no) were introduced into multivariate Cox regression analysis, uwGRS9 ≥ 16 was still an independent predictor for ESRD in IgAN. The relative risks for uwGRS9 ≥ 16, ACEI/ARB use, and steroid use were 2.52 (95% CI, 1.29–4.91, p = 6.68 × 10−3), 0.09 (95% CI, 0.04–0.23, p = 2.85 × 10−7), and 3.75 (95% CI, 1.90–7.41, p = 1.42 × 10−4). Regarding clinical parameters at the time of disease onset, including blood pressure, hematuria, proteinuria and renal pathology, the uwGRS9 ≥ 16 group also showed no significant difference compared with the uwGRS9 < 16 group (p > 0.05). Similarly, using standardized GRS ≥ mean + SD as the cut-off value, a marginally significant 10-year ESRD rate was observed in IgAN patients with standardized GRS ≥ mean + SD compared with that of standardized GRS < mean + SD (21.3% vs. 12.1%, p = 0.06).
Table 7. Comparison of different genetic risk scores in disease prediction.
| Genetic risk score | R2 | C (95% CI) | p |
|---|---|---|---|
| uwGRS5 | 4.5% | 0.61 (0.58–0.63) | 1.77 × 10−16 |
| uwGRS7 | 6.4% | 0.62 (0.60–0.65) | 2.97 × 10−22 |
| uwGRS9 | 7.3% | 0.63 (0.61–0.66) | 6.83 × 10−25 |
| uwGRS4 | 3.0% | 0.59 (0.56–0.61) | 3.28 × 10−11 |
| wGRS5 | 4.2% | 0.61 (0.58–0.63) | 1.41 × 10−16 |
| wGRS7 | 6.1% | 0.62 (0.60–0.65) | 4.35 × 10−22 |
| wGRS9 | 6.8% | 0.63 (0.61–0.66) | 6.83 × 10−25 |
| wGRS4 | 3.7% | 0.59 (0.56–0.61) | 3.28 × 10−11 |
| Standardized GRS | 7.7% | 0.64 (0.61–0.66) | 1.72 × 10−26 |
As reported, the percentage of the total variance in the disease state explained by the risk score was estimated by Nagelkerke's pseudo R2 from the logistic regression model, with the risk score as a quantitative predictor and disease state as an outcome.
The C-statistic was estimated as an area under the receiver operating characteristic curve provided by the above logistic model.
Figure 3. Kaplan-Meier survival curves without ESRD/dialysis/death event, with time zero set at kidney biopsy and uwGRS9 ≤ 2 in IgAN patients.
Using the Kaplan-Meier survival method with the optimal derived cut-off values, we observed a worse renal prognosis rate of 26.3% only in IgAN patients with uwGRS9 ≥ 16 at 10 years ESRD, compared with 12.1% with uwGRS9 < 16.
Discussion
We tested previously established SNPs associated with IgAN in a large collection of Chinese patients. Although the two SNPs from a southern China GWAS were marginally associated with IgAN in our cohort, we validated that all nine SNPs could be replicated as associated with IgAN, suggesting their real genetic effect12,27.
To determine their cumulative effect, we constructed two genetic risk scores24, uwGRS and wGRS, with different portfolios of different SNPs, HLA alleles or non-HLA alleles or in combination, for IgAN and tested their correlation to ESRD events and their potential for disease prediction. Compared with single SNP, GRSs were more significantly associated with susceptibility to IgAN. All GRSs were associated with IgAN, in which the prediction power increased with increasing numbers of SNPs selected. However, IgAN cases could have one more copy of a risk allele compared with controls, mainly HLA alleles. Although a non-HLA uwGRS model (uwGRS4) could also discriminate IgAN from controls, the difference of uwGRS count between IgAN and controls were smaller than that of the HLA GRS model (uwGRS5). Compared with the HLA GRS model (GRS5), with increase of non-HLA alleles (GRS7 and GRS9), the prediction power increased only slightly. This suggested that HLA allele-based GRSs might have larger power in disease prediction than non-HLA allele-based GRSs. The other issue was the GRS calculation method24,25,26,27. Similar to previous reports21,22,23, we did not observe a highly significant difference between the two methods (uwGRS and wGRS) in disease prediction power, as shown by slightly different AUC (difference < 0.05) and Nagelkerke's pseudo R2 (difference < 0.01) scores using the same number of risk alleles. The data was consistent with previous reports: it mattered little in terms of discriminative accuracy whether genetic scores were constructed using the count method or the log odds procedure for most complex diseases with ORs for disease risk alleles similar and close to 121,22,23. uwGRS showed slightly lower p values compared with wGRS, suggesting that uwGRS might be chosen for risk stratification in IgAN. However, it may not be true for other disease. The ORs for IgAN risk alleles were similar and close to 1(<1.5); therefore, the weighted index seemed to have a marginal effect. If great discrepancies of ORs for IgAN risk alleles and risk alleles with larger effects were identified, the strategy may need change5,24,25,26,27. However, the seven–SNP genetic risk score showed a better fit in disease prediction than GRS7 and, occasionally, GRS9. There are several possible explanations: it was based on more samples (5 times larger than ours); it included a second kind of genetic interaction information; and the model was constructed using a stepwise logistic regression algorithm. Future evaluation of the seven–SNP genetic risk score in sub-phenotypes and disease prognosis in more widespread populations are still warranted.
For clinical parameter or sub-phenotype associations, we observed highly significant associations between GRS and serum IgA/IgA1 levels. The risk genetic group was consistently associated with increased IgA1 level. The data also validated associations between rs3803800 and serum IgA level, noted in a previous GWAS of IgA level conducted in a Chinese population12,28. Our data supported the notion that genetically deregulated IgA play a key role in the pathogenesis in IgAN4,6,10,11,29. However, less concordant or significant associations for other sub-phenotypes of IgAN were observed as associated with single or cumulative gene effects.
When the weak effects of the individual SNPs are considered together, we observed a strong and consistent effect on ESRD because of the GRS. The effect was independent of therapy with ACEI/ARB and corticosteroids. Consistent with a recent GRS study conducted in hypertension, which suggested that a blood pressure genetic risk score could be a significant predictor of incident cardiovascular events, the current data may further support the idea of prospects for genetic risk prediction in clinical practice17,18,19,24. We speculated that the genetic variants have cumulative effects on IgA deregulation involved in disease susceptibility and progression. Although power analysis indicated that we had about a 0.6–0.8 power to detect a two-fold increased risk considering clinical parameters and disease progression, assuming an α-level of 0.05 and allele frequency of 10%–30% (http://biostat.mc.vanderbilt.edu/PowerSampleSize), the effect size identified was far smaller than two and all the associations did not survive multiple testing. Thus, the data requires further widespread replications and functional investigations.
The strengths of the current study include the large sample size, the availability of complete genetic information and relevant covariates, the comparatively long follow-up period with a certain number of ESRD outcomes available for prospective analyses and adoptions of different GRS methods. Limitations include its single center experience, and the inability to generalize to Southern Chinese Han and non-Chinese ancestry groups. The current GRS modeling was mainly based on genotyping data from a previous GWAS cohort; therefore, we cannot rule out the possibility of bias of GRS from over-fitted association, requiring future evaluation of GRS in more widespread cohorts and in prospective studies. We lacked the ability to adjust for time-varying clinical factors in disease progression. The proportion of variation explained by the SNPs remained low, and the level of prediction for events was also relatively small. We lacked power to demonstrate associations with moderate genetic effects reported in a previous Southern Chinese Han GWAS.
In conclusion, we observed that GRSs comprising nine SNPs identified in a GWAS of IgAN were strongly associated with susceptibility to IgAN, in which HLA alleles contributed more than non-HLA alleles and uwGRS calculation was simpler than wGRS for prediction. The high risk GRS9 group (uwGRS9 ≥ 16) had a high risk of ESRD in follow-up, suggesting a need for early and positive intervention.
Methods
Study population
The case-control cohort analyzed in this study was the same as the previous Chinese Han cohort included in the GWAS27: 1,194 IgAN cases and 902 healthy controls recruited in the renal division of Peking University First hospital. Quality control was performed as described27. All cases carried a biopsy diagnosis of IgAN defined by typical light microscopy features and predominant IgA staining on kidney tissue immunofluorescence, in the absence of liver disease, vasculitis, Henoch–Schoenlein purpura, or other autoimmune diseases. This investigation was conducted according to the Declaration of Helsinki. All subjects provided informed consent to participate in genetic studies and the ethic review committee of Peking University First Hospital approved the study protocol.
Baseline and follow-up clinical phenotypes
Detailed phenotypic data from the patients, including degree of renal dysfunction, hematuria, and proteinuria at presentation, total serum IgA, and detailed biopsy findings (Haas staging), were collected at the time of renal biopsy at enrollment. Among the patients involved in the GWAS, 297 patients were followed for a mean of 5 years (range 1 to 15 years). An enzyme-linked immunosorbent assay quantified Serum IgA and IgA16. All patients received the same therapy regimen, including optimal blood pressure control target to less than 130/80 mmHg, RAS inhibition and steroids or other immunosuppressive agents for patients with persistent proteinuria. The blood pressure and proteinuria controls were expressed as time-average mean artery pressure or time-average proteinuria. The endpoint in this study was defined by diagnosis of ESRD or death. ESRD was defined as eGFR < 15 ml/min/1.73 m2 or need for renal replacement therapy (hemodialysis, peritoneal dialysis or renal transplantation). The eGFR was calculated using the Modification of Diet in Renal Disease (MDRD) formula30,31.
SNP selection
We firstly selected seven SNPs (Table 1), including five HLA SNPs and two non-HLA SNPs at five independent loci, which were independently associated with IgAN in the GWAS and they were selected in the GRS calculated in the previous report5,27.Another large GWAS conducted in a different Chinese Han population from Southern China identified additional IgAN associated non-HLA alleles; therefore, they were also selected for the current study. The additional non-HLA SNPs were rs2738048 (8p23), rs3803800 (17p13), rs4227 (17p13), and rs12537 (22q12)12. As the D′ between rs3803800 and rs4227 was 0.92, and that between rs3803800 and rs4227 was 0.91, indicating high linkage disequilibrium and possibly non-independent genetic effects, a seven-SNP model at the five independent loci and a nine-SNP model at the seven independent loci were constructed. The nine-SNP model included the novel IgAN associated variant rs2738048 and the missense variant rs3803800.
Genetic risk score
Two GRSs were constructed on an a priori basis. The first GRS using an unweighted approach (uwGRS) was the simple counts of the total number of risk alleles rather than weighting by the effect of each SNP, as the current data available may be insufficient to provide stable estimates for each effect of small magnitude24. The second GRS was the weighted-GRS (wGRS) that utilized the allelic odds ratios (OR) to account for the strength of the genetic association within each allele, because different IgAN alleles may have different odds ratios. The wGRS was the weighted sum of risk allele counts, where the weight for each SNP was the natural log of the OR25,26. Different ORs may be observed in different populations for the same allele; therefore, we adopted ORs observed in our current dataset. For comparison and cross-validation, we also directly calculated standardized genetic risk based on the seven SNPs associated with IgA nephropathy in the previous analysis of 10,755 individuals from 12 international case-control cohorts. A coded allele is an allele coded 0, 1, or 2 according to the number of copies of the target allele, as reported5,27. Individuals with 100% non-missing genotypes across all the scored loci were analyzed. Ultimately, 1190 cases and 899 controls were included in the current study.
Statistical analysis
We used logistic regression to study the association of each allele with the risk of IgAN, according to an additive log-odds model. We calculated a GRS5 that included five HLA alleles, a GRS4 that included four non-HLA alleles, a GRS7 that included five HLA alleles and two non-HLA risk alleles, and a GRS9 that included five HLA alleles and four non-HLA risk alleles.
The difference in the distribution of uwGRSs between IgAN cases and controls was tested using the chi-squared test. To explore the observed patterns in more detail, we also divided the subjects into quartiles based on the GRS of controls, and computed the proportions of cases and controls in each quartile. To assess whether risk was significantly different according to quartile, we performed logistic regressions that modeled the risk of disease as a function of each GRS quartile compared with the reference quartile. Finally, we calculated the odds for the top group (group 4) compared with the bottom group (group 1) as the referent group25,26.
Linear regression was applied for correlation analysis of natural log-transformed serum IgA levels, natural log-transformed proteinuria and natural log-transformed eGFR. Binary logistic regression was carried out for the correlation analysis of history of gross hematuria. Ordinal logistic regression was performed for the correlation analysis of clinical subtype, microscopic hematuria, CKD stage at the time of biopsy, and Hass biopsy grade12,27.
To set the cut-off values between patients and controls, and within the cohort of IgAN patients grouped as progressive cases versus non-progressive cases, we used ROC curve analyses to find the best compromise value between sensitivity and specificity; we also generated ROC curves by plotting the sensitivity of the GRS score against 1-specificity and calculated the area under the curve (AUC). As reported, the percentage of the total variance in the disease state explained by the risk score was estimated by Nagelkerke's pseudo R2 from the logistic regression model, with the risk score as a quantitative predictor and disease state as an outcome. The C-statistic was estimated as an area under the receiver operating characteristic curve provided by the above logistic model. The AUC statistics were compared using a non-parametric approach, as described previously4. The Kaplan-Meier survival method and Cox proportional hazards models were used to generate estimates of predicted risk of ESRD.
Descriptive statistics included mean (SD) and median (with range values).These analyses were carried out with SPSS Statistics version 16.0.
Author Contributions
Z.X.J., Q.Y.Y., H.P., L.J.C., S.S.F., L.L.J., Z.N. and Z.H. are co-authors on this manuscript. Z.H. is the senior author on this manuscript. Z.X.J. and Z.H. conceived and designed experiments. Z.X.J., Q.Y.Y., H.P., S.S.F., L.L.J. and Z.N. performed the experiments and analyzed the data; Z.X.J. and Z.H. wrote the manuscript. All authors reviewed the manuscript.
Acknowledgments
We thank our collaborators of Ali G. Gharavi and Krzysztof Kiryluk from the Department of Medicine, Columbia University College of Physicians and Surgeons, New York, USA for providing GWAS data. The authors also thank all the members of the laboratory for technical assistance and the patients and their families for their cooperation and for giving consent to participate in this study. This work was supported by grants from the Major State Basic Research Development Program of China (973 program, No. 2012CB517700), the National Natural Science Foundation of China (No. 81200524), the Research Fund of Beijing Municipal Science and Technology for the Outstanding PhD Program (20121000110), The Foundation of Ministry of Education of China (20120001120008), and the Natural Science Fund of China to the Innovation Research Group (81021004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Wyatt R. J. & Julian B. A. IgA nephropathy. N Engl J Med 368, 2402–14 (2013). [DOI] [PubMed] [Google Scholar]
- Suzuki H. et al. The pathophysiology of IgA nephropathy. J Am Soc Nephrol 22, 1795–803 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J. K., Cheung C. K., Molyneux K., Feehally J. & Barratt J. An update on the pathogenesis and treatment of IgA nephropathy. Kidney Int 81, 833–43 (2012). [DOI] [PubMed] [Google Scholar]
- Berthoux F. et al. Autoantibodies targeting galactose-deficient IgA1 associate with progression of IgA nephropathy. J Am Soc Nephrol 23, 1579–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryluk K. et al. Geographic differences in genetic susceptibility to IgA nephropathy: GWAS replication study and geospatial risk analysis. PLoS Genet 8, e1002765 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N. et al. The level of galactose-deficient IgA1 in the sera of patients with IgA nephropathy is associated with disease progression. Kidney Int 82, 790–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryluk K., Novak J. & Gharavi A. G. Pathogenesis of immunoglobulin A nephropathy: recent insight from genetic studies. Annu Rev Med 64, 339–56 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J. et al. Predicting progression of IgA nephropathy: new clinical progression risk score. PLoS One 7, e38904 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S. I., Ramirez S. B., Winn M. P., Bonventre J. V. & Owen W. F. Evidence for genetic factors in the development and progression of IgA nephropathy. Kidney Int 57, 1818–35 (2000). [DOI] [PubMed] [Google Scholar]
- Kiryluk K. et al. Aberrant glycosylation of IgA1 is inherited in both pediatric IgA nephropathy and Henoch-Schonlein purpura nephritis. Kidney Int 80, 79–87 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M. C. et al. Galactose-deficient IgA1 in African Americans with IgA nephropathy: serum levels and heritability. Clin J Am Soc Nephrol 5, 2069–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. Q. et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet 44, 178–82 (2012). [DOI] [PubMed] [Google Scholar]
- Xie J., Shapiro S. & Gharavi A. Genetic studies of IgA nephropathy: what have we learned from genome-wide association studies. Contrib Nephrol 181, 52–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W. et al. Polymorphisms in the nonmuscle myosin heavy chain 9 gene (MYH9) are associated with the progression of IgA nephropathy in Chinese. Nephrol Dial Transplant 26, 2544–9 (2011). [DOI] [PubMed] [Google Scholar]
- Feehally J. et al. HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol 21, 1791–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. J. et al. FCGR2B and FCRLB gene polymorphisms associated with IgA nephropathy. PLoS One 8, e61208 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havulinna A. S. et al. A blood pressure genetic risk score is a significant predictor of incident cardiovascular events in 32,669 individuals. Hypertension 61, 987–94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S. Prospects for genetic risk prediction in hypertension. Hypertension 61, 961–3 (2013). [DOI] [PubMed] [Google Scholar]
- Lieb W. et al. Genetic predisposition to higher blood pressure increases coronary artery disease risk. Hypertension 61, 995–1001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens A. C., Ioannidis J. P., van Duijn C. M., Little J. & Khoury M. J. Strengthening the reporting of genetic risk prediction studies: the GRIPS statement. BMJ 342, d631 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens A. C. et al. The impact of genotype frequencies on the clinical validity of genomic profiling for predicting common chronic diseases. Genet Med 9, 528–35 (2007). [DOI] [PubMed] [Google Scholar]
- Evans D. M., Visscher P. M. & Wray N. R. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum Mol Genet 18, 3525–31 (2009). [DOI] [PubMed] [Google Scholar]
- Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet 9, e1003348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paynter N. P. et al. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA 303, 631–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahalad S. et al. Susceptibility to childhood-onset rheumatoid arthritis: investigation of a weighted genetic risk score that integrates cumulative effects of variants at five genetic loci. Arthritis Rheum 65, 1663–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson E. W. et al. Cumulative association of 22 genetic variants with seropositive rheumatoid arthritis risk. Ann Rheum Dis 69, 1077–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharavi A. G. et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43, 321–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. et al. Genome-wide association study identifies TNFSF13 as a susceptibility gene for IgA in a South Chinese population in smokers. Immunogenetics 64, 747–53 (2012). [DOI] [PubMed] [Google Scholar]
- Gharavi A. G. et al. Aberrant IgA1 glycosylation is inherited in familial and sporadic IgA nephropathy. J Am Soc Nephrol 19, 1008–14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X. et al. Evaluation of the Chronic Kidney Disease Epidemiology Collaboration equation for estimating glomerular filtration rate in the Chinese population. Nephrol Dial Transplant 28, 641–51 (2013). [DOI] [PubMed] [Google Scholar]
- Ma Y. C. et al. Modified glomerular filtration rate estimating equation for Chinese patients with chronic kidney disease. J Am Soc Nephrol 17, 2937–44 (2006). [DOI] [PubMed] [Google Scholar]