Abstract
Advances in adeno-associated virus (AAV)-mediated gene therapy have brought the possibility of commercial manufacturing of AAV vectors one step closer. To realize this prospect, a parallel effort with the goal of ever-increasing sophistication for AAV vector production technology and supporting assays will be required. Among the important release assays for a clinical gene therapy product, those monitoring potentially hazardous contaminants are most critical for patient safety. A prominent contaminant in many AAV vector preparations is vector particles lacking a genome, which can substantially increase the dose of AAV capsid proteins and lead to possible unwanted immunological consequences. Current methods to determine empty particle content suffer from inconsistency, are adversely affected by contaminants, or are not applicable to all serotypes. Here we describe the development of an ion-exchange chromatography-based assay that permits the rapid separation and relative quantification of AAV8 empty and full vector particles through the application of shallow gradients and a strong anion-exchange monolith chromatography medium.
Lock and colleagues describe the development of an ion-exchange (IEX) chromatography-based assay that permits the rapid separation and relative quantification of AAV8 empty and full vector particles. Unlike previously published particle assays, the IEX-based method described here requires little in the way of sample preparation or special reagents and eliminates interpretative counting and compounded errors.
Introduction
Adeno-associated virus (AAV)-based gene therapy vectors have long been considered to have high potential for correcting single-gene defects for a number of human diseases. Although most early work in this regard was based on the AAV2 serotype, many other serotypes have been discovered and shown to confer considerable advantages over AAV2 in terms of gene delivery to various tissues. Success in the clinic with AAV2 gene therapy vectors for the treatment of Parkinson's disease and the hereditary eye defect, Leber's congenital amaurosis (Kaplitt et al., 2007; Bainbridge et al., 2008; Eberling et al., 2008; Hauswirth et al., 2008; Maguire et al., 2008, 2009; Christine et al., 2009; Simonelli et al., 2010), has led to an upsurge of interest and high expectations for increased benefits using other AAV serotypes in the treatment of a variety of diseases. Such a positive outlook raises the probability that commercial AAV vector manufacturing will become a reality and, while considerable progress in the large-scale manufacture of AAV vectors has been described (Clement et al., 2009; Smith et al., 2009; Thorne et al., 2009; Virag et al., 2009; Zhang et al., 2009; Wright, 2011a,b), certain aspects of the manufacturing platforms are in need of improvement. One such area is the development of AAV-specific assays for the monitoring of production processes and the release of final vector product, to a level of sophistication compatible with the rigorous demands of current good manufacturing practices.
AAV contains a single-stranded DNA genome that is packaged into a self-assembled, viral particle through the action of the AAV replicase (Rep) protein (Timpe et al., 2005). During AAV vector manufacturing this process is often inefficient, leading to the presence in the preparation of “empty” particles that lack the vector genome (Grimm et al., 1999). The degree of contamination of AAV vector preparations with empty particles varies depending on the manufacturing process used, but can be as high as a 20- to 30-fold excess over full particles for transfection-based procedures (Lock et al., 2010b). The presence of empty particles effectively increases the dose of the AAV capsid proteins given during therapy and therefore increases the potential for unwanted immune consequences against the vector capsid. One example of such a reaction was observed during a clinical trial for the treatment of hemophilia with an AAV2 vector bearing the human factor IX gene (Mingozzi et al., 2007). An asymptomatic and reversible transaminitis was observed during this trial with the concomitant generation of T cells against the AAV2 capsid, which presumably led to the observed deletion of AAV2-transduced liver cells. The removal of empty capsids from vector preparations has since become a priority and purification methods such as cesium chloride and iodixanol gradients have been used for this purpose (Ayuso et al., 2010; Lock et al., 2010a). Although effective, such methods are not suitable to cope with the vector production demands of late-stage clinical trials and commercial manufacturing and more scalable processes are required. Scalable methods of empty particle removal have been described whereby the separation of empty and full AAV vector particles of serotypes 1, 2, and 6 was achieved on the basis of charge, using ion-exchange chromatography (Urabe et al., 2006; Qu et al., 2007; Okada et al., 2009). This type of approach is compatible with large-scale AAV vector production and if sufficiently robust, is likely to supplant gradient-based separation technologies in commercial manufacturing.
Equally important as methods to remove empty particles are assays to detect their presence, both during production and in the final released product. Perhaps the most widely used particle assay is the electron microscope (EM) assay wherein vector particles are negatively stained, directly visualized at high magnification, and counted. Empty particles are distinguished on these images by an electron-dense central region of the capsid in comparison with full particles, which exclude the negative stain. This technique is highly dependent on the procedure used and also on the purity of the preparation. In the wrong hands, images can be produced where it is difficult to distinguish empty and full particles and this situation is greatly exacerbated if contaminating protein or other material is present. Even in the sharpest images, some particles appear only partially stained and this creates ambiguity in characterizing them as empty or full. In general, therefore, quantification of electron micrographs can be problematic and lead to inconsistent results (Allay et al., 2011). A second approach to empty particle determination is to combine a total particle assay (usually an ELISA) with a genome copy (GC) titration (qPCR) and to determine empty particle content by subtracting the GC titer from the particle titer (Grimm et al., 1999). The problems with this type of approach include the compounded error of the two assays, the additional time and labor involved in performing two separate assays, and the lack of readily available reagents to perform the particle titration for certain AAV serotypes including AAV8. Last, a 260 nm/280 nm absorbance-based assay has been published for AAV2, in which extinction coefficients were determined in order to calculate total genome, total particle, and empty particle content (Sommer et al., 2003). Although this assay can be useful for highly pure preparations of AAV2, a major drawback is that protein contaminants in the vector preparation or the presence of excipients that absorb in the 260 nm/280 nm range can lead to artificial increases in the determined titers. An additional limitation with this method at present is that extinction coefficients for serotypes other than AAV2 have yet to be published.
In the current study, we present a new approach to AAV serotype 8 particle analysis, based on the chromatographic separation of empty and full particles by charge. A prominent feature of the chromatography medium used in this study is the ability to produce high resolution at fast flow rates, which allows for a simple, rapid, and reproducible assay.
Materials and Methods
Preparation of CsCl gradient-purified vector
CsCl gradient-purified vector was purified as previously described (Xiao et al., 1999). Forty 15-cm plates of HEK293 cells were transfected by the calcium phosphate method with vector genome, rep/cap, and adenovirus helper plasmids. For empty particle preparations, the vector genome plasmid was omitted from the transfection cocktail. Cell lysates were prepared at 72 hr posttransfection by three successive freeze–thaw cycles and were purified by two rounds of cesium chloride centrifugation. Gradient fractions were measured by refractometry and those with refractive indices between 1.3660 and 1.3740 were pooled as full vector, whereas those with refractive indices between 1.3740 and 1.3800 were taken as empty vector. All fractions were concentrated and desalted in phosphate-buffered saline (PBS)–35 mM NaCl, using Ultra 15 centrifugal concentrator devices (Amicon; Millipore, Bedford, MA). Glycerol was added to 5% before aliquoting and freezing.
Preparation of iodixanol gradient-purified vector
Large-scale vector preparations were made as described by Lock et al. (2010a). Polyethylenimine (PEI)-based transfections were performed in 10-layer cell stacks containing 75% confluent monolayers of HEK293 cells. Ten liters of feedstock culture medium from the cell stacks was clarified and then concentrated by tangential flow filtration. The concentrated clarified feedstock was purified over iodixanol (Optiprep; Sigma-Aldrich, St. Louis, MO) gradients. All fractions directly below a visible contaminating protein band were collected and pooled. Pooled fractions were combined, diafiltered against 10 volumes of PBS–35 mM NaCl, and then concentrated 4-fold to ∼10 ml by tangential flow filtration. Glycerol was added to the diafiltered, concentrated product to 5% final and the preparation was aliquoted and stored at −80°C.
Vector genome titration (qPCR)
DNase I-resistant vector genomes present in purified vector preparations and fast protein liquid chromatography (FPLC) fractions were titered by TaqMan PCR amplification (Applied Biosystems, Foster City, CA), using primers and probes directed against the polyadenylation signal encoded in the transgene cassette.
Vector purity
The purity of gradient fractions and final vector lots was evaluated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and proteins were visualized with SYPRO ruby stain (Invitrogen, Carlsbad, CA) and ultraviolet excitation. Purity relative to nonvector impurities visible on stained gels was determined with GeneTools software (Syngene, Frederick, MD).
Electron microscopy
Particle content of vector preparations was assessed by negative staining and electron microscopy. Copper grids (400-mesh coated with a formvar–thin carbon film; Electron Microscopy Sciences, Hatfield, PA) were pretreated with 1% Alcian blue (Electron Microscopy Sciences) and loaded with 5 μl of vector preparation. The grids were then washed, stained with 1% uranyl acetate (Electron Microscopy Sciences), and viewed with a Philips CM100 transmission electron microscope. Empty-to-full particle ratios were determined by direct counting of the electron micrographs.
Ion-exchange particle assay
All chromatography studies were performed with an ÄKTAFPLC system (GE Healthcare Life Sciences, Piscataway, NJ) fitted with a 10-ml Superloop, Unicorn software, and a 0.34-ml CIM-QA monolithic disk (Bia Separations, Ajdovscina, Slovenia). An amount equivalent to 1×1012 to 5×1012 genome copies (GC) of full AAV8 vector preparations (lot nos. V0083 and CS0010) and/or 75 μl of an empty AAV8 vector preparation (lot no. Z4033) were brought to 10 ml in 20 mM Bis-Tris propane (BTP, pH 9.0)–50 mM NaCl and injected into the Superloop. The sample was loaded to the CIM-QA disk at 3 ml/min and washed through with 10 column volumes of 20 mM BTP (pH 9.0)–50 mM NaCl. In the initial binding studies, elution was performed over 20 column volumes with a 50–150 mM NaCl gradient in 20 mM BTP (pH 9.0), but thereafter was achieved with an 80–115 mM NaCl gradient. All elutions were followed by a 10 column volume high-salt wash (1 M NaCl) in the 20 mM BTP (pH 9.0) buffer. One-milliliter fractions were collected throughout the runs and were analyzed for vector genome copies by qPCR. Elution peaks were integrated, using the Unicorn software evaluation module (GE Healthcare Life Sciences), and the resulting peak areas were used to determine particle ratios. Baselines were calculated according to a morphological algorithm, the default algorithm for the Unicorn software. This algorithm can be described as a line that follows the chromatogram parallel to the x axis. Data points for the baseline are created whenever the line touches the curve, and the points are joined at the end to create the baseline. The algorithm searches for all parts of the source curve where the curve parts come into contact at both ends of a horizontal line of a defined length and the center point of the line defines the data point.
Results
Differential elution of empty and full AAV8 vector preparations from CIMQA monolith media using shallow gradients
Conditions to bind and elute purified AAV8 preparations to and from various quaternary amine (QA) strong anion exchangers were initially determined by FPLC scouting runs (data not shown). The consensus of these experiments was that relatively high pH (pH 9.0) and low ionic strength (50 mM NaCl) facilitated binding while a shallow gradient (50–150 mM NaCl over 20 column volumes) was optimal for elution. Monolith-based convective interaction chromatography media (CIM; Bia Separations) have been specifically designed to separate large biomolecules such as viruses at high flow rates while retaining a high degree of resolution. The strong anion-exchange version of this type of medium (CIM-QA) was therefore a logical choice to attempt the robust separation of empty and full AAV8 particles at an analytical scale.
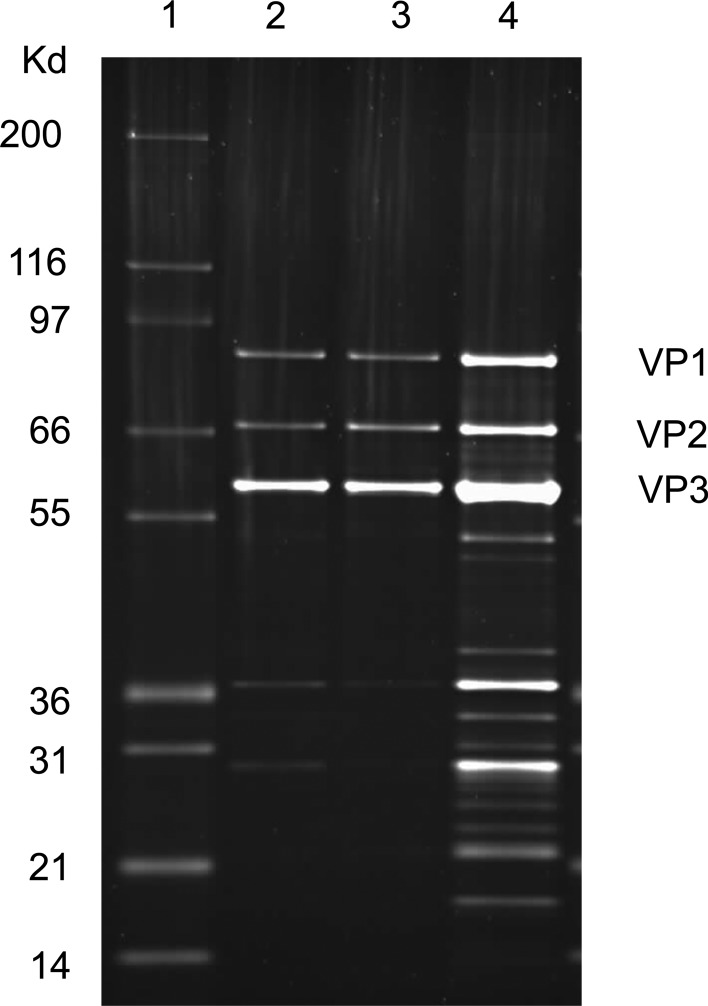
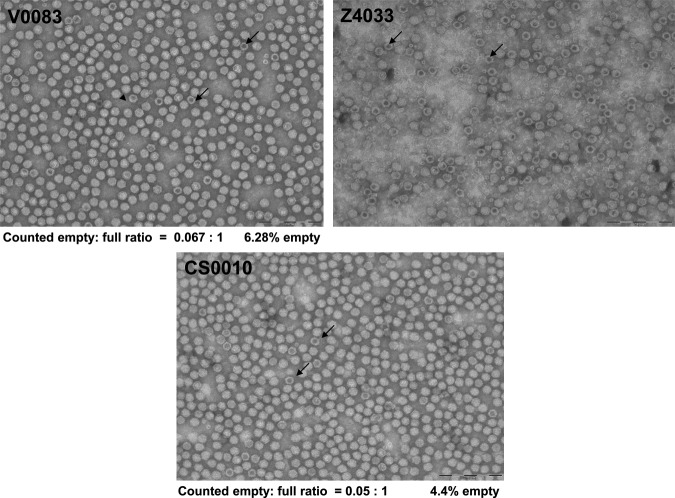
For the initial AAV8 vector binding experiments using CIM-QA, two cesium chloride gradient-purified preparations were employed. The first preparation (lot no. V0083) was produced by a standard calcium phosphate-based triple transfection procedure and was therefore considered a “full” vector preparation with the majority of particles expected to contain the vector genome. The second preparation (lot no. Z4033) was produced in a similar way except that the vector genome (cis) plasmid was omitted, and this lot was expected to contain mostly empty particles. A high degree of purity is required for FPLC analysis such that contaminant proteins do not overly complicate the chromatograms and significantly contribute to vector peak volumes. SDS–PAGE analysis of the two vector preparations showed that each preparation was relatively pure although the empty vector (Z4033) did show some minor low molecular weight contaminant bands (Fig. 1). To confirm the empty/full nature of the particles, the preparations were negatively stained and analyzed by electron microscopy (Fig. 2). By this analysis, lot no. V0083 was estimated to contain 93.7% full particles. The electron micrograph of lot no. Z4033, on the other hand, was less easily interpreted; a majority of the particles appeared to be empty, as would be expected considering the omission of the vector genome plasmid during production; however, some particles appeared to exclude the negative stain and were therefore considered full, and a subset of particles was incompletely stained, making them difficult to characterize. One explanation for the presence of what appeared to be full particles in lot no. Z4033 is that extraneous DNA (e.g., plasmid) was packaged at a low level as has been published previously (Chadeuf et al., 2005; Halbert et al., 2011). A further subset, also present, consisted of empty particles that were considerably smaller than the true empty particles, but had apparently migrated to the same position on isopycnic CsCl gradients; these may represent self-assembly intermediates. The general difficulty encountered in characterizing the particle types in the Z4033 electron micrograph can be a recurrent problem with EM-based determination of empty-to-full particle ratios and is one reason why this method is considered to be inconsistent.
FIG. 1.
Purity of AAV vector stocks. 1.6×1010 GC of AAV8 vector preparations (full: lot nos. V0083 and CS0010) and approximately 2.6×1010 empty particles (lot no. Z4033) were loaded onto an SDS–polyacrylamide gel; proteins were visualized by SYPRO ruby staining. The AAV capsid proteins (VP1, VP2, and VP3) are indicated. Lane 1, molecular weight marker; lane 2, lot no. V0083 (CsCl purified, full); lane 3, lot no. CS0010 (iodixanol purified, full); lane 4, lot no. Z4033 (CsCl, empty).
FIG. 2.
AAV8 vector particle analysis by electron microscopy. rAAV8 vector preparations were negatively stained with uranyl acetate and examined with a transmission electron microscope. The lot number is indicated on each image. Empty particles can be distinguished on the basis of the electron-dense center and are indicated by arrows. Where possible, the ratio of empty to full particles and the percentage of empty particles are shown below the images.
Using the binding/elution conditions previously determined for other strong anion exchangers, 1×1012 GC of the “full” vector (lot no. V0083) was loaded onto a 0.34-ml CIM-QA disk and subsequently eluted as described in Materials and Methods; the chromatographic trace of this experiment is shown in Fig. 3b. A large, prominent peak observed in the elution gradient corresponded with collected fractions containing greater than 99% of the vector genome load; this peak was therefore considered to represent full particles (Fig. 3b). A similar peak was also observed in the elution gradient when the “empty” vector (lot no. Z4033) was run under the same conditions (Fig. 3a). Overlaying the two chromatograms, however (Fig. 3c), demonstrated that the empty vector peak preceded the full vector peak and indicated chromatographic separation of the empty and full particle species. Interestingly, small peaks were seen either preceding the full peak or tailing the empty peak; these minor peaks directly coincided with the relative retention volumes of the major empty and full peaks, respectively, and indicated contamination of each type of particle preparation with low levels of the other particle type. Other minor peaks, especially in the case of the empty vector preparation, were observed during the high-salt wash and most likely represented the low molecular weight contaminant bands visible on SDS–polyacrylamide gels (Fig. 1).
FIG. 3.
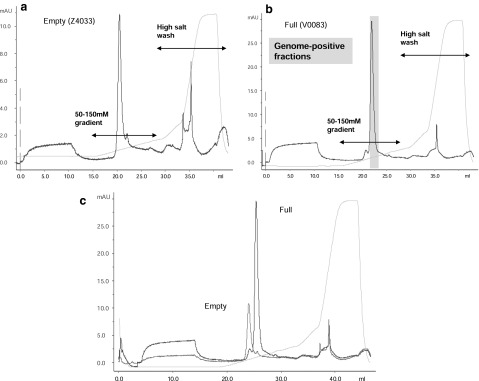
IEX particle assay. (a) Seventy-five microliters of an empty particle AAV8 preparation (lot no. Z4033) was loaded onto a 0.34-ml CIM-QA disk, using FPLC, and eluted with a 50–150 mM salt gradient. The y axis shows the absorbance (mAU) at 280 nm and the x axis the elution volume (ml). The detected conductivity and absorbance are represented by solid light and dark blue lines, respectively. The vertical dashed pink line represents the point of vector injection. (b) A full AAV8 vector preparation (lot no. V0083, 1×1012 GC) was run under the same binding/elution conditions as used for the empty particle preparation. Fractions were quantified for vector GC content and those fractions containing >99% of the loaded material are indicated (shaded box). (c) An overlay of the elution profiles of the empty and full AAV8 vector preparations is shown.
Separation of a mixture of empty and full AAV8 vectors and quantification of relative particle amounts
Having demonstrated differential retention of what were considered to be full and empty AAV8 vector particles on CIM-QA medium, our next questions were, first, whether a mixture of these two types of particle could be readily separated and, second, whether the relative amounts of each could be quantified. On the basis of the retention volumes of the two particle types in our initial runs, an attempt was made to improve resolution of the peaks by further shallowing the elution gradient to 80–115 mM NaCl over 20 column volumes. Lot no. V0083 (full) was run under these conditions and a chromatographic profile similar to the previous run with the steeper elution gradient was observed (Fig. 4a). Despite the employment of a shallower gradient, no dramatic improvement in peak resolution was observed. Nevertheless, the separation was judged sufficient for the purpose of relative quantification and the area under these peaks was determined by integration. An empty (minor peak)-to-full (major peak) ratio of 0.016 was calculated and both this ratio and the calculated empty particle percentage (1.6%) were slightly lower than the corresponding ratio and empty percentage determined by EM analysis (0.067 and 6.28%, respectively).
FIG. 4.
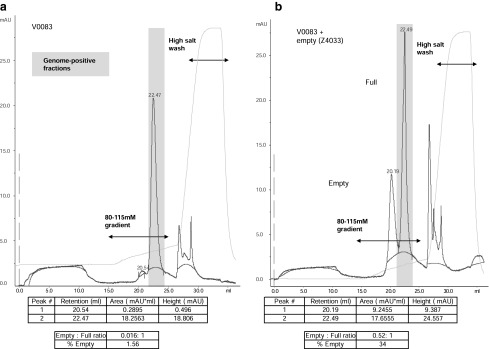
Quantification of relative empty-to-full particle content in a CsCl gradient-purified AAV8 vector preparation. (a) A full AAV8 vector preparation (lot no. V0083, 1×1012 GC) was loaded onto a 0.34-ml CIM-QA disk and eluted with an 80–115 mM salt gradient. The y axis shows the absorbance (mAU) at 280 nm and the x axis the elution volume (ml). The detected conductivity and absorbance are represented by solid light and dark blue lines, respectively. Peak retention volumes are indicated by numbers on the dark blue trace on the chromatograph and in the table (bottom). Peak areas and heights in the table were determined by integration, using the Unicorn evaluation module; baselines calculated according to a morphological algorithm are shown as a red line. The peak areas were used to calculate the ratio of empty to full particles and percentage of empty particles. Fractions were quantified for vector GC content and those fractions containing >99% of the loaded material are indicated (shaded box). (b) A mixture of 75 μl of empty AAV8 particles (lot no. Z4033) and 1×1012 GC of a full AAV8 vector preparation (lot no. V0083) was run under the same conditions and the peaks were evaluated identically.
An arbitrary volume (75 μl) of the “empty” preparation (lot no. Z4033) was then mixed with 1×1012 GC of V0083, bound to CIM-QA, and eluted with the 80–115 mM NaCl gradient (Fig. 4b). Two clearly distinguishable peaks were obtained with retention values that mirrored those obtained when the two lots were run separately on CIM-QA medium. The second peak, which had a retention volume similar to that of the full vector preparation (22.5 ml), contained >99% of the loaded vector genomes as determined by qPCR. This result confirmed the chromatographic elution position of the empty particle peak as preceding the full peak and demonstrated separation of a mixture of empty and full AAV8 vector particles in a case in which the relative amount of empty particles was high (calculated as 34%).
Analysis of AAV8 vector preparations purified from supernatant by an iodixanol gradient
To demonstrate the utility of the new analytical ion-exchange (IEX) particle assay for AAV8 vector produced by alternative processes, a set of experiments was initiated with vector purified from production culture supernatant by tangential flow filtration and iodixanol gradients (Lock et al., 2010a) (Fig. 5). Lot no. CS0010 was produced in this way and was shown to be greater than 95% pure by SDS–PAGE analysis (Fig. 1). An amount equivalent to 5×1012 GC was loaded onto the CIM-QA disk and eluted with the same gradient previously used for the CsCl gradient-purified material (Fig. 5a). Despite loading five times more material than in the previous runs, major and minor peaks with retention volumes (20.6 and 22.2 ml, respectively) strikingly similar to those obtained previously (20.54 and 22.47 ml) were seen. The major (full) peak was broader than in previous runs (in which less material was loaded) and encroached on the minor peak to some extent. Nevertheless, integration of peak areas was possible and the calculated empty-to-full ratio and empty particle percentage were close to those values determined by EM (Figs. 2 and 5a: empty-to-full ratios, 0.05 [EM] and 0.026 [CIM-QA]; percentage empty, 4.5% [EM] and 2.5% [CIM-QA]). Finally, in an experiment similar to that run with the CsCl gradient-purified vector, 1×1012 GC of lot no. CS0010 was mixed with the empty particle preparation (Z4033) and run on the CIM-QA disk (Fig. 5b). The empty and full peak retention volumes remained constant with respect to those observed previously with the mixture of V0083 and Z4033 (Fig. 4b) and with CS0010 alone (Fig. 5a). These results confirmed the consistency of the IEX assay and the applicability to vector preparations made by separate production processes.
FIG. 5.
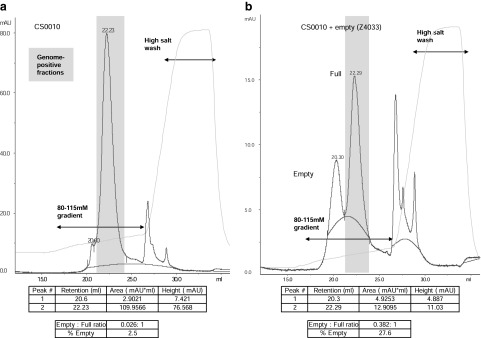
Quantification of relative empty-to-full particle content in an iodixanol gradient-purified AAV8 vector preparation. (a) A full AAV8 vector preparation (5×1012 GC) purified by iodixanol gradients (lot no. CS0010) was loaded onto a 0.34-ml CIM-QA disk and eluted with an 80–115 mM salt gradient. The y axis shows the absorbance (mAU) and the x axis the elution volume (ml). The detected conductivity and absorbance are represented by solid light and dark blue lines, respectively. Calculated baselines are represented as a red line. Peak retention volumes, peak areas, and GC content of fractions were determined as described in the legend to Fig. 4. (b) A mixture of 75 μl of empty AAV8 particles (lot no. Z4033) and 1×1012 GC of lot no. CS0010 was run under the same conditions and the peaks were evaluated identically.
Discussion
As the use of AAV vectors for gene therapy and other applications progresses to late-stage clinical trials and beyond, the need for highly robust, reproducible, and sensitive assays for production monitoring and final release will escalate. Many AAV-specific assays date from the early days of research in the field and may not be best suited for the rigorous standards of GMP production. With these limitations in mind, we have sought new and more sophisticated approaches to vector release assays.
In this study, we have employed the resolution properties of monolith anion-exchange chromatography media to analyze particle content of AAV8 vector preparations by separating empty and full AAV8 particles on the basis of charge. The use of automated FPLC equipment has resulted in reproducible assay runs and consistent retention volumes such that the two particle species can be readily identified and the relative amounts of each precisely quantified through integration of peak areas. The empty-to-full particle ratios obtained by the new method are in close agreement with ratios determined by EM analysis. Unlike previously published particle assays, the IEX-based method described here requires little in the way of sample preparation or special reagents and eliminates interpretative counting and compounded errors.
Although not directly addressed in the current study, the ability to quantify particle numbers in AAV8 vector preparations, using the IEX particle assay, should be entirely feasible through the use of a known standard such as the AAV8 reference material currently undergoing characterization (Moullier and Snyder, 2008). In addition, adaptation of the new assay to serotypes other than AAV8 is possible because the separation of empty and full particles of different serotypes (AAV2 and AAV6) has been described (Qu et al., 2007) and the purification of most AAV serotypes by ion-exchange chromatography has been demonstrated (Brument et al., 2002; Kaludov et al., 2002; Davidoff et al., 2004; Okada et al., 2009; Zhou et al., 2011). A wide range of CIM monolith media with different chemistries is available and should permit the adaptation of published methods for the purpose of developing IEX particle assays for other serotypes.
Although the advantages of the IEX particle assay are numerous, some limitations exist. Chief among these is the ability of protein contaminants to influence particle peak size and hence distort the particle ratios obtained. Separation of protein contaminants in the high-salt wash was noted, especially in the case of lot no. Z4033 (Figs. 3–5), but the presence of additional minor contaminants migrating with the AAV particles in the elution gradient is difficult to rule out. For this reason, the assay is not ideally suited for in-process monitoring but is best applied to final products with a high degree of purity. Another issue with the IEX assay is the degree of resolution between empty and full peaks, especially if higher quantities of material are assayed, leading to broadening of peaks. Although some attempt was made to improve resolution through the application of increasingly shallow gradients, complete separation was not obtained (Figs. 4 and 5). Because we were concerned mostly with relative quantification of the two peak sizes and not absolute quantification, the peak overlap was not considered detrimental to our purposes. However, when absolute quantification of particle numbers is desired, further optimization by standard chromatographic practices will be necessary in order to achieve complete peak resolution.
In summary, an anion exchange-based particle assay for the relative quantification of empty and full particle content in an AAV8 vector preparation has been developed. The reproducibility, simplicity, and speed of the IEX particle assay are distinct improvements on currently available methods and render it suitable as a release assay for clinical-quality AAV8 vectors.
Acknowledgments
The authors thank Julie Johnston, Arbans Sandhu, and other members of the Penn Vector Core for providing vector; and Peter Bell for providing the electron microscope images. This work was performed with funding from the NHLBI Gene Therapy Resource Program (HHSN268200748202C), a grant from the Bill & Melinda Gates Foundation, and a sponsored research agreement from ReGenX Biosciences.
Author Disclosure Statement
M.L., M.R.A., and J.M.W. are inventors on patents licensed to various biopharmaceutical companies, including ReGenX. J.M.W. is a consultant to ReGenX Holdings, and is a founder of, holds equity in, and receives a grant from affiliates of ReGenX Holdings.
References
- Allay J.A. Sleep S. Long S., et al. Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum. Gene Ther. 2011;22:595–604. doi: 10.1089/hum.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso E. Mingozzi F. Montane J., et al. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 2010;17:503–510. doi: 10.1038/gt.2009.157. [DOI] [PubMed] [Google Scholar]
- Bainbridge J.W. Smith A.J. Barker S.S., et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N. Engl. J. Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- Brument N. Morenweiser R. Blouin V., et al. A versatile and scalable two-step ion-exchange chromatography process for the purification of recombinant adeno-associated virus serotypes 2 and 5. Mol. Ther. 2002;6:678–686. doi: 10.1006/mthe.2002.0719. [DOI] [PubMed] [Google Scholar]
- Chadeuf G. Ciron C. Moullier P. Salvetti A. Evidence for encapsidation of prokaryotic sequences during recombinant adeno-associated virus production and their in vivo persistence after vector delivery. Mol. Ther. 2005;12:744–753. doi: 10.1016/j.ymthe.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Christine C.W. Starr P.A. Larson P.S., et al. Safety and tolerability of putaminal AADC gene therapy for Parkinson disease. Neurology. 2009;73:1662–1669. doi: 10.1212/WNL.0b013e3181c29356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement N. Knop D.R. Byrne B.J. Large-scale adeno-associated viral vector production using a herpesvirus-based system enables manufacturing for clinical studies. Hum. Gene Ther. 2009;20:796–806. doi: 10.1089/hum.2009.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidoff A.M. Ng C.Y. Sleep S., et al. Purification of recombinant adeno-associated virus type 8 vectors by ion exchange chromatography generates clinical grade vector stock. J. Virol. Methods. 2004;121:209–215. doi: 10.1016/j.jviromet.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Eberling J.L. Jagust W.J. Christine C.W., et al. Results from a phase I safety trial of hAADC gene therapy for Parkinson disease. Neurology. 2008;70:1980–1983. doi: 10.1212/01.wnl.0000312381.29287.ff. [DOI] [PubMed] [Google Scholar]
- Grimm D. Kern A. Pawlita M., et al. Titration of AAV-2 particles via a novel capsid ELISA: Packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 1999;6:1322–1330. doi: 10.1038/sj.gt.3300946. [DOI] [PubMed] [Google Scholar]
- Halbert C.L. Metzger M.J. Lam S.L. Miller A.D. Capsid-expressing DNA in AAV vectors and its elimination by use of an oversize capsid gene for vector production. Gene Ther. 2011;18:411–417. doi: 10.1038/gt.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W.W. Aleman T.S. Kaushal S., et al. Treatment of Leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: Short-term results of a phase I trial. Hum. Gene Ther. 2008;19:979–990. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaludov N. Handelman B. Chiorini J.A. Scalable purification of adeno-associated virus type 2, 4, or 5 using ion-exchange chromatography. Hum. Gene Ther. 2002;13:1235–1243. doi: 10.1089/104303402320139014. [DOI] [PubMed] [Google Scholar]
- Kaplitt M.G. Feigin A. Tang C., et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: An open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- Lock M. Alvira M. Vandenberghe L.H., et al. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum. Gene Ther. 2010a;21:1259–1271. doi: 10.1089/hum.2010.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock M. McGorray S. Auricchio A., et al. Characterization of a recombinant adeno-associated virus type 2 Reference Standard Material. Hum. Gene Ther. 2010b;21:1273–1285. doi: 10.1089/hum.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire A.M. Simonelli F. Pierce E.A., et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire A.M. High K.A. Auricchio A., et al. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: A phase 1 dose-escalation trial. Lancet. 2009;374:1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F. Maus M.V. Hui D.J., et al. CD8+ T-cell responses to adeno-associated virus capsid in humans. Nat. Med. 2007;13:419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- Moullier P. Snyder R.O. International efforts for recombinant adeno-associated viral vector reference standards. Mol. Ther. 2008;16:1185–1188. doi: 10.1038/mt.2008.125. [DOI] [PubMed] [Google Scholar]
- Okada T. Nonaka-Sarukawa M. Uchibori R., et al. Scalable purification of adeno-associated virus serotype 1 (AAV1) and AAV8 vectors, using dual ion-exchange adsorptive membranes. Hum. Gene Ther. 2009;20:1013–1021. doi: 10.1089/hum.2009.006. [DOI] [PubMed] [Google Scholar]
- Qu G. Bahr-Davidson J. Prado J., et al. Separation of adeno-associated virus type 2 empty particles from genome containing vectors by anion-exchange column chromatography. J. Virol. Methods. 2007;140:183–192. doi: 10.1016/j.jviromet.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Simonelli F. Maguire A.M. Testa F., et al. Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol. Ther. 2010;18:643–650. doi: 10.1038/mt.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.H. Levy J.R. Kotin R.M. A simplified baculovirus–AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol. Ther. 2009;17:1888–1896. doi: 10.1038/mt.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer J.M. Smith P.H. Parthasarathy S., et al. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol. Ther. 2003;7:122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- Thorne B.A. Takeya R.K. Peluso R.W. Manufacturing recombinant adeno-associated viral vectors from producer cell clones. Hum. Gene Ther. 2009;20:707–714. doi: 10.1089/hum.2009.070. [DOI] [PubMed] [Google Scholar]
- Timpe J. Bevington J. Casper J., et al. Mechanisms of adeno-associated virus genome encapsidation. Curr. Gene Ther. 2005;5:273–284. doi: 10.2174/1566523054065011. [DOI] [PubMed] [Google Scholar]
- Urabe M. Xin K.Q. Obara Y., et al. Removal of empty capsids from type 1 adeno-associated virus vector stocks by anion-exchange chromatography potentiates transgene expression. Mol. Ther. 2006;13:823–828. doi: 10.1016/j.ymthe.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Virag T. Cecchini S. Kotin R.M. Producing recombinant adeno-associated virus in foster cells: Overcoming production limitations using a baculovirus-insect cell expression strategy. Hum. Gene Ther. 2009;20:807–817. doi: 10.1089/hum.2009.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J.F. Adeno-associated viral vector manufacturing: Keeping pace with accelerating clinical development. Hum. Gene Ther. 2011a;22:913–914. doi: 10.1089/hum.2011.2514. [DOI] [PubMed] [Google Scholar]
- Wright J.F. New adeno-associated virus strategies to support momentum in the clinic. Hum. Gene Ther. 2011b;22:519–521. doi: 10.1089/hum.2011.4080. [DOI] [PubMed] [Google Scholar]
- Xiao W. Chirmule N. Berta S.C., et al. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 1999;73:3994–4003. doi: 10.1128/jvi.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Xie J. Xie Q., et al. Adenovirus-adeno-associated virus hybrid for large-scale recombinant adeno-associated virus production. Hum. Gene Ther. 2009;20:922–929. doi: 10.1089/hum.2009.125. [DOI] [PubMed] [Google Scholar]
- Zhou J. Yang X. Wright J.F., et al. PEG-modulated column chromatography for purification of recombinant adeno-associated virus serotype 9. J. Virol. Methods. 2011;173:99–107. doi: 10.1016/j.jviromet.2011.01.013. [DOI] [PubMed] [Google Scholar]