Abstract
Methylation of DNA CpG sites is a major mechanism of epigenetic gene silencing and plays important roles in cell division, development and carcinogenesis. One of its regulators is the 64-residue C-terminal Transcriptional Repressor Domain (the TRD) of MBD1, which recruits several repressor proteins such as MCAF1, HDAC3 and MPG that are essential for the gene silencing. Using NMR spectroscopy, we have characterized the solution structure of the C-terminus of MBD1 (MBD1-c, residues D507 to Q605), which included the TRD (A529 to P592). Surprisingly, the MBD1-c is intrinsically disordered. Despite its lack of a tertiary folding, MBD1-c could still bind to different partner proteins in a selective manner. MPG and MCAF1Δ8 showed binding to both the N-terminal and C-terminal residues of MBD1-c but HDAC3 preferably bound to the C-terminal region. This study reveals how MBD1-c discriminates different binding partners, and thus, expands our understanding of the mechanisms of gene regulation by MBD1.
Methylation of CpG sites at gene promoters and in other genomic regions by the DNA methyltransferases is an important epigenetic modification in vertebrate genomes and is necessary for the regulation of gene expression and the stability of the chromatin1,2,3. This fundamental process is a cornerstone of carcinogenesis and embryonic development, involving genomic imprinting in the latter4,5,6. Notably, aberrant CpG methylation of tumor suppressor genes has been implicated in tumor progression7,8,9.
CpG methylation sites are recognized by five methyl-CpG binding domain (MBD) proteins, MeCp2 and MBD1-410, which bind to the methylated DNA11,12 and act as transcriptional repressors by recruiting various inhibitory protein complexes to block the CpG sites from the transcriptional activators. For instance, MBD1 isoforms contain two or three cysteine-rich CXXC domains, bind to methylated CpG sites in the promoters of tumor suppressors such as p16, VHL and E-cadherin13,14, and prevent gene expression through the synergistic co-operation of the two key structural components of MBD1: the CXXC domain and the transcriptional repressor domain (TRD)15. So far, the α/β sandwich structure of MBD1 complexed with methylated DNA and the α/loop-rich structure of CXXC domain of CFP1 bearing 63% sequence identity to the CXXC domain of MBD1, have been determined16,17,18. However, the structure of the TRD of MBD1 still remains unresolved.
The TRD plays a critical role in at least some of MBD1 functional activities. MBD1 has been shown to interact with MBD1 Chromatin Associated Factor 1 (MCAF1) through its TRD and recruit Histone-lysine N-methyltransferase SETDB1 to form heterochromatin protein 1 (HP1) condensed heterochromatin19,20,21. Furthermore, the MBD1-TRD acts as a repressor of the RARβ2 tumor suppressor gene in the promyelocytic leukemia (PML); PML-RARα, a t(15;17)(q24;q21) translocation gene product, is an oncoprotein that promotes methylation of the RARβ2 gene promoter and recruits MBD1 to the gene's methylated CpGs through histone deacetylase 3 (HDAC3)22. MBD1 is also known to bind several other repressor proteins to form a tight heterochromatin complex. For example, 3-methyl purine DNA glycosylase (MPG), a DNA repair enzyme is recruited by the MBD1-TRD to the methylated gene promoters to form a tight repressor complex, which potentiates the gene silencing23. MeCp2, another MBD family member, when bound to methylated CpG, recruits the Sin3-histone deacetylase repressor complex though its TRD domain and promotes silencing of the target genes24. Similarly, other MBD proteins such as MBD2, MBD3 and MBD4 are also known to recruit repressor complexes and silence genes expression25.
All MBD proteins share a common DNA binding domain but differ in other domains. For example, MBD4 has a distinct DNA glycosylase domain that is involved in DNA repair26,27, Similarly, both the hydrophilic TRD of MeCp2 and the hydrophobic TRD of MBD1 significantly differ in protein sequence from the TRD of MBD214. Importantly, the degree of gene repression by MBD1 decreases by several folds when the TRD region is deleted14. Furthermore, the MBD1-TRD, when fused to the GAL4 DNA binding domain is sufficient to silence target genes28. Hence the MBD1-TRD has the ability to bind and recruit multiple repressor proteins such as MCAF1, HDAC3, and MPG20,22,23. However, the molecular basis of the MBD1-TRD interactions with different binding partners remains largely unknown. In this study, we have identified the solution structure of MBD1-c (D507-Q605) comprised mostly of the TRD (A529 to P592) using NMR spectroscopy. Based on the structure, we have found MBD1-c to be intrinsically disordered. Our studies also reveal how an intrinsically disordered MBD1-c is capable of interacting with its different partners (MCAF1Δ8, HDAC3 and MPG) in a specific manner.
Results
Transcriptional Repressor Domain (TRD) Sequence Conservation among MBD1 isoforms
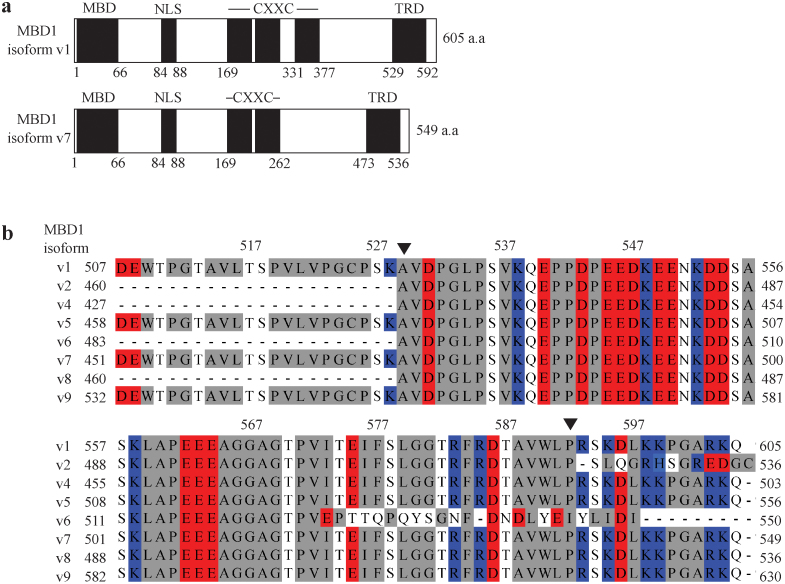
MBD1 is a transcriptional repressor containing a methylated DNA binding (MBD) domain, a putative nuclear localization signal (NLS) domain, a cysteine rich CXXC domain and a C-terminal transcriptional repressor domain (TRD) (Figure 1A). Sequence alignment of the C-terminal end of eight MBD1 isoforms (Figure 1B) shows that MBD1-c consists of an acidic cluster (E540 to E564) and a hydrophobic cluster (A565 to G581), mostly comprising the TRD (A529 to P592). The TRD is highly conserved among all MBD1 isoforms except the v6 isoform, where only its first 44 amino-acid residues, but not the last 20 residues, are conserved. There is another hydrophobic cluster (W509 to P535) that is adjacent to the TRD. A 22 amino-acid residue stretch (D507 to K528), almost completely residing in this hydrophobic cluster, is conserved among only four of the MBD1 isoforms: v1, v5, v7 and v9. An additional 13-residue stretch (R593 to Q605) located directly after the TRD domain is also highly conserved among the MBD1 isoforms, except of v2 and v6.
Figure 1. Transcriptional Repressor Domain (TRD) Sequence Conservation among MBD1 isoforms.
(A) The domain architecture of MBD1 contains a methyl-CpG binding domain (MBD), putative nuclear localization signal (NLS), two or three cysteine-rich CXXC domains and transcriptional repression domain (TRD) shown as black boxes. The white boxes correspond to unestablished domains. (B) Multiple sequence alignment of eight isoforms from the human MBD1 family showing, only isoform 6 varies from the other isoforms in the region from I573 - P592 (numbering based on isoform v1). MBD1 isoform v3 is not available at present. The residues are colored red (acidic), blue (basic), grey (neutral) and white (polar).
MBD1-c is intrinsically disordered
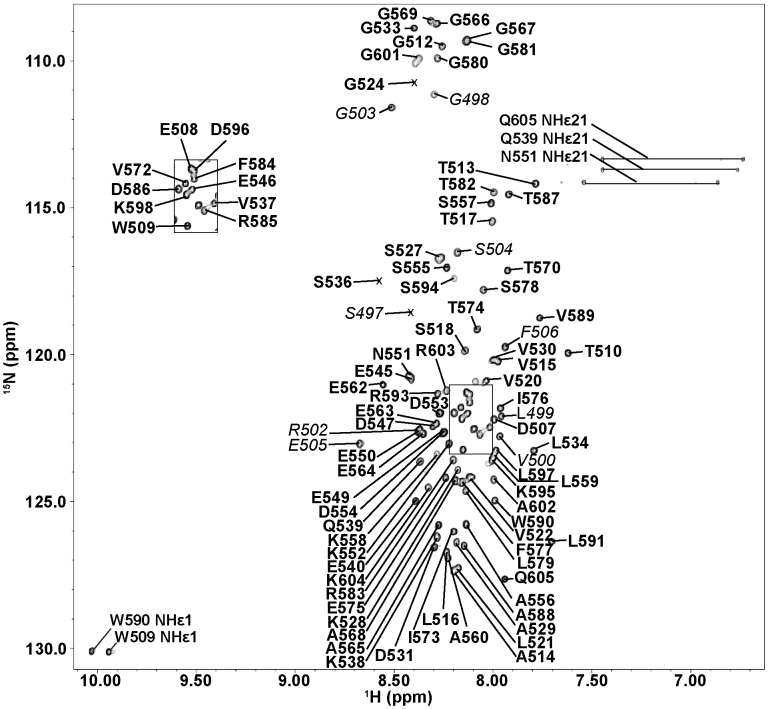
Using NMR spectroscopy, we have obtained the 2D 1H-15N Heteronuclear Single Quantum Coherence (HSQC) spectrum of MBD1-c, which shows a narrow dispersion of the amide peak resonances between 1H: 7.6–8.7 ppm which is typically of an unfolded protein. Nonetheless, the HSQC spectrum remains overall well-resolved for virtually complete residue assignment (Figure 2).
Figure 2. 1H-15N HSQC of MBD1-c.
Assigned residues in boxed region between 1H: 8.0–8.2 ppm and 15N: 121.0–123.3 ppm. Peak resonances of residues belonging to the His6-tag are italicized and to MBD1 are in bold. The peak resonances of residues are numbered according to MBD1 isoform 1.
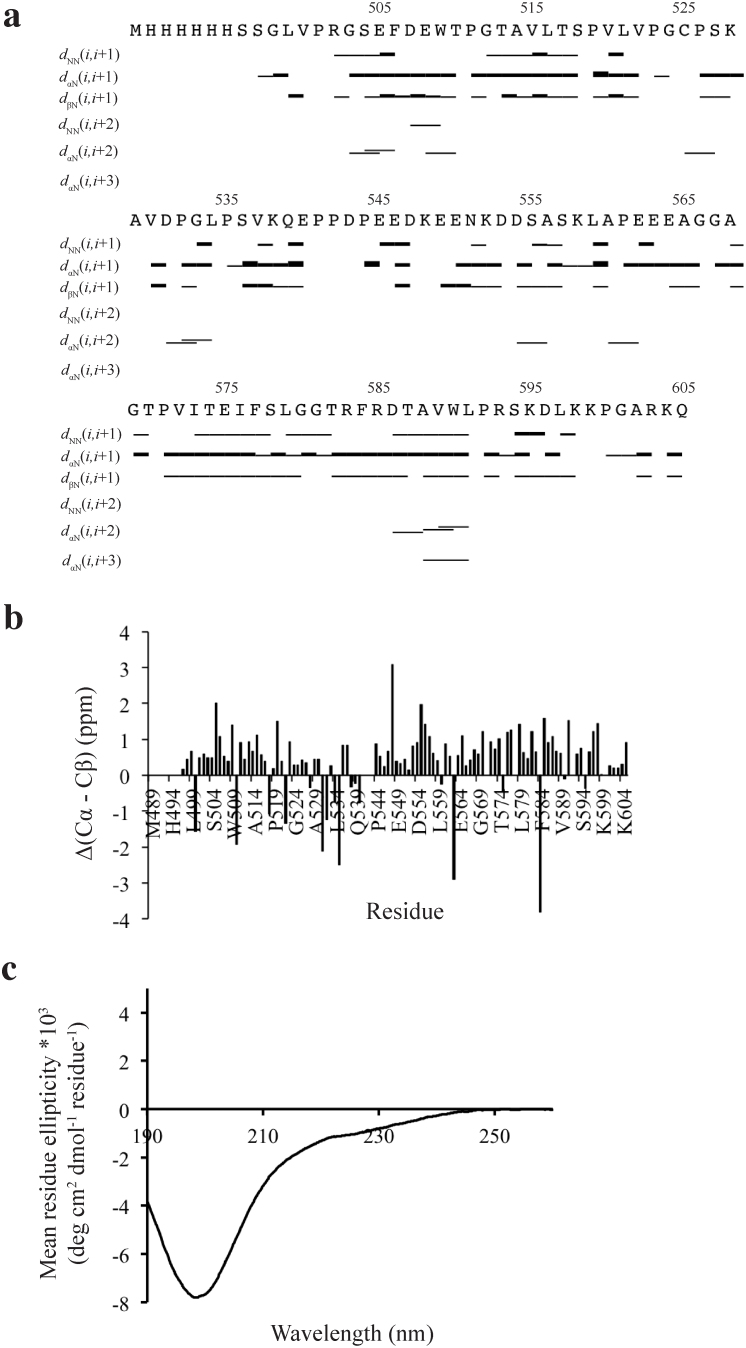
Near-complete backbone assignments were obtained and the intrinsically disordered state is represented by only a few medium and long range nuclear overhauser effects (NOEs) (Figure 3A). Also the standard chemical shift index for individual amino acid (based on random coil values in the BioMagResBank) does not reveal any well-defined secondary structure due to Δ(Cα–Cβ) values < ±1 ppm (Figure 3B). The mainly disordered conformation is due to its NOEs pattern of only a few and less intense dNN (i,i + 2) or dαN (i,i + 3) NOEs in the residues D507 to W509 and A588 to L591, which do not suffice for any fully formed secondary structures (Figure 3A, B and S2). Circular dichroism also confirmed that MBD1-c is intrinsically disordered in the native solution. In conclusion, the TRD protein is highly disordered in native solution, implying that the key transcriptional repressor role of the TRD does not require a well-defined protein fold for its interaction with partner proteins.
Figure 3. NMR analysis of MBD1-c.
(A) NOE connectivity plot of MBD1-c. The bar thickness reflects of the relative inter-proton distance. A thicker bar denotes a closer inter-proton distance. (B) Traditional chemical shift index (CSI) plot based on Δ(Cα − Cβ) = (Cα, MBD1-c − Cα, random coil − Cβ, MBD1-c − Cβ, random coil). The residues are numbered according to MBD1 isoform 144,45. (C) The CD spectra of MBD1-c.
Per-residue NMR mapping of MBD1-c when it binds to target proteins
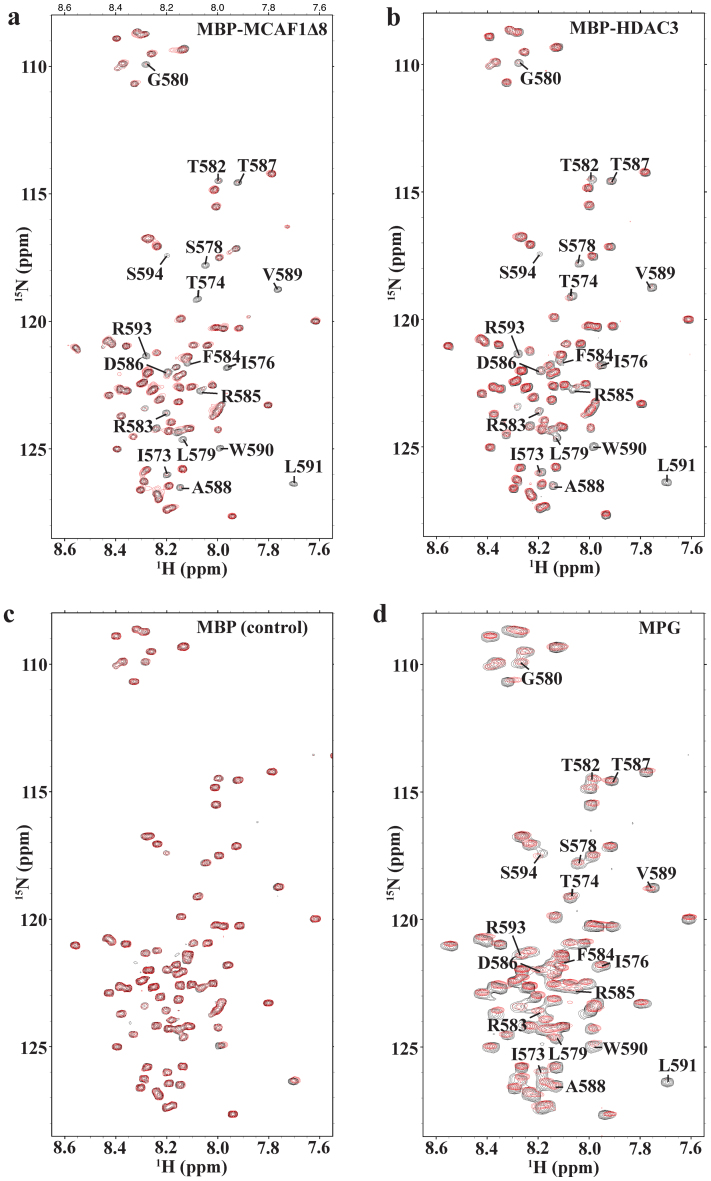
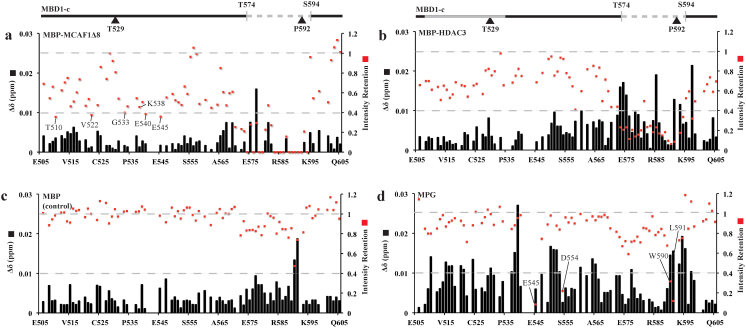
As mentioned, the TRD is indispensable for MBD1 interactions with its binding partners such as MCAF1Δ8, HDAC3 and MPG. To have a more detailed understanding of these TRD-dependent interactions, we have collected a series of 2D 1H-15N HSQCs to probe the interactions at a per residue level. Unlabeled MBP-MCAF1Δ8, MBP-HDAC3, MPG or MBP was added to 15N-labeled MBD1-c at the protein molar ratios of 1:0 to 1:3. The combined chemical shift perturbation and peak intensities of the residues were compared before and after the titration. There were only negligible chemical shift changes (<0.03 ppm), even at the molar ratio 1:3 of MBD1-c to its ligands (Figure 4A–D). This suggests that MBD1-c undergoes only minimal structural changes upon binding. We therefore analyzed the effects of the protein binding to MBD1-c on the basis of retention of peak intensities for each residue. Apart from an overall loss in peak intensity across the MBD1-c protein sequence, we observed drastic loss in intensities within a common 21-residue region between T574 and S594, which contains the distal fragment of the TRD (Figure 5A–D). The peak intensity loss is caused by forming a weak MBD1-c: protein complex and conformational change of the complex, as compared to the free MBD1-c on μs-ms timescales. Moreover, the intensity loss is relatively larger for MBP-MCAF1Δ8 and MBP-HDAC3 compared to MBP, serving as a control. This is consistent with the earlier reports on the interaction of the TRD with MCAF1 through the I576 and L579 residues20.
Figure 4. 1H-15N HSQCs of MBD1-c titrated with different partners.
(A) MBP-MCAF1Δ8 (B) MBP-HDAC3 (C) MBP as a control and (D) MPG. Selected region 1H: 7.55–8.65 ppm and 15N: 108.0–128.5 ppm showing MBD1-c before (black) and after titration till excess molar ratio of 1:3 (red). The peak resonances of residues which disappeared in (A) are also labeled in (B) and (D) for comparison.
Figure 5. 1H-15N HSQC-TROSY of MBD1-c titrated with different partners.
The titration was performed at 15N MBD1-c protein molar ratios of 1:0 (black) and 1:3 (red) with (A) MBP-MCAF1Δ8 (B) MBP-HDAC3 (C) MBP as a control and (D) MPG. Chemical shift perturbation (Δδ) and intensity retention are based on the peak resonance changes in the 15N-HSQC-TROSY. Gain/loss of interaction will lead to decrease/increase of peak intensity retention, respectively. For any absence or similar interaction in free and bound MBD1-c, the peak intensity retention would be 1. A 0.4-cutoff is also set based on 30% of the peak resonances of residues with significant intensity loss upon interacting with MCAFΔ8. Two gray dashed lines are marked at intensity retention values of 1 and 0.4. The two hydrophobic regions are colored grey for D507-A536 (bold) and T574-S594 (dashed), and marked above panels (A) and (B). The residues are numbered according to MBD1 isoform 1.
Furthermore, we have identified two new highly conserved residues, R583 and R585 of the TRD whose peak resonances either disappear or show weaker intensities in the presence of MBP-MCAF1Δ8 and MBP-HDAC3, respectively. Importantly, with MBP-MCAF1Δ8, besides the major disappeared region between T574 and S594, peak resonances of residues in another acidic and hydrophobic clusters between W509 and E564 were also found to show a distinct but less uniform pattern of intensity loss. Based on an intensity retention cutoff of <0.4, two residues E540 and E545 in the acidic-rich cluster of MBD1-c, residues T510, V522 and G533 in the hydrophobic-rich cluster (W509 to P535) are also visibly affected upon binding to MBP-MCAF1Δ8 (Figure 5A). Next, we have also characterized the binding between MBD1-c and MPG, an important DNA repair enzyme, to identify the specific residues of MBD1-c interacting with MPG. Interestingly, we found a general attenuation in peak intensities per residue that was less pronounced, when compared to the ones induced by the MBP-MCAF1Δ8 and MBP-HDAC3 interactions (Figure 5E). This relatively lower intensity loss or line width broadening is likely due to formation of the relatively smaller and/or weaker MBD1-c: MPG complex formed. Nonetheless, significant intensity losses (intensity retention cutoff of <0.4) are still observed but only for peak resonances of residues E545, D554, W590 and L591 in the acidic and hydrophobic clusters in the TRD domain. These data indicate that MBD1-c interacts with different proteins via different residues, without evident structural change to the intrinsically disordered TRD.
MBD1-c interaction with its binding partners
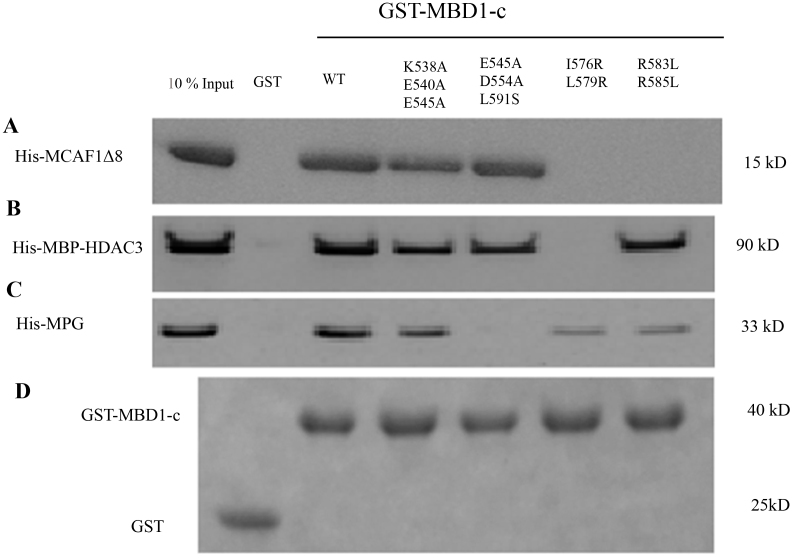
To further confirm these interactions, we performed GST pull-down assays. Indeed MBD1-c successfully binds to MCAF1Δ8, MBP-HDAC3 and MPG. Next, based on the NMR titration results, we made several MBD1-c mutants, such as K538A/E540A/E545A (KEE), E545A/D554A/L591S (EDL), I576R/L579R and R583L/R585L. The KEE MBD1-c mutant showed significantly weaker binding to MCAF1Δ8 as compared to the wild-type MBD1-c and the interaction was completely abolished when either double mutant I576R/L579R and R583L/R585L was examined (Figure 6A). These results are in full agreement with our above mentioned NMR titration data. Of note, only the double mutant I576R/L579R showed complete loss of binding to MBP-HDAC3 and the other charged mutations do not significantly affect the binding, suggesting a hydrophobic preference in the TRD for this interaction (Figure 6B). In contrast, only the EDL mutant (and neither I576R/L579R nor R583L/R585L) showed a complete loss of binding to MPG (Figure 6C), also consistent with our NMR titration findings (Figure 5C). As shown in the control gel, the proteins were loaded in equal amount in the pull-down assay (Figure 6D). Hence both NMR titration and pull-down assay show that these MBD1-c bindings are residue-specific and without a need for a globular protein fold. Thus MBD1-c can recognize MCAF1Δ8, HDAC3 and MPG in a highly partner protein-specific manner that is highly dependent on the MBD1-c structure.
Figure 6. GST pull down of MBD1-c with different partners.
GST and GST-fused MBD1-c wild type (wt) and mutants were immobilized on glutathione-agarose beads and incubated with (A) His6-tag MCAF1Δ8 (B) His6-tag MBP-HDAC3 (C) His6-tag MPG and (D) 10% of the incubated proteins are used as reference. Full length blots are presented in Supplementary Figure S4.
Isothermal titration calorimetry (ITC)
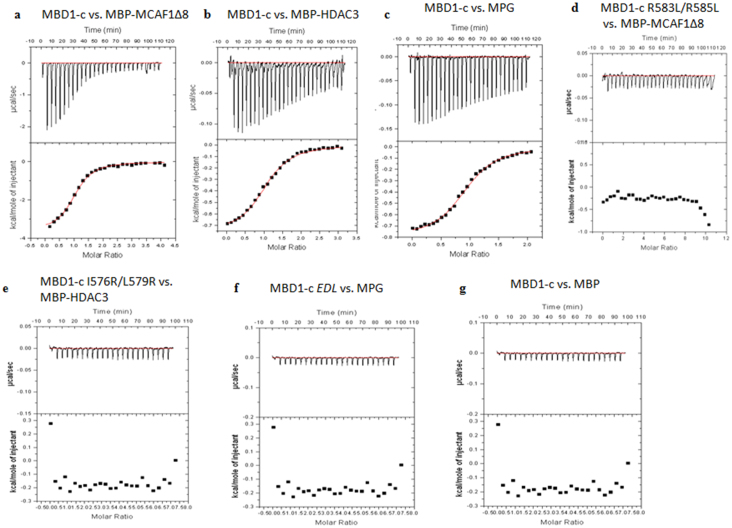
ITC was next performed to determine the thermodynamics of MBD1-c with MBP-MCAF1Δ8, MBP-HDAC3 and MPG. MBD1-c was found to bind to MBP-MCAF1Δ8, MBP-HDAC3 and MPG with Kd value of 6.40, 2.35 and 2.29 μM, respectively with experimental stoichiometry close to 1 (Figure 7A–C and Table 1). In agreement with the results of the pull-down assay, these mutants also did not bind their partners in the ITC experiments (Figure 7D–F). To exclude the possibility of the MBP-tag binding to MBD1-c, standalone MBP control was also titrated against MBD1-c and showed no significant binding (Figure 7G).
Figure 7. Isothermal Titration Calorimetry of MBD1-c and its mutants with MBP-MCAF1Δ8, MBP-HDAC3, MPG and MBP.
(A) MBD1-c titrated against MBP-MCAF1Δ8 until saturation, show single binding site with molar ratio of 1. (B) MBD1-c titrated against MBP-HDAC3 until saturation, show single binding site with molar ratio of 1. (C) MBD1-c titrated against MPG until saturation, show single binding site with molar ratio of 1. (D–F) MBD1-c mutants titrated against MBP-MCAF1Δ8, MBP-HDAC3 and MPG, respectively, did not show any significant binding. (G) MBD1-c titrated against MBP did not show any significant binding.
Table 1. Binding Affinity Measurements with Isothermal Titration Calorimetry. Affinities and thermodynamic parameters of the MBD1-c interaction with MBP-MCAF1Δ8, MBP-HDAC3 and MPG at 298 K.
| Protein | Binding Partner | N | Ka (×106 M−1) | Kd (μM) | ΔH (kcal/mol) | −TΔS (kcal/mol) | ΔG (kcal/mol) | c | ΔCp (kcal/mol/K) |
|---|---|---|---|---|---|---|---|---|---|
| MBD1-c | MBP-MCAF1Δ8 | 1.06 ± 0.15 | 0.16 ± 0.03 | 6.4 ± 1.22 | −3.92 ± 1.2 | −3.14 ± 1.37 | −7.06 ± 1.82 | 8.48 ± 1.2 | −0.28 ± 0.08 |
| MBD1-c | MBP-HDAC3 | 0.98 ± 0.06 | 0.43 ± 0.07 | 2.35 ± 0.37 | −1.75 ± 0.78 | −5.85 ± 0.77 | −7.61 ± 1.096 | 21.22 ± 3.7 | −0.13 ± 0.05 |
| MBD1-c | MPG | 1.1 ± 0.11 | 0.45 ± 0.09 | 2.29 ± 0.49 | −0.87 ± 0.09 | −6.37 ± 0.75 | −7.24 ± 0.76 | 24.76 ± 6.19 | −0.05 ± 0.02 |
In addition, the isobaric heat capacity, ΔCp values obtained for MBD1-c binding with MBP-MCAF1Δ8, MBP-HDAC3 and MPG were −0.28, −0.13 and −0.05 kcal/mol/K, respectively, by measuring the enthalpy of binding (ΔH) at the temperatures: 288, 298 and 308 K (Supplementary Table 2). The decreasing negative ΔCp values suggest that the amount of MBD1-c buried surface in protein-protein complexes decreases from MBP-MCAF1Δ8, to MBP-HDAC3, to MPG, consistent with the number of residues affected in the NMR titration experiments (Figure S3). In summary, the combined NMR spectroscopy, pull-down assay, and ITC experiments, indicate that the interactions between MBD1-c and its protein partners are highly MBD1-TRD residue-specific.
Discussion
We have demonstrated that the functional MBD1-c is intrinsically disordered, similar to the previously reported Transactivation Domain (TAD) of Myc and the regulatory region of CFTR29,30. We also describe how the intrinsically disordered MBD1-c interacts through its TRD with different binding partners.
MBD1 is an essential for the effective gene silencing and, consequently, plays a key role in cell division and differentiation and other fundamental cell functions31. Not surprisingly, its over expression is often linked to carcinogenesis due to its recruitment of transcriptional repressor complexes and the ensuing silencing of tumor suppressor genes15,32. It is now well recognized that MBD1 requires the TRD to interact with various transcriptional repressor proteins in either a histone deacetylase-dependent or -independent manner. These repressor proteins include HDAC3 in the N-CoR complex and the PML-RARα oncoprotein in acute promyelocytic leukemia22 as well as proteins from the MCAF family19,20. MBD1 also recruits MPG to enhance heterochromatin formation23. Previous studies have demonstrated that MBD1 lacking the TRD (MBD1ΔTRD) is inefficient in transcriptional gene silencing and, conversely, the TRD by itself is effective enough in suppressing gene expression14,28. Thus, the TRD is a key component of MBD1 and its molecular structure and how it can recognize different binding partners are of substantial significance.
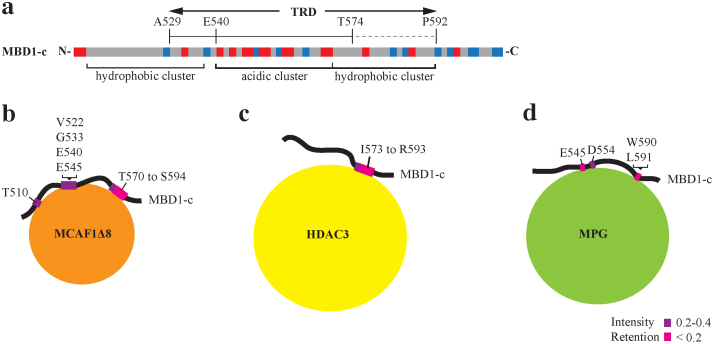
The 64-residue TRD of MBD1 is composed of hydrophobic and charged residues (Figure 8A). We have used NMR spectroscopy to elucidate the solution structure of MBD1-c and subsequently explored in detail its interactions with MBP-MCAF1Δ8, MBP-HDAC3 and MPG. Upon titration of MBD1-c with MBP-MCAF1Δ8, MBP-HDAC3 and MPG, there is no significant peak resonance shift to suggest even the minimal structural changes. Subsequently, we have identified a 21-residue region, T574 to S594, as a common binding site with peak resonances showing significant intensity losses in the presence of MBP-MCAF1Δ8 or MBP-HDAC3. This 21-residue region is almost completely located within the TRD and is enriched in hydrophobic residues with a few highly conserved acidic (E575 and D586) and basic (R583 and R585) residues.
Figure 8. Biological functionalities of MBD1-c.
(A) The electrostatic architecture of MBD1-c is shown as hydrophobic or acidic clusters. Identified interacting residues, based on NMR and ITC studies, are mapped onto MBD1-c while interacting with (B) MCAF1Δ8, (C) HDAC3 and (D) MPG. Residues with peak resonances which disappeared and strongly disappeared are colored magenta and pink, respectively. The greater the disappearance, the lower is the intensity retention value.
Ichimura and co-workers have previously shown that point mutations of two hydrophobic residues I576 and L579 within the TRD domain to basic Arg residues abolished MBD1 binding to MCAF20. We found that the point mutations of two previously non-characterized basic residues to hydrophobic residues, R583L and R585L, did not enhance MBD1-c binding to MCAF1Δ8, MBP-HDAC3 and MPG. On the contrary, we observed significant loss of binding to MCAF1Δ8 and MPG. The loss in binding is likely due to key residue loss and not structural changes as the 2D 1H-15N HSQC spectrum of the R583L/R585L double mutant is largely similar to the native MBD1-c. The key difference is the appearance of two new peak resonances in the place of the mutated R583L and R585L, and peak shifts of residues I576 and L579 which were only minimally perturbed due to local chemical changes near the point mutations (Supplemental Figure S1). Additionally, both R583 and R585 are conserved in all but one (v6) isoforms, suggesting unique recognition role of these residues within the primary TRD domain of MBD1. The need for charge-charge interactions of the TRD with the protein partners may be vital to possibly compensate for the lack of a conformational fit. It is perhaps this lack of globular fold which may enable the disordered TRD domain to act through its specific residues as a more universal recognition domain to bind a wider array of proteins fueled by specific residues (Figure 8B–D). Notably, the TRD-protein interactions are weak and no binding-induced conformational changes could be detected in MBD1-c. At the same time, the presence of a large MBD1-c-MCAF/HDAC3/MPG complex was further confirmed by GST pull-down assay.
NMR titration and ITC analysis also highlight that MBD1-c interacts with MBP-MCAF1Δ8, MBP-HDAC3 and MPG in a different manner with distinct binding affinities and the amount of the MBD1 hydrophobic region buried being higher in the interactions with MBP-MCAF1Δ8 than with MBP-HDAC3 or MPG. The TRD in MBD1-c is composed of three clusters arranged in the hydrophobic, acidic and hydrophobic order. NMR titrations show that MBP-MCAF1Δ8, MBP-HDAC3 and MPG, interact with a small but common hydrophobic cluster, described as the 21-residue region. Based on the NMR titrations, we have designed mutants and showed specific loss of binding to the selected protein partners in both pull-down assays and ITC experiments. As we have shown in this study, MCAF1Δ8 interaction with MBD1-c involves both the N-terminal residues predicted to be a sumoylation motif (SUMOsp 2.0) and the C-terminal stretch of 21 amino acids. A previous study has reported that sumoylation, mediated by the PIAS proteins, impairs the binding of SETDB1 complex to MBD133 and MCAF1 is known to be part of the SETDB1 complex20. Thus MCAF1, in the absence of MBD1 sumoylation, appears to directly interact with the MDB1-c region. With regard to the MBD1 KEE mutant, MCAF1Δ8 partially lost interaction with the mutant in the pull-down assay. Similarly, in the pull-down assay, the MBD1-c I576R/L579R mutant showed loss of interaction with both MCAF1Δ8 and MBP- HDAC3 but not with MPG, whereas the R583L/R585L mutant showed loss of interaction with MCAF1Δ8 but not with MBP-HDAC3 and MPG. Interestingly, MPG showed complete loss of interaction only with the EDL mutant, which retained binding to both MCAF1Δ8 and MBP-HDAC3. Thus, our results suggest that MBD1 interacts differently, yet highly specifically with MCAF1Δ8, MBP-HDAC3 and MPG and likely, the disordered nature of the TRD playing the critical role in the process.
Previous studies on p53 have shown that most of its regions remain unstructured and become folded once it binds to its interacting proteins34. However, MBD1-c remains unstructured even after interacting with its binding partners and this feature might be a unique prerequisite to be able to bind partner proteins without a definite tertiary fold. Apart from the known hydrophobic residues, the previously uncharacterized charge-charge interactions may also compensate for the lack of a tertiary fold that is required for specific interactions with different binding partners. This unique mode of recognition through an amphiphatic combinations of hydrophobic and charge-charge interactions will enable MBD1 to sample several proteins for regulating a cobweb of cell functions, primarily through the intrinsically disordered TRD domain.
Because of the oncogenic role of MBD122,25,35,36, development of small molecule inhibitors of MBD1, facilitated by the understanding of its structure and interactions, should have important therapeutic implications for cancer and, likely, other diseases. In principle, such inhibitors could be highly specific when compared to the currently available DNA methyl transferases (DNMTs) inhibitors37,38 by targeting the minimal region of interaction of the TRD with its partner protein(s).
Methods
Protein expression and purification
The gene encoding the C-terminus of MBD1 (MBD1-c, residues D507-Q605) was cloned into the pET-M vector [modified pET-32a (+) vector, Novagen] with an N-terminal His6-tag and transformed into BL21 (DE3) cells. The transformed cells were incubated at 37°C until OD600 of 0.6 and induced with 0.3 mM IPTG at 20°C for 16 h. Harvested cells were resuspended in lysis buffer: 50 mM Tris–HCl (pH 7.0), 200 mM NaCl, 5% glycerol and 1 mM DTT, sonicated on ice for 10 mins and centrifuged at 25,000 g for 30 min. The supernatant was allowed to bind to Ni-NTA beads for an hour at 4°C and was subsequently washed twice with wash buffer: 50 mM Tris–HCl (pH 7.0), 200 mM NaCl, 5% glycerol, 1 mM DTT and 5 mM imidazole. Finally the protein was eluted with 250 mM of imidazole in the lysis buffer. The eluted protein was loaded onto a Superdex75 size exclusion column (GE Healthcare) equilibrated with buffer containing 20 mM sodium phopshate (pH 6.5), 50 mM NaCl and 3 mM DTT. The eluted protein was concentrated to 10 mg/ml. A similar protocol was adopted to purify the 15N or 13C/15N labeled MBD1-c using M9 medium39. For 15N and 13C/15N labeled media, the 15N ammonium chloride and 13C glucose were purchased from Cambridge Isotopes Laboratories. Similar protein preparations were used for MBP-MCAF1Δ8, MBP-HDAC3 and MPG, and provided in supplementary data.
NMR spectroscopy
All NMR data were acquired at 298 K on an 800 MHz NMR spectrometer (Bruker Avance) equipped with a TXI cryogenic probe. All NMR samples, MBD1-c, MPG, MBP-MCAF1Δ8, MBP-HDAC3 and MBP (control), were prepared in 20 mM sodium phosphate buffer (pH 6.5), 50 mM NaCl, 3 mM DTT, 0.1 mM NaN3 and 5% 2H2O. For structure determination, 1.0 mM 13C and 15N-labelled MBD1-c was used to collect the following NMR spectra: 2D 1H-15N HSQC, 2D 1H-13C HSQC, 3D HN(CO)CA, 3D HNCA, 3D MQ-(H)CCH TOCSY and 4D timeshared 13C/15N edited-NOESY. NOE distance restraints were obtained from the timeshared 4D 13C/15N-edited NOESY (containing three 13C, 15N-edited, 13C, 13C-edited and 15N, 15N-edited sub-spectra) using NMRspy40 and an extension XYZ4D (yangdw.science.nus.edu.sg/Software&Scripts/XYZ4D/index.htm). The sequence-specific assignments have been deposited under BMRB accession number 19171.
NMR titration
For titration experiments using 1H-15N HSQC-TROSY, an initial concentrated unlabelled MBP-MCAF1Δ8/MBP-HDAC3/MBP/MPG was minimally added to 0.2 mM 15N-labelled MBD1-c (cleaved His6-tag) until a final protein: MBD1-c molar ratio of 3:1. The combined 1H and 15N chemical shift difference, before and after titration was calculated with equation 1,
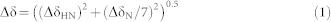 |
where ΔδHN and ΔδN are the respective differences of 1H and 15N chemical shifts of a residue in the free and bound MBD1-c. For intensity analysis, the intensity of each residue from the 2D 1H-15N HSQC spectra was extracted using NMRspy. The intensity retention is based on the intensity ratio of the bound and the free MBD1-c, and corrected with a scaling factor to account for dilution.
Isothermal titration calorimetry (ITC)
The interactions of MBD1-c with MBP-MCAF1Δ8, MBP-HDAC3 and MPG, were verified using a VP-ITC microcalorimeter (MicroCal) at 25°C in 20 mM sodium phosphate buffer (pH 6.5), 50 mM NaCl and 3 mM DTT. The cell contained 50 μM of MBP-MCAF1Δ8, MBP-HDAC3, MPG and MBP as a control for each titration, separately. The above experiments were also carried out for MBD1-c mutants in the same condition. The syringe containing 500 μM of MBD1-c, was prepared in the same buffer. Samples were first degassed for 15 min and titration was performed using a stirring speed of 500 rpm. The initial injection for MBD1-c was 2 μl for 10 s and subsequent injections were 10 μl for 10 s with 240 s spacing. The amount of heat in μJ is plotted against the injection number to gives us the raw data and shown as peaks corresponding to each injection. Thus obtained raw data peaks were converted using the Origin software to make a plot of enthalpy change per mole of injectant (ΔH0, kJ mol−1) against molar ratio. To obtain exact thermodynamic parameters, this depends on the use of nonlinear least square fitting and also using appropriate model to explain the interaction under study. The simplest model which explain the independent single site binding where ligand to macromolecule ratio 1:1 is
 |
The objective of fitting data is to obtain the value of important parameters which gives better explanation of the data. The fitting procedure is done using single site model based on Wisemen isotherm41,42,43
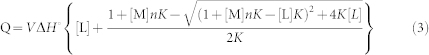 |
where V is the volume of the calorimeter cell, ΔH0 is enthalpy, [L] is ligand concentration, [M] is macromolecule concentration, n is the molar ratio of interacting species, and K is the equilibrium binding constant.
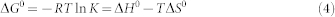 |
To calculate the isobaric heat capacity, ΔCp, the experiment is repeated at three different temperatures such as 288, 298 and 308 K and then ΔH values are plotted against the temperature, and the slope of the plot gives the ΔCp value given that
 |
GST pull-down
Recombinant GST fusion proteins were expressed in E. coli strain BL21 (DE3) and purified with the use of glutathione agarose beads (GE Healthcare). His6-tag MCAF1Δ8, MBP-HDAC3 and MPG proteins were expressed as recombinant proteins. GST and GST-tag MBD1-c were loaded onto glutathione agarose beads for 1 hour at 4°C in lysis buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, and 5% glycerol) containing 1 mM DTT. The beads were washed with the lysis buffer and incubated overnight at 4°C with recombinantly expressed proteins. The beads were isolated and washed three times, before the addition of SDS-PAGE sample buffer. The binding partners are observed using Ant-His antibodies from Santacruz.
Circular dichroism
Circular dichroism (CD) spectra were recorded on a Jasco J-810 spectropolarimeter equipped with a thermal controller. A final concentration of 30 μM of MBD1-c protein was prepared in in 20 mM sodium phosphate buffer (pH 6.5) at 25°C.
Author Contributions
U.F.S.H., Q.Z., M.W. and K.S. designed the project. U.F.S.H. and L.J.W. performed the experiments, Figures 1, 2,3,4,5 and 8 prepared by L.J.W. and the Figures 6, 7 prepared by U.F.S.H. Manuscript written and edited by U.F.S.H., L.J.W., M.W., Y.D. and K.S. All authors reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We appreciate the support of Profs. Rabindra Roy from Georgetown University, USA and Luciano DiCroce from Center for Genomic Regulation, Spain for providing the MPG and HDAC3 expression constructs, respectively. The authors thank the BioScience core Lab of King Abdullah University of Science and Technology (KAUST), Saudi Arabia for their support in performing ITC experiments. This project was supported by Academic Research Fund (FRC) of the Ministry of Education, Singapore to K.S. and National University of Singapore research scholarship to U.F.S.H.
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 16, 6–21, 10.1101/gad.947102 (2002). [DOI] [PubMed] [Google Scholar]
- Rottach A., Leonhardt H. & Spada F. DNA methylation-mediated epigenetic control. J Cell Biochem 108, 43–51, 10.1002/jcb.22253 (2009). [DOI] [PubMed] [Google Scholar]
- Chen R. Z., Pettersson U., Beard C., Jackson-Grusby L. & Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature 395, 89–93, 10.1038/nature.25779 (1998). [DOI] [PubMed] [Google Scholar]
- Illingworth R. et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 6, e22, 10.1371/journal.pbio.0060022 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello J. F. et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet 24, 132–138, 10.1038/nat.gen.72785 (2000). [DOI] [PubMed] [Google Scholar]
- Maunakea A. K. et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257, 10.1038/nature.09165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D. et al. Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proc Natl Acad Sci U S A 108, 4364–4369, 10.1073/pnas.1013224108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoph F. et al. A gene expression profile of tumor suppressor genes commonly methylated in bladder cancer. J Cancer Res Clin Oncol 133, 343–349, 10.1007/s00432-006-0174-9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M., Corn P. G., Baylin S. B. & Herman J. G. A gene hypermethylation profile of human cancer. Cancer Res 61, 3225–3229 (2001). [PubMed] [Google Scholar]
- Ballestar E. & Wolffe A. P. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem 268, 1–6, ejb1869 (2001). [DOI] [PubMed] [Google Scholar]
- Clouaire T., de Las Heras J. I., Merusi C. & Stancheva I. Recruitment of MBD1 to target genes requires sequence-specific interaction of the MBD domain with methylated DNA. Nucleic Acids Res 38, 4620–4634, 10.1093/nar/gkq228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Ma W., Solov'yov I. A., Chipot C. & Schulten K. Recognition of methylated DNA through methyl-CpG binding domain proteins. Nucleic Acids Res 40, 2747–2758, 10.1093/nar/gkr1057 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N. et al. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol 19, 6415–6426 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Jeppesen P. & Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol 20, 1394–1406 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y. et al. Overlapping roles of the methylated DNA-binding protein MBD1 and polycomb group proteins in transcriptional repression of HOXA genes and heterochromatin foci formation. J Biol Chem 282, 16391–16400, 10.1074/jbc.M700011200 (2007). [DOI] [PubMed] [Google Scholar]
- Ohki I., Shimotake N., Fujita N., Nakao M. & Shirakawa M. Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1. EMBO J 18, 6653–6661, 10.1093/emboj/18.23.6653 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki I. et al. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell 105, 487–497, S0092-8674(01)00324-5 (2001). [DOI] [PubMed] [Google Scholar]
- Cierpicki T. et al. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol 17, 62–68, 10.1038/nsmb.1714 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N. et al. MCAF mediates MBD1-dependent transcriptional repression. Mol Cell Biol 23, 2834–2843 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T. et al. Transcriptional repression and heterochromatin formation by MBD1 and MCAF/AM family proteins. J Biol Chem 280, 13928–13935, 10.1074/jbc.M413654200 (2005). [DOI] [PubMed] [Google Scholar]
- Sarraf S. A. & Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell 15, 595–605, 10.1016/j.molcel.2004.06.043 (2004). [DOI] [PubMed] [Google Scholar]
- Villa R. et al. The methyl-CpG binding protein MBD1 is required for PML-RARalpha function. Proc Natl Acad Sci U S A 103, 1400–1405, 10.1073/pnas.0509343103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S. et al. Methylated DNA-binding domain 1 and methylpurine-DNA glycosylase link transcriptional repression and DNA repair in chromatin. Proc Natl Acad Sci U S A 100, 12859–12864, 10.1073/pnas.2131819100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. L. et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19, 187–191, 10.1038/561 (1998). [DOI] [PubMed] [Google Scholar]
- Patra S. K., Patra A., Zhao H., Carroll P. & Dahiya R. Methyl-CpG-DNA binding proteins in human prostate cancer: expression of CXXC sequence containing MBD1 and repression of MBD2 and MeCP2. Biochem Biophys Res Commun 302, 759–766, S0006291X03002535 (2003). [DOI] [PubMed] [Google Scholar]
- Moréra S. et al. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res 40, 9917–9926, 10.1093/nar/gks714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manvilla B. A., Maiti A., Begley M. C., Toth E. A. & Drohat A. C. Crystal structure of human methyl-binding domain IV glycosylase bound to abasic DNA. J Mol Biol 420, 164–175, 10.1016/j.jmb.2012.04.028 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N. et al. Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1. Mol Cell Biol 20, 5107–5118 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen C. et al. Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding. Nucleic Acids Res 40, 6353–6366, 10.1093/nar/gks263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J. M. et al. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol 14, 738–745, 10.1038/nsmb1278 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 6, 433–444, 10.1016/j.stem.2010.02.017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao M. et al. Regulation of transcription and chromatin by methyl-CpG binding protein MBD1. Brain Dev 23 Suppl 1, S174–176, S0387760401003485 (2001). [DOI] [PubMed] [Google Scholar]
- Lyst M. J., Nan X. & Stancheva I. Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. EMBO J 25, 5317–5328, 10.1038/sj.emboj.7601404 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bell S., Klein C., Müller L., Hansen S. & Buchner J. p53 contains large unstructured regions in its native state. J Mol Biol 322, 917–927 (2002). [DOI] [PubMed] [Google Scholar]
- Luo G. et al. RNA interference of MBD1 in BxPC-3 human pancreatic cancer cells delivered by PLGA-poloxamer nanoparticles. Cancer Biol Ther 8, 594–598 (2009). [DOI] [PubMed] [Google Scholar]
- Jang J. S. et al. Methyl-CpG binding domain 1 gene polymorphisms and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev 14, 2474–2480, 10.1158/1055-9965.EPI-05-0423 (2005). [DOI] [PubMed] [Google Scholar]
- Foulks J. M. et al. Epigenetic drug discovery: targeting DNA methyltransferases. J Biomol Screen 17, 2–17, 10.1177/1087057111421212 (2012). [DOI] [PubMed] [Google Scholar]
- Lyko F. & Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst 97, 1498–1506, 10.1093/jnci/dji311 (2005). [DOI] [PubMed] [Google Scholar]
- Marley J., Lu M. & Bracken C. A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR 20, 71–75 (2001). [DOI] [PubMed] [Google Scholar]
- Xu Y., Zheng Y., Fan J. S. & Yang D. A new strategy for structure determination of large proteins in solution without deuteration. Nat Methods 3, 931–937, 10.1038/nmeth938 (2006). [DOI] [PubMed] [Google Scholar]
- Brown A. Analysis of cooperativity by isothermal titration calorimetry. Int J Mol Sci 10, 3457–3477, 10.3390/ijms10083457 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman T., Williston S., Brandts J. F. & Lin L. N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem 179, 131–137 (1989). [DOI] [PubMed] [Google Scholar]
- Li X., Wang G., Chen D. & Lu Y. Binding of ascorbic acid and α-tocopherol to bovine serum albumin: a comparative study. Mol Biosyst 10, 326–337, 10.1039/c3mb70373h (2014). [DOI] [PubMed] [Google Scholar]
- Wishart D. S., Bigam C. G., Holm A., Hodges R. S. & Sykes B. D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR 5, 67–81 (1995). [DOI] [PubMed] [Google Scholar]
- Schwarzinger S. et al. Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc 123, 2970–2978, ja003760i (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information