Abstract
Type 1 diabetes is etiologically a multifactorial disease caused by a complex interaction of genetic and environmental factors, with the former consisting of multiple susceptibility genes. Identification of genes conferring susceptibility to type 1 diabetes would clarify etiological pathways in the development and progression of type 1 diabetes, leading to the establishment of effective methods for prevention and intervention of the disease. Among multiple susceptibility genes, HLA and INS are particularly important because of their contribution to tissue specificity in the autoimmune process. DRB1*04:05‐DQB1*04:01 is associated with autoimmune type 1 diabetes, idiopathic fulminant type 1 diabetes and anti‐islet autoimmunity in autoimmune thyroid diseases, suggesting that this haplotype is associated with beta‐cell specificity in autoimmune diseases. Genes involved in the expression of insulin in the thymus contribute to beta‐cell‐specific autoimmune mechanisms in type 1 diabetes. These genes and pathways are important targets for tissue‐specific prevention and intervention of type 1 diabetes. (J Diabetes Invest, doi: 10.1111/j.2040‐1124.2011.00176.x, 2011)
Keywords: Autoimmune disease, Genetics, Type 1 diabetes
Introduction
Type 1 diabetes is caused by destruction of insulin‐producing beta‐cells of the pancreas in genetically susceptible individuals. Etiologically, type 1 diabetes is classified into two major subtypes, autoimmune (type 1A) and idiopathic (type 1B). The etiologic factors and pathogenesis of idiopathic type 1 diabetes are still unknown, but recent studies suggested that fulminant type 1 diabetes may belong to this subtype1,2. Type 1A diabetes is an organ‐specific autoimmune disease in which beta‐cells of the pancreas are the target organ of the autoimmune attack.
Type 1 diabetes is a multifactorial disease caused by a complex interaction of genetic and environmental factors, with the former consisting of multiple susceptibility genes. Identification of genes conferring susceptibility to type 1 diabetes would clarify the etiological pathways in the development and progression of type 1 diabetes, leading to the establishment of effective methods for prevention and intervention of the disease. In this review, clinical problems in the treatment of type 1 diabetes are summarized in order to help understand the reason why identification of genes conferring susceptibility to type 1 diabetes is necessary, and then the current status of the molecular genetics of type 1 diabetes is reviewed with special emphasis on genes that contribute to tissue specificity of autoimmune mechanisms.
Why Genes?
Among patients with type 1 diabetes, heterogeneity of residual beta‐cell function is observed. Some patients completely lack endogenous insulin secretion, while others have preservation of minimal insulin secretory capacity. Complete lack of endogenous insulin secretion in type 1 diabetes is associated with unstable glycemic control, so‐called brittle diabetes, as evidenced by our previous studies showing an inverse correlation between unstable glycemic control and minimal residual beta‐cell function in type 1 diabetes3. These data have recently been confirmed in fulminant diabetes4. This can be explained by the buffering action of endogenous insulin, whose secretion, even in a small amount, is automatically adjusted to the body’s need on a minute‐to‐minute basis. Excess exogenous insulin can be adjusted by a decrease in endogenous insulin, whereas deficiency of insulin can be adjusted by a small increase in endogenous insulin. Type 1 diabetic patients with no residual beta‐cell function lack this buffering action of endogenous insulin, and therefore have difficulty maintaining stable glycemic control, even with continuous subcutaneous insulin infusion (CSII). At the moment, pre‐programmable CSII may be the only way to achieve glycemic control in such patients. Figure 1 shows the basal insulin infusion rate of pre‐programmable CSII to achieve stable glycemic control in five patients with type 1 diabetes with complete lack of endogenous insulin. To achieve stable glycemic control, very dynamic adjustment of basal insulin infusion was required, with a decrease in infusion rate to avoid nocturnal hypoglycemia and an increase in infusion rate to overcome the dawn phenomenon. This in turn suggests that type 1 diabetic patients, particularly those with complete lack of endogenous insulin, are at high risk of nocturnal hypoglycemia when treated with bedtime NPH insulin or a long‐acting insulin analogue. In fact, when plasma glucose level was measured at 3:00 am in diabetic inpatients (n = 87) who had relatively stable glycemic control with multiple insulin injections of bedtime NPH insulin or a long‐acting insulin analogue, marked variation in the glucose level was noticed (Figure 2), with nocturnal hypoglycemia occurring in 18% of patients. Nocturnal hypoglycemia was significantly more frequent in type 1 diabetic patients than in type 2 diabetic patients (31 vs 12%, P = 0.03). These data indicate the importance of preservation, or possibly regeneration, of beta‐cells in type 1 diabetes. At the clinical onset of type 1 diabetes, beta‐cells are not completely destroyed, and low, but significant, secretory capacity of insulin still remains in most patients. To preserve residual beta cells at an early stage of type 1 diabetes and protect regenerating beta cells from recurrent autoimmune attack, the molecular mechanisms of autoimmune beta‐cell destruction must be clarified in order to establish effective methods for prevention and intervention. Identification of genes conferring susceptibility to type 1 diabetes is thus important because molecular pathways can be clarified by studying the function of genes identified.
Figure 1.
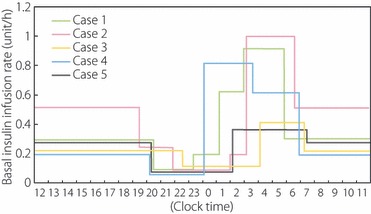
Basal insulin infusion rate in type 1 diabetics with complete lack of endogenous insulin. C‐peptide‐negative patients with type 1 diabetes (n = 5) were treated with continuous subcutaneous insulin infusion (CSII) with a pre‐programmable insulin pump. The infusion rate of basal insulin at night was adjusted to achieve target glycemic control with near‐normal glycemia at bedtime and before breakfast, and no hypoglycemia (<70 mg/dL) at 3:00 am. Very dynamic changes in basal insulin infusion rate were required. To avoid nocturnal hypoglycemia, a decrease in infusion rate to a level as low as 1/5 the daytime infusion rate was required, whereas an increase to as great as four times the daytime infusion rate was required to overcome the ‘dawn phenomenon’.
Figure 2.
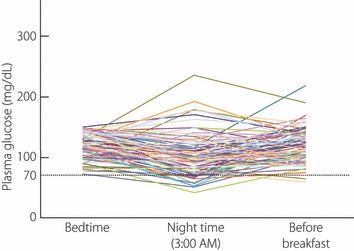
Plasma glucose level at 3:00 am compared with bedtime and before breakfast. Plasma glucose levels were monitored at bedtime (9:00 pm), during the night (3:00 am) and before breakfast (7:00 am) in diabetic inpatients (n = 87) treated with bedtime NPH insulin or a long‐acting insulin analogue, with relatively stable glycemic control (bedtime glucose level 70–150 mg/dL). It is evident that the night time glucose level cannot be estimated from glucose levels at bedtime and/or before breakfast in some patients. Nocturnal hypoglycemia is seen in some patients, while in others an increase in glucose level with a nocturnal peak is observed.
Which Genes?
Initially, susceptibility genes for type 1 diabetes were studied by the candidate gene approach, and several important genes, such as HLA and insulin gene (INS), have been identified5–9. The random marker approach was then adopted initially in multiplex families, and more recently in a large number of cases and controls with hundreds of thousands of single nucleotide polymorphisms (SNPs), termed genome‐wide association studies (GWAS) (10‐12). By using these approaches, more than 40 susceptibility loci have been mapped in Caucasian populations (Figure 3). Most of them, however, were loci, and responsible genes are yet to be identified. To clarify the etiological pathway in order to develop effective methods for prevention and intervention, responsible genes must be identified.
Figure 3.
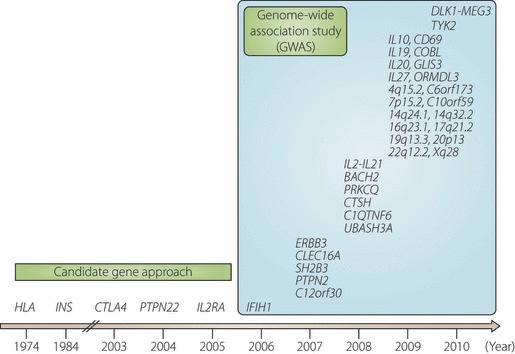
Susceptibility genes or loci for type 1 diabetes identified by candidate gene approach and/or genome‐wide association study (GWAS). Genes or loci for type 1 diabetes are shown relative to the year when convincing evidence was reported. HLA, INS, CTLA4, PTPN22 and IL2RA (CD25) were identified by candidate gene approach. IFIH1 was identified by genome‐wide association study (GWAS) with non‐synonymous SNPs, and the rest were identified by GWAS. Note that most gene symbols reported by GWAS are markers associated with the disease, but not necessarily actual genes or causal variants responsible for the disease.
Among multiple susceptibility genes, at least five genes, HLA, INS, CTLA4, PTPN22 and IL2RA (CD25), have been shown to be responsible for type 1 diabetes susceptibility in Caucasian populations5–12. Although the incidence of type 1 diabetes is markedly different between Japanese and Caucasian populations, the association of candidate genes with type 1 diabetes is generally similar in both populations, and there are good reasons for the apparent differences in the genes associated with type 1 diabetes between Japanese and Caucasians13‐19.
Among these genes, HLA shows particularly strong susceptibility in both Japanese and Caucasian populations20–22. The contribution of insulin gene (INS) to susceptibility to type 1 diabetes is well established in Caucasian populations, but its contribution in Japanese is not as clear as that in Caucasians due to the very high frequency of risk haplotype in the Japanese general population23. Recent studies demonstrated that INS is associated with type 1 diabetes in Japanese18. These two genes appear to contribute to type 1 diabetes at different steps in the etiological pathway, but as discussed below, several lines of evidence suggest that not only INS, but also class II HLA may contribute to tissue specificity of autoimmune destruction.
Major Susceptibility Gene: HLA
Class II HLA, DRB1 and DQB1, have been consistently reported to be associated with type 1 diabetes in almost all ethnic groups. Differences, however, in alleles and haplotypes associated with type 1 diabetes have been reported among different ethnic groups. The DR3 (DRB1*03:01‐DQB1*02:01) and DR4 (DRB1*04:01‐DQB1*03:02) haplotypes are positively associated with type 1 diabetes in Caucasian populations, whereas the DR4 (DRB1*04:05‐DQB1*04:01) and DR9 (DRB1*09:01‐DQB1*03:03) haplotypes are associated with the disease in Japanese and most east‐Asian populations20–22. The difference in HLA haplotypes associated with type 1 diabetes between Japanese and Caucasian populations can be explained by the presence or absence of haplotypes in each population13,14.
HLA in Rare Multiplex Family
Type 1 diabetes clusters in families, not only in Western countries24 but also in Japan, as evidenced by the much higher frequency in siblings of type 1 diabetic probands than in the general population13,14,25. Although the incidence of type 1 diabetes is much lower in Japan26, the frequency of type 1 diabetes in siblings of type 1 diabetic probands is similar to that in white populations of European descent13,14,25. As a consequence, the ratio of frequencies in siblings and the general population, termed λs, which is often used to express the degree of familial clustering of a disease, is much higher in Japanese than in Caucasian populations13,14,24,25. A high λs value with low incidence in the general population suggests two possibilities: rare variants, that is susceptibility variants with low frequencies, but with high penetrance, cluster in families, and the shared environment within families contributes to familial clustering.
To study the former possibility, we studied a rare multiplex family in which three out of four sisters developed type 1 diabetes and the fourth sister was found to be positive for anti‐GAD and anti‐IA‐2 antibodies27, suggesting that the type 1 disease process existed in all four sisters, with three of them having developed the disease clinically. In this family, all four sisters shared the same HLA genotypes, DRB1*04:05‐DQB1*04:01/DRB1*08:02‐DQB1*03:0227. This genotype has a very low frequency in the Japanese general population, but was previously reported to show a very high odds ratio for type 1 diabetes21,28, suggesting that this HLA genotype is a rare variant and is one of the reasons for clustering of type 1 diabetes in this family. Both the DRB1*04:05‐DQB1*04:01 and DRB1*08:02‐DQB1*03:02 haplotypes are very rare in Caucasian populations, and as a consequence, the DRB1*04:05‐DQB1*04:01/DRB1*08:02‐DQB1*03:02 genotype is almost absent in Caucasian populations. However, a rare DRB1*04:05‐DQB1*03:02 haplotype does exist in Caucasian populations and was reported to be the highest risk haplotype for type 1 diabetes in Caucasians29, although its frequency is low in the general population. These data suggest that the combination of DRB1*04:05 and DQB1*03:02 in either trans (Japanese) or cis (Caucasians) acts as a kind of rare variant and confers very high susceptibility to type 1 diabetes (Table 1).
Table 1. Combination of DRB1*0405 and DQB1*0302 as rare variant with low frequency, but high penetrance for type 1 diabetes.
HLA in Fulminant Type 1 Diabetes
In contrast to autoimmune type 1A diabetes, idiopathic type 1B diabetes is not well characterized, but recent studies suggest that fulminant type 1 diabetes belongs to this subtype1,2. Fulminant type 1 diabetes is characterized by a markedly acute onset of diabetes and an absence of islet‐related autoantibodies1, accounting for up to 20% of type 1 diabetes in Japan2 and 7% in Korea30. In contrast to the relatively high frequencies in Asian populations, fulminant type 1 diabetes appears to be very rare in Caucasian and other non‐Asian populations. The reason for the difference is still unknown, but one possibility may be the difference in the frequencies of risk HLAs in general populations. DRB1*04:05‐DQB1*04:01, in particular in the homozygous form, is strongly associated with fulminant type 1 diabetes28. The frequency of fulminant type 1 diabetes appears to correlate with the frequency of the DRB1*04:05‐DQB1*04:01 haplotype, in that fulminant type 1 diabetes is common in Japanese and most East Asian populations, where the DRB1*04:05‐DQB1*04:01 haplotype is common in the general population, but is absent or extremely rare in Caucasian populations, where the DRB1*04:05‐DQB1*04:01 haplotype is also absent or very rare.
HLA in Autoimmune Thyroid Diseases Complicated with Islet Autoimmunity
Patients with type 1 diabetes frequently develop other organ‐specific autoimmune diseases, of which autoimmune thyroid diseases (AITD) are the most frequent disorder31,32. In contrast to the large number of studies on autoimmunity against the thyroid gland in patients with type 1 diabetes, little is known about the anti‐islet autoimmune status in patients with AITD. We recently studied the anti‐islet autoimmune status in patients with AITD, and the clinical and genetic characteristics of AITD patients with anti‐islet autoimmunity33. The prevalence of anti‐islet autoimmunity as assessed by GAD Ab was significantly higher in patients with AITD than in normal control subjects. AITD patients with GAD Ab showed a significantly higher frequency of diabetes than did those without GAD Ab, and this was more pronounced in patients with a high titer of GAD Ab. Diabetes in AITD patients with GAD Ab was characterized by younger age‐at‐onset, lower BMI, higher HbA1c and higher frequency of insulin treatment than that in patients without GAD Ab, suggesting that diabetes in AITD patients positive for GAD Ab shows the clinical features of type 1 diabetes.
The DRB1*04:05‐DQB1*04:01 haplotype, which confers susceptibility to type 1 diabetes, was associated with AITD positive for GAD Ab, but not with AITD negative for GAD Ab33, suggesting that the DRB1*04:05‐DQB1*04:01 haplotype is associated with anti‐islet autoimmunity in subjects with as well as without AITD. In contrast, the DRB1*08:03‐DQB1*06:01 haplotype was associated with AITD without GAD Ab, but not with AITD with GAD Ab33, suggesting that the DRB1*08:03‐DQB1*06:01 haplotype confers susceptibility to autoimmunity against the thyroid gland, but not anti‐islet autoimmunity. These data suggest the contribution of HLA haplotypes not only to immune regulation, but also to organ specificity in autoimmune diseases, with DRB1*04:05‐DQB1*04:01 contributing to beta‐cell specificity of the destructive process by an autoimmune mechanism in type 1A diabetes and in AITD with anti‐islet autoimmunity, as well as an idiopathic mechanism in type 1B (fulminant) diabetes.
Insulin Gene‐Related Pathway
Cis Regulatory Region: INS‐VNTR
Accumulating lines of evidence suggest that insulin is a primary autoantigen in type 1 diabetes34‐37. Association of the insulin gene region with type 1 diabetes has been repeatedly reported in Caucasian populations6,38‐41. Allelic variation in the variable number of tandem repeats (VNTR) located in the 5′ upstream region of INS has been suggested to be responsible for disease susceptibility39,40. In the Japanese population, the markedly high frequency (>90%) of disease‐susceptible haplotype in the general population made it difficult to demonstrate the contribution of INS to disease susceptibility23. Recent large scale studies, however, demonstrated that INS‐VNTR is associated with type 1 diabetes in Japanese as well as in Caucasian populations18. INS‐VNTR is thought to contribute to type 1 diabetes susceptibility through reduced expression of insulin in the thymus, leading to impaired negative selection of insulin‐specific autoreactive T‐cells41,42.
Trans‐acting factor
In contrast to the association of the cis‐regulatory region of INS with type 1 diabetes in humans, such variants have not been identified in the NOD mouse, an animal model of type 1 diabetes. Since expression of insulin is regulated not only by cis‐regulatory elements, but also trans‐acting factors, we studied the expression of beta‐cell specific transcription factors in the thymus43. Among beta‐cell‐specific transcription factors, such as Pdx‐1, Neurod 1, and MafA, only MafA was expressed in the thymus43. Functional polymorphisms of MafA were newly identified in the NOD mouse, which were associated with reduced expression of insulin in the thymus and susceptibility to type 1 diabetes in the NOD mouse43. Functional polymorphisms of human MAFA were also identified and shown to be associated with type 1 diabetes43, suggesting that antigen‐specific transcriptional factors play a critical role in induction of central tolerance to self antigens, and abnormality in such regulation may lead to organ‐specific autoimmune diseases (Figure 4).
Figure 4.
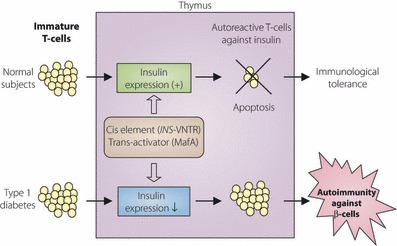
Expression of insulin in thymus and negative selection of autoreactive T‐cells against insulin. In subjects whose expression level of insulin is normal in the thymus, apoptosis is induced in insulin‐specific T‐cells upon recognition of insulin in the thymus, leading to negative selection of autoreactive T‐cells and induction of central tolerance to insulin. In subjects with reduced expression of insulin in the thymus, negative selection of insulin‐specific T cells is impaired, resulting in autoimmune attack against insulin‐producing beta‐cells of the pancreas and development of type 1 diabetes. Intra‐thymic expression of insulin is regulated by cis‐regulatory elements, such as INS‐VNTR, and trans‐acting factors, such as MafA. Functional variants in these elements or factors thus cause autoimmunity against pancreatic beta‐cells through impaired negative selection of insulin‐specific T‐cells.
Conclusions
Identification of genes conferring susceptibility to type 1 diabetes is important, even if the effect of each gene is small, because each gene contributes to a step or steps in the etiological pathway, and modification of the function of the gene or gene product could contribute to prevention and intervention of the disease. In particular, genes that contribute to tissue specificity of the autoimmune process, as in the case of genes involved in the regulatory pathway of intra‐thymic expression of insulin, are important because they are targets for tissue‐specific prevention and intervention of autoimmunity in type 1 diabetes. Although a large number of loci have been mapped in Caucasian populations by GWAS, most of them are still loci, but not responsible genes. It will be a formidable challenge to identify genes responsible for susceptibility loci mapped by GWAS because of the relatively small effect of each locus and the multifactorial nature of the disease. To overcome this, GWAS must be performed in a population possessing haplotypes different from those in Caucasians, and genes should be identified by trans‐racial studies. Such studies are now underway as a nationwide effort by the Committee on Type 1 Diabetes of the Japan Diabetes Society.
Acknowledgments
This study was supported by grants‐in‐aid for scientific research (C) (to H.I, S.N., Y.K.), a grant‐in‐aid for young scientists (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N.B.), Health and Labor Sciences Research Grants (to H.I), a grant from the Ministry of Health, Labor and Welfare (to H.I), a grant for research on intractable diseases (to H.I), and a Grant of National Center for Global Health and Medicine (to H.I). No potential conflict of interest relevant to this article is reported.
References
- 1.Imagawa A, Hanafusa T, Miyagawa J, et al. A novel subtype of type 1 diabetes mellitus characterized by a rapid onset and an absence of diabetes‐related antibodies. N Engl J Med 2000; 342: 301–307 [DOI] [PubMed] [Google Scholar]
- 2.Imagawa A, Hanafusa T. Fulminant type 1 diabetes: a novel clinical entity requiring special attention by all medical practitioners. Nat Clin Pract Endocrinol Metab 2007; 3: 36–45 [DOI] [PubMed] [Google Scholar]
- 3.Fukuda M, Tanaka A, Tahara Y, et al. Correlation between minimal secretory capacity of pancreatic beta‐cells and stability of diabetic control. Diabetes 1988; 37: 81–88 [DOI] [PubMed] [Google Scholar]
- 4.Shibasaki S, Imagawa A, Terasaki J, et al. Endogenous insulin secretion even at a very low level contributes to the stability of blood glucose control in fulminant type 1 diabetes. J Diabetes Invest 2010; 1: 283–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nerup J, Platz P, Andersen OO, et al. HL‐A antigens and diabetes mellitus. Lancet 1974; 2: 864–866 [DOI] [PubMed] [Google Scholar]
- 6.Bell GI, Horita S, Karam JH. A polymorphic locus near the human insulin gene is associated with insulin‐dependent diabetes mellitus. Diabetes 1984; 33: 176–183 [DOI] [PubMed] [Google Scholar]
- 7.Ueda H, Howson JMM, Esposito L, et al. Association of the T‐cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003; 423: 506–511 [DOI] [PubMed] [Google Scholar]
- 8.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type 1 diabetes. Nat Genet 2004; 36: 337–338 [DOI] [PubMed] [Google Scholar]
- 9.Lowe CE, Cooper JD, Brusko T, et al. Large‐scale genetic fine mapping and genotype‐phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet 2007; 39: 1074–1082 [DOI] [PubMed] [Google Scholar]
- 10.Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome‐wide analyses of type 1 diabetes. Nat Genet 2007; 39: 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakonarson H, Grant SFA, Bradfield JP, et al. A genome‐wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature 2007; 448: 591–595 [DOI] [PubMed] [Google Scholar]
- 12.Barrett JC, Clayton DG, Concannon P, et al. Genome‐wide association study and meta‐analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009; 41: 703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikegami H, Kawabata Y, Noso S, et al. Genetics of type 1 diabetes in Asian and Caucasian populations. Diabetes Res Clin Pract 2007; 77 (Suppl. 1): S116–S121 [DOI] [PubMed] [Google Scholar]
- 14.Ikegami H, Noso S, Babaya N, et al. Genetic basis of Type 1 diabetes: similarities and differences between East and West. Rev Diabet Stud 2008; 5: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikegami H, Awata T, Kawasaki E, et al. The association of CTLA4 polymorphism with type 1 diabetes is concentrated in patients complicated with autoimmune thyroid disease: a multi‐center collaborative study in Japan. J Clin Endocrinol Metab 2006; 91: 1087–1092 [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki E, Awata T, Ikegami H, et al. Systematic search for single nucleotide polymorphisms in a lymphoid tyrosine phosphatase (PTPN22) gene: association between promoter polymorphism and type 1 diabetes in Asian populations. Am J Med Genet 2006; 140: 586–593 [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki E, Awata T, Ikegami H, et al. Genetic association between the IL2RA and mode of onset of type 1 diabetes in the Japanese population. J Clin Endocrinol Metab 2009; 94: 947–952 [DOI] [PubMed] [Google Scholar]
- 18.Awata T, Kawasaki E, Ikegami H, et al. Insulin gene/IDDM2 locus in Japanese type 1 diabetes: contribution of class I alleles and influence of class I subdivision in susceptibility to type 1 diabetes. J Clin Endocrinol Metab 2007; 92: 1791–1795 [DOI] [PubMed] [Google Scholar]
- 19.Noso S, Ikegami H, Fujisawa T, et al. Genetic heterogeneity in association of SUMO4 M55V variant with susceptibility to type 1 diabetes. Diabetes 2005; 54: 3582–3586 [DOI] [PubMed] [Google Scholar]
- 20.Ikegami H, Kawaguchi Y, Yamato E, et al. Analysis by the polymerase chain reaction of histocompatibility leukocyte antigen‐DR9‐linked susceptibility to insulin‐dependent diabetes mellitus. J Clin Endocrinol Metab 1992; 75: 1381–1385 [DOI] [PubMed] [Google Scholar]
- 21.Kawabata Y, Ikegami H, Kawaguchi Y, et al. Asian‐specific HLA haplotypes reveal heterogeneity of the contribution of HLA‐DR and ‐DQ haplotypes to susceptibility to type 1 diabetes. Diabetes 2002; 51: 545–551 [DOI] [PubMed] [Google Scholar]
- 22.Thomson G, Valdes AM, Noble JA, et al. Relative predispositional effects of HLA class II DRB1‐DQB1 haplotypes and genotypes on type 1 diabetes: a meta‐analysis. Tissue Antigens 2007; 70: 110–127 [DOI] [PubMed] [Google Scholar]
- 23.Kawaguchi Y, Ikegami H, Shen GQ, et al. Insulin gene region contributes to genetic susceptibility to, but may not to low incidence of, insulin‐dependent diabetes mellitus in Japanese. Biochem Biophys Res Commun 1997; 233: 283–287 [DOI] [PubMed] [Google Scholar]
- 24.Rish N. Assessing the role of HLA‐linked and unlinked determinants of disease. Am J Hum Genet 1987; 40: 1–14 [PMC free article] [PubMed] [Google Scholar]
- 25.Ikegami H, Ogihara T. Genetics of insulin‐dependent diabetes mellitus. Endocr J 1996; 43: 605–613 [DOI] [PubMed] [Google Scholar]
- 26.Karvonen M, Viik‐Kajander M, Moltchanova E, et al. Incidence of childhood type 1 diabetes worldwide. Diabetes Mondiale (DiaMond) Project Group. Diabetes Care 2000; 23: 1516–1526 [DOI] [PubMed] [Google Scholar]
- 27.Kishi A, Kawabata Y, Ugi S, et al. The onset of diabetes in three out of four sisters: a Japanese family with type 1 diabetes. a case report. Endocr J 2009; 56: 767–772 [DOI] [PubMed] [Google Scholar]
- 28.Kawabata Y, Ikegami H, Awata T, et al. Differential association of HLA with three subtypes of type 1 diabetes: fulminant, slowly progressive and acute‐onset. Diabetologia 2009; 52: 2513–2521 [DOI] [PubMed] [Google Scholar]
- 29.Erlich H, Valdes AM, Noble J, et al. HLA DR‐DQ haplotypes and genotypes and Type 1 diabetes risk: analysis of the Type 1 diabetes genetics consortium families. Diabetes 2008; 57: 1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho YM, Kim JT, Ko KS, et al. Fulminant type 1 diabetes in Korea: high prevalence among patients with adult‐onset type 1 diabetes. Diabetologia 2007; 50: 2276–2279 [DOI] [PubMed] [Google Scholar]
- 31.Kawasaki E, Abiru N, Yano M, et al. Autoantibodies to glutamic acid decarboxylase in patients with autoimmune thyroid disease: relation to competitive insulin autoantibodies. J Autoimmun 1995; 8: 633–643 [DOI] [PubMed] [Google Scholar]
- 32.Maugendre D, Verite F, Guilhem I, et al. Anti‐pancreatic autoimmunity and Graves’ disease: study of a cohort of 600 Caucasian patients. Eur J Endocrinol 1997; 137: 503–510 [DOI] [PubMed] [Google Scholar]
- 33.Moriguchi M, Noso S, Kawabata Y, et al. Clinical and genetic characteristics of patients with autoimmune thyroid disease with anti‐islet autoimmunity. Metabolism 2011; 60: 761–766 [DOI] [PubMed] [Google Scholar]
- 34.Nakayama M, Abiru N, Moriyama H, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 2005; 435: 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alleva DG, Paul D, Jin L, et al. A disease‐associated cellular immune response in type 1 diabetics to an immunodominant epitope of insulin. J Clin Invest 2001; 107: 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kent SC, Chen Y, Bregoli L, et al. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature 2005; 435: 224–228 [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Nakayama M, Eisenbarth GS. Insulin as an autoantigen in NOD/human diabetes. Curr Opin Immunol 2008; 20: 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Julier C, Hyer RN, Davies J, et al. Insulin‐IGF2 region on chromosome 11p encodes a gene implicated in HLA‐DR4‐dependent diabetes susceptibility. Nature 1991; 354: 155–159 [DOI] [PubMed] [Google Scholar]
- 39.Lucassen AM, Julier C, Beressi JP, et al. Susceptibility to insulin dependent diabetes mellitus maps to a 4.1kb segment of DNA spanning the insulin gene and associated with VNTR. Nat Genet 1993; 4: 305–310 [DOI] [PubMed] [Google Scholar]
- 40.Bennett ST, Lucassen AM, Gough SC, et al. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet 1995; 9: 284–292 [DOI] [PubMed] [Google Scholar]
- 41.Vafiadia P, Bennett ST, Todd JA, et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet 1997; 15: 289–292 [DOI] [PubMed] [Google Scholar]
- 42.Pugliese A, Zeller M, Fernandez A, et al. The insulin gene is transcribed in the human thymus and transcription levels correlate with allelic variation at the INS VNTR‐IDDM2 susceptibility locus for type 1 diabetes. Nat Genet 1997; 15: 293–296 [DOI] [PubMed] [Google Scholar]
- 43.Noso S, Kataoka K, Kawabata Y, et al. Insulin transactivator MafA regulates intra‐thymic expression of insulin and affects susceptibility to type 1 diabetes. Diabetes 2010; 59: 2579–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]


