Abstract
Adeno-associated virus (AAV)-based vectors have gained increasing attention as gene delivery vehicles in basic and preclinical studies as well as in human gene therapy trials. Especially for the latter two—for both safety and therapeutic efficacy reasons—a detailed characterization of all relevant parameters of the vector preparation is essential. Two important parameters that are routinely used to analyze recombinant AAV vectors are (1) the titer of viral particles containing a (recombinant) viral genome and (2) the purity of the vector preparation, most commonly assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by silver staining. An important, third parameter, the titer of total viral particles, that is, the combined titer of both genome-containing and empty viral capsids, is rarely determined. Here, we describe a simple and inexpensive method that allows the simultaneous assessment of both vector purity and the determination of the total viral particle titer. This method, which was validated by comparison with established methods to determine viral particle titers, is based on the fact that Coomassie Brilliant Blue, when bound to proteins, fluoresces in the infrared spectrum. Viral samples are separated by SDS–PAGE followed by Coomassie Brilliant Blue staining and gel analysis with an infrared laser-scanning device. In combination with a protein standard, our method allows the rapid and accurate determination of viral particle titers simultaneously with the assessment of vector purity.
Kohlbrenner and colleagues report on a simple and inexpensive method, based on SDS-PAGE and Coomassie Brilliant Blue staining, that allows the simultaneous assessment of AAV vector purity and viral particle titer. According to the authors, this method can be used regardless of the method of vector production, the vector genome or the AAV serotype used.
Introduction
As encouraging studies that use adeno-associated virus (AAV) as a gene therapy vector accumulate (e.g., for Leber's congenital amaurosis [Maguire et al., 2009], hemophilia B [Nathwani et al., 2011], and heart failure [Jessup et al., 2011]), researchers have become interested in using recombinant AAV vectors as a gene delivery vehicle in a large number of preclinical animal disease models and in clinical trials. In particular, for in vivo experiments, consistent and accurate dosing is critical for data interpretation and for the design of future studies. Accurate titers are particularly important in studies with cohorts that are injected with vectors carrying different transgenes or vectors of different serotypes with the same transgene. Today, one of the most widely used characterization assays to determine the vector dose for in vitro and in vivo studies is quantitative real-time PCR (qPCR), which measures encapsidated viral genomes (Rohr et al., 2002). Predictably, the numerous qPCR protocols that have been published vary in important parameters such as primer sequences, amplicon length, brand of enzyme, type of machine, and cycling parameters. It is not difficult to imagine that these factors lead to variability in vector titers both within a single laboratory and, more prominently, among different laboratories (Lock et al., 2010). Several efforts to improve traditional qPCR protocols have been made. These methods include the development of a qPCR assay that is more suitable for self-complementary genomes (Fagone et al., 2012) and the development of a general qPCR protocol that allows the genome titer determination of vectors with any transgene because the primers anneal within the inverted terminal repeat (ITR) sequences that flank the recombinant genome (Aurnhammer et al., 2012).
Another commonly used approach to vector characterization is the visualization of capsid proteins with a protein dye after sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). This method is used to assess vector purity, which is important for both efficiency of transduction (Ayuso et al., 2010) and safety purposes (Wright, 2008). Typically, this is a qualitative assay to ensure a highly purified virus preparation, in which the only proteins visible in the gel are the VP1, VP2, and VP3 proteins that make up the capsid particle.
In the past we have required defined quantities of empty AAV particles for some of our studies (Rapti et al., 2012), and the inability to employ qPCR necessitated alternative quantification methods. An elegant approach to quantify capsids, based on measuring the optical density at 260 and 280 nm, has been described previously (Sommer et al., 2003). Regrettably, this method is problematic for AAV vectors purified by iodixanol gradient centrifugation (Zolotukhin et al., 1999), a commonly used purification approach for small-scale AAV production. Because of its high extinction coefficient at 260 and 280 nm,* even minute amounts of iodixanol can interfere with the determination of viral particle titers based on measuring the optical density at these wavelengths. ELISAs have been described previously for AAV2 and AAV3 (Grimm et al., 1999) and for AAV1, AAV4, AAV5, and AAV6 (Kuck et al., 2007) but not for other serotypes, and only ELISA kits for AAV1, AAV2, AAV3, and AAV5 are commercially available. ELISAs have the advantage of measuring intact particles, but they are dependent on binding of the capsid to usually serotype-specific antibodies. Although it is likely that for most of the commonly used serotypes ELISA kits will eventually become commercially available, much work is currently being done in engineering the viral capsid to create novel mutant and chimeric AAVs that, for instance, target specific tissues (Asokan et al., 2012) or show improved resistance to neutralizing antibodies (Maheshri et al., 2006). The development of ELISAs for many of these novel capsids would likely depend on the generation of new antibodies directed against these novel capsids, which is a labor-intensive task. As a consequence, an easy and inexpensive method to quantify AAV particle titers, regardless of the specific capsid proteins of the vector, is highly desirable.
Two developments led us to hypothesize that a simple method—based on SDS–PAGE and staining of the gel with Coomassie Brilliant Blue—could be developed that would allow the simultaneous determination of vector purity and viral particle titers. First, Luo and colleagues reported that protein bands in SDS–polyacrylamide gels that are stained with Coomassie could be accurately quantified with an infrared scanner (Luo et al., 2006). Second, the production and commercial availability of a thoroughly characterized AAV reference standard (Lock et al., 2010) would allow the rigorous validation of such a method. Here, we show that our newly developed quantification protocol can be used to determine viral particle titers accurately regardless of the method of vector production, the vector genome, or the AAV serotype used.
Materials and Methods
Virus preparations
The AAV2 Reference Standard Stock (AAV2-RSS) was obtained from the American Type Culture Collection (ATCC, Manassas, VA; cat. no. VR-1616). The wild-type AAV preparations used in this study have been described previously (Zeltner et al., 2010). All in-house preparations of recombinant AAVs were produced by transfection of 293T cells with two plasmids, one providing the AAV2 Rep proteins, the serotype-specific capsid proteins, and the adenoviral helper functions, and the second plasmid carrying the transgene expression cassette flanked by two AAV2 inverted terminal repeats (Grimm et al., 1998). A detailed protocol for the production of the recombinant AAVs used in this study can be found in Supplementary Materials and Methods (supplementary data are available online at http://www.liebertpub.com/hgtb).
qPCR titration
Titers of the AAV2-RSS and wild-type AAVs were reported previously (Lock et al., 2010; Zeltner et al., 2010). For all other samples reported, an Applied Biosystems 7500 real-time PCR system with SDS software version 1.4 (Life Technologies, Carlsbad, CA) was used for data acquisition and analysis. iTaq fast SYBR green supermix with ROX (Bio-Rad, Hercules, CA) was used according to the manufacturer's instructions, except that total reaction volumes were 10 μl with sample volumes of 2 μl. For all assays, except for the titration of the AAV1-LacZ preparation, the primers amplified a region of the simian virus 40 (SV40) poly(A) sequence (Fwd, TTG GAC AAA CCA CAA CTA GAA; Rev, AAC CTC CCA CAC CTC CC). Because the lacZ viral cassette did not have a poly(A) sequence matching the AAV2-RSS, but did have matching cytomegalovirus (CMV) promoter sequences, the primers used amplified a region of the CMV promoter (Fwd, TCA ATT ACG GGG TCA TTA GTT C; Rev, ACT AAT ACG TAG ATG TAC TGC C). The conditions for PCR were 2 min at 95°C (1 cycle); followed by 15 sec at 95°C, 15 sec at 58°C, 40 sec at 72°C (40 cycles). Melting curve analysis showed one peak, indicating a single product species.
Amino acid analysis of virus samples
Amino acid analysis of virus samples was conducted at the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, CT).
Electron microscopy
Negative staining microscopy of viral samples was performed as described previously (Zeltner et al., 2010). Full and empty particles were counted, using the “cell counter” function in the software program ImageJ (Abramoff et al., 2004).
SDS–PAGE, Coomassie blue staining, gel scanning, and capsid titer calculation
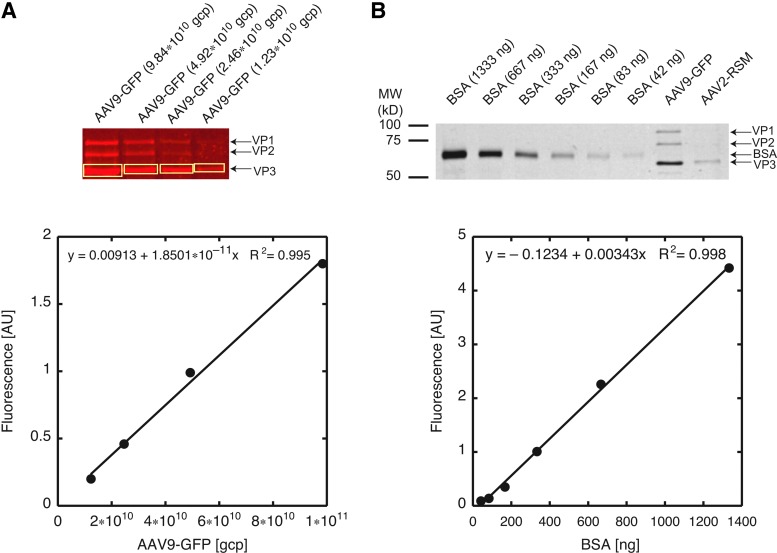
All polyacrylamide gels (Laemmli, 1970) used a 4% stacking gel and a 12% separating gel. The gels were loaded with a total volume of 36 μl, which contained 30 μl of sample solution (bovine serum albumin [BSA, cat. no. 23209; Pierce Biotechnology/Thermo Fisher Scientific, Rockford, IL] dilution or AAV sample) and 6 μl of 6×loading buffer (per 10 ml: 0.8 g of SDS; 5 ml of 1 M Tris, pH 6.8; 5 ml of glycerol; 5 mg of bromophenol blue). 2-Mercaptoethanol (5%, v/v) was added to the loading buffer immediately before adding it to the 30-μl samples. The AAV samples were incubated for 5 min and the BSA standards for 3 min at 95°C before loading (boiling the virus samples for only 3 min works equally well, but boiling BSA samples for 5 min results in partial degradation of the protein). The gels were run first at 70 V and then, after the loading dye moved past the stacking gel, at 120 V. The gel was then run until the loading dye reached the bottom of the gel. The gel was immersed in prestaining solution (50% methanol, 10% acetic acid) and incubated twice for 5 min each. The gel was then placed into staining solution (50% methanol, 10% acetic acid, 0.03% Coomassie Brilliant Blue R-250 [cat. no. 191491000; Acros Organics/Thermo Fisher Scientific, Pittsburgh, PA]) and incubated overnight with gentle agitation. The next day, the gel was destained with 40% methanol–8% acetic acid solution (a Kimwipe was placed in the container to absorb dye that eluted from the gel). Whenever the destaining solution or the Kimwipe appeared blue, they were removed and fresh destaining solution and/or a clean Kimwipe was added. The time to completely destain the gels varied, but the background was usually removed by 4 hr. Before scanning, the gel was soaked two times in water for 10 min each. Gels were then scanned with an Odyssey imaging system (LI-COR Biosciences, Lincoln, NE) and analyzed with Odyssey software version 3.0. Before scanning, the important non-default variable parameters were set as follows: 169 μm resolution, medium quality, and scan type “protein gel.” After scanning, the image was adjusted by selecting Filter>Noise Removal, and setting the image display in the 700 channel to “Auto Linear” to obtain optimal images. BSA and VP3 bands were then enclosed within rectangles. The size of the rectangle was allowed to change depending on band size, as the higher BSA concentrations resulted in bands with larger areas (Fig. 1). The background was set to “top/bottom.” All the rectangles on the gel image were selected, and a report, including integrated intensity values, was exported to the spreadsheet software Numbers (Apple, Cupertino, CA) or Excel (Microsoft, Redmond, WA). A standard curve was generated by plotting the amount of BSA versus the integrated fluorescence intensity. Linear regression analysis yielded a formula that allowed us to calculate the nanograms of VP3 loaded in each lane with a virus sample, based on the integrated fluorescence intensity of the particular VP3 band (Fig. 1).
FIG. 1.
Quantification of viral particle titers by Coomassie Brilliant Blue-stained SDS–PAGE and infrared-fluorescence scanning with an Odyssey scanner. (A) The fluorescence intensity is linear over a wide range of viral particles loaded onto the gel. Top: The indicated amount of AAV9-GFP particles was separated on a 12%, reducing SDS–polyacrylamide gel, stained with Coomassie Brilliant Blue, followed by scanning of the wet gel with an Odyssey (LI-COR) scanner, using the 700-nm channel. v, viral particles. Brightness was adjusted uniformly across the entire image. Positions of the viral capsid proteins (VP1, VP2, and VP3) are indicated with arrows. Bottom: The number of viral particles loaded in each lane is plotted against integrated fluorescence of the boxed VP3 bands (top). Quantification was performed before adjusting the brightness. Regression analysis demonstrates the linearity of the integrated fluorescence intensity over the entire particle range covered. (B) The fluorescence intensity is linear over a wide range of BSA loaded onto the gel. Top: The indicated amounts of BSA were separated on a 12%, reducing SDS–polyacrylamide gel, stained with Coomassie Brilliant Blue, followed by scanning of the wet gel with an Odyssey (LI-COR) scanner, using the 700-nm channel. The picture was converted to grayscale and inverted. Brightness was adjusted uniformly across the entire image. Bottom: Quantification of the gel (top) demonstrates the linearity of the integrated fluorescence intensity over the entire range of BSA loaded. Regression analysis of the BSA standard curve allowed determination of the number of viral particles loaded in the two right-most lanes: AAV9-GFP (1.32×1011 viral particles) and AAV2-RSS (2.41×1010 viral particles). Positions of the viral capsid proteins (VP1, VP2, and VP3) and BSA are indicated with arrows, and the position and size of the molecular weight standards are shown on the left.
Although we did not investigate this in detail, similar results could be obtained when the wet, Coomassie-stained gels were scanned with a flatbed scanner followed by quantification with ImageJ (Abramoff et al., 2004) or Photoshop (Adobe Systems, San Jose, CA). However, anecdotally, the R2 values obtained by infrared scanning tended to be somewhat higher.
Results and Discussion
To be able to use infrared fluorescence scanning of Coomassie-stained SDS–polyacrylamide gels to quantify AAV particles, the fluorescence signal must be linearly correlated with the number of viral particles across a sufficiently broad range of viral particle amounts loaded onto the gel. Hence, in a first experiment, we ran a four-point, 2-fold dilution series of an in-house preparation of AAV9-GFP and the newly available AAV2 Reference Standard Stock (Lock et al., 2010) from the ATCC on a 12% SDS–polyacrylamide gel followed by Coomassie staining and infrared scanning with an Odyssey scanner (LI-COR). Because VP3 is the most abundant capsid protein and, consequently, yields the highest band intensity, we chose VP3 over VP1 or VP2 for analysis. When we plotted the amount of virus loaded (ranging from 7.07×109 to 1.11×1011 capsid particles per lane) against the fluorescence intensity of the VP3 band and performed a linear regression analysis we found that the R2 values for both virus preparations were >0.99, thus proving the linearity of the fluorescence signal over a sufficiently broad range of vector particle titers (Fig. 1 and data not shown).
It is noteworthy that the lowest amount of virus (7.07×109) was just above the detection limit. The loading of ≥1.5×1010 viral particles, on the other hand, always yielded robust signals. These titration results with viral samples suggested that, as long as a well-defined reference material for use in a standard curve was available, an accurate absolute particle titer determination was possible by this method. It could be argued that the AAV2-RSS represents the ideal reference material, at least for AAV2 vectors. However, its relatively low protein content combined with the limited loading capacity per well inevitably restricts a standard curve to a relatively narrow range of viral particles. Because of this and because of the comparatively high cost of the AAV2-RSS, we sought to test whether accurate viral particle titers could be determined with another widely available protein standard with a higher initial protein concentration, the rigorously defined BSA standard from Pierce Biotechnology Pierce Biotechnology/Thermo Fisher Scientific, which is calibrated against the standard reference material 927d (7% BSA solution) from the National Institute of Standards and Technology.
To this end, we analyzed a dilution series of the BSA standard together with single quantities of the AAV2- RSS and an AAV9-GFP preparation produced in-house. After SDS–PAGE, Coomassie staining, and infrared fluorescence scanning (Fig. 1), we plotted the amount of BSA loaded (in nanograms) versus the background-corrected fluorescence intensity of the BSA band (Fig. 1). Linear regression analysis yielded an equation that allowed the calculation of nanograms of VP3 in each lane with virus sample (Fig. 1B). Using the peptide sequence of VP3 and the generally accepted presence of 50 VP3 molecules per viral capsid (Johnson et al., 1971; Becerra et al., 1988), we calculated that each viral particle contains 4.987×10–9 ng of VP3 for AAV2 and 4.967×10–9 ng for AAV9. Division of the nanograms of VP3 determined by the gel method by the nanograms of VP3 per viral particle yields the number of AAV particles loaded onto the gel. Dividing the viral particle number loaded by the loading volume yields the viral titer. For the ATCC AAV2-RSS, we performed a total of nine analyses (five at King's College London School of Medicine [London, UK] and four at Mount Sinai School of Medicine [New York, NY]). The average of the viral particle titers obtained was 1.03×109 particles/μl (standard deviation, 3.30×108 particles/μl). When we compared our titers with the ELISA titers reported for the reference standard (Lock et al., 2010), our titers corresponded to 111.6% (95% confidence interval, 87.9–135.2%; Table 1) of the ELISA values (the raw numbers for all of our titer determinations are listed in Supplementary Table S1). These results indicated that it is possible by our method to determine viral particle titers accurately and that our protocol would be useful for the characterization of all our recombinant AAV preparations.
Table 1.
Viral Particle Titers and Percentage of Reference Values of the AAV2-RSS, Wild-Type AAVs, and In-House-Prepared Recombinant AAVs
| AAV sample | Reference value | Mean (CBB/SDS–PAGE) | Stdv (CBB/SDS–PAGE) | Number of quantifications | Percent reference value | 95% Confidence interval (percent reference value) |
|---|---|---|---|---|---|---|
| AAV2-RSS (at KC) | 9.18×108a | 1.08×109 | 3.99×108 | 5 | 118.1 | 80.0–156.1 |
| AAV2-RSS (at MSSM) | 9.18×108a | 9.50×108 | 2.62×108 | 4 | 103.5 | 75.5–131.4 |
| AAV2-RSS (KC and MSSM) | 9.18×108a | 1.03×109 | 3.30×108 | 9 | 111.6 | 87.9–135.2 |
| AAV2 wild-type A | 3.50×109b | 2.72×109 | 4.47×108 | 4 | 77.6 | 65.1–90.1 |
| AAV2 wild-type B | 5.80×109b | 5.34×109 | 1.46×109 | 4 | 92.0 | 67.3–116.8 |
| AAV2 wild-type C | 6.00×109b | 5.29×109 | 1.17×109 | 4 | 88.2 | 69.1–107.2 |
| AAV2 wild-type (A–C) | N/A | N/A | N/A | 12 | 85.9 | 75.2–96.7 |
| AAV9.GFP | 3.28×109b | 3.46×109 | 7.25×108 | 4 | 105.4 | 83.8–127.1 |
| AAV1-SERCA | 2.34×109b | 2.48×109 | 1.89×108 | 3 | 106.1 | 97.0–115.3 |
| AAV1-LacZ | 3.91×109b | 3.69×109 | 2.10×108 | 3 | 94.3 | 88.2–100.4 |
| In house AAVs | N/A | N/A | N/A | 10 | 102.3 | 93.2–111.4 |
| All AAVs | N/A | N/A | N/A | 31 | 98.7 | 89.6–107.7 |
AAV, adeno-associated virus; AAV2-RSS, AAV2 reference standard stock; CBB, Coomassie Brilliant Blue; GFP, green fluorescent protein; KC, virus analyzed at King's College London School of Medicine; LacZ, galactosidase; MSSM, virus analyzed at Mount Sinai School of Medicine; N/A, not applicable; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; SERCA, sarco/endoplasmic reticulum calcium ATPase; stdv, standard deviation.
Note: Titers are expressed as viral particles per microliter. Rows in boldface represent the average of “Percent reference value” of all virus preparations of a particular type of virus, that is, AAV2-RSS, all wild-type AAVs, all recombinant AAVs prepared in-house, or all the virus preparations.
Titer determined by ELISA.
Titer determined by quantitative amino acid analysis.
A priori, the availability and thorough characterization of the AAV2-RSS would seem to make it an ideal candidate to be used as a community-wide standard curve for all future AAV qPCR quantifications as well as the characterization of other important parameters of viral preparations. However, the amount of AAV2-RSS is ultimately finite. Hence, we posit that—where possible—the best use of the AAV2-RSS is for the calibration of in-house reference standards. This rationale led us to create our own in-house reference standard for vectors containing either the CMV immediate enhancer, the GFP-coding region, and/or an SV40 p(A) sequence. For this, we ran qPCR assays of an AAV9-GFP preparation in parallel with the AAV2-RSS in triplicates of six dilution points for each virus. With the AAV2-RSS as standard points of “known” genome-containing particle (GCP) titer, we determined the GCP titer of our in-house reference standard to be 1.70×109 GCP/μl. The determination of GCP titers, combined with the determination of the ratio of genome-containing to empty viral particles by negative-staining electron microscopy (Zeltner et al., 2010), furthermore allowed us to independently calculate a total viral particle titer.† With this approach, we found the viral particle titer of our AAV9-GFP preparation determined by either our newly developed SDS–PAGE method or by combining qPCR titers with the ratio of empty to full capsids to be in close accordance (3.46×109 particles/μl [gel method] vs. 3.16×109 particles/μl [qPCR and electron microscopy]). It must be pointed out, however, that we did not observe such a close correlation for all recombinant virus preparations, likely a result of the inherent variability of qPCR methods to determine genomic titers. Hence, we decided to validate our method by an additional technique. Because ELISA kits are not commercially available for AAV9, we could not use this method. Therefore, we decided to use quantitative amino acid analysis to determine accurately the particle titers of our AAV9-GFP preparation. Although this approach is comparatively expensive, it is a precise method of analyzing amino acid content and has been used by us (Zeltner et al., 2010) and others (Sommer et al., 2003) to determine AAV particle titers. The titers obtained by our method closely matched the titers obtained by amino acid analysis; the mean titer determined by our gel method was 105.4% (95% confidence interval, 83.8–127.1%) of the reference value (see Table 1).
In addition, our method to determine particle titers was further validated by determining particle titers of three wild-type AAV2 preparations that have been rigorously characterized previously—including amino acid analysis (Zeltner et al., 2010). Compared with the reported amino acid analysis titers, the mean titers were 85.9% (95% confidence interval, 75.2–96.7%; see Table 1). Last, we quantified the viral particle titers of two additional recombinant AAV preparations of a third serotype, AAV1, and our titers closely matched those obtained by amino acid analysis (Table 1). When the data of all our analyses are aggregated we obtained mean titers of 98.7% (95% confidence interval, 89.6–107.7%) of the reference titers (ELISA or amino acid analysis titers), demonstrating the high precision, accuracy, and reproducibility of our approach. It is worth pointing out, that although, in principle, our method can be used for semipurified virus preparations, the purity of the virus preparation must be high enough to distinguish unambiguously the VP3 band from potential contaminations with similar molecular weight. In principle, it should also be possible to determine the VP1:VP2:VP3 ratio by our method. We noticed, however, that because of the low abundance of VP1 and VP2 this proved to be challenging, at least with the titers of our viral samples.
In summary, in the present study we describe a simple and inexpensive method that allows the simultaneous evaluation of the purity of AAV vector preparations together with an accurate determination of total viral particle titers (i.e., particle titers reflecting both virions with genomes and empty viral particles). We propose that, especially if combined with negative-staining electron microscopy, the routine use of this method will greatly increase experimental intra- and interlaboratory reproducibility and should be adopted as a general method of vector characterization, particularly in preclinical animal disease models where even modest variations in vector genome and total vector particle titers can substantially affect therapeutic outcome.
Supplementary Material
Footnotes
The molar extinction coefficients of iodixanol at 260 and 280 nm are approximately 2000 and 400, respectively.
We used the following formula to calculate total particle titer: total particle titer=qPCR titer divided by the fraction of full (i.e., genome-containing) particles.
Acknowledgments
This work was supported by U.S. National Institutes of Health grants HL100396 (R.J.H.) and HL088434 (R.J.H.) and by U.K. Medical Research Council grant G1001764 (R.M.L.).
Author Disclosure Statement
Dr. Hajjar is the scientific cofounder of Celladon, which plans to commercialize AAVI.SERCA2a for the treatment of heart failure. All other authors declare no competing financial interest.
References
- Abramoff M. Magalhaes P. Ram S. Image processing with Image. J. Biophot. Int. 2004;11:36–42. [Google Scholar]
- Asokan A. Schaffer D.V. Samulski R.J. The AAV vector toolkit: Poised at the clinical crossroads. Mol. Ther. 2012;20:699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurnhammer C. Haase M. Muether N., et al. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods. 2012;23:18–28. doi: 10.1089/hgtb.2011.034. [DOI] [PubMed] [Google Scholar]
- Ayuso E. Mingozzi F. Montane J., et al. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 2010;17:503–510. doi: 10.1038/gt.2009.157. [DOI] [PubMed] [Google Scholar]
- Becerra S.P. Koczot F. Fabisch P. Rose J.A. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J. Virol. 1988;62:2745–2754. doi: 10.1128/jvi.62.8.2745-2754.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagone P. Wright J.F. Nathwani A.C., et al. Systemic errors in quantitative polymerase chain reaction titration of self-complementary adeno-associated viral vectors and improved alternative methods. Hum. Gene Ther. Methods. 2012;23:1–7. doi: 10.1089/hgtb.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D. Kern A. Rittner K. Kleinschmidt J.A. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 1998;9:2745–2760. doi: 10.1089/hum.1998.9.18-2745. [DOI] [PubMed] [Google Scholar]
- Grimm D. Kern A. Pawlita M., et al. Titration of AAV-2 particles via a novel capsid ELISA: Packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 1999;6:1322–1330. doi: 10.1038/sj.gt.3300946. [DOI] [PubMed] [Google Scholar]
- Jessup M. Greenberg B. Mancini D., et al. Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): A phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation. 2011;124:304–313. doi: 10.1161/CIRCULATIONAHA.111.022889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F.B. Ozer H.L. Hoggan M.D. Structural proteins of adenovirus-associated virus type 3. J. Virol. 1971;8:860–863. doi: 10.1128/jvi.8.6.860-863.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck D. Kern A. Kleinschmidt J.A. Development of AAV serotype-specific ELISAs using novel monoclonal antibodies. J. Virol. Methods. 2007;140:17–24. doi: 10.1016/j.jviromet.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lock M. McGorray S. Auricchio A., et al. Characterization of a recombinant adenoassociated virus type 2 Reference Standard Material. Hum. Gene Ther. 2010;21:1273–1285. doi: 10.1089/hum.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S. Wehr N.B. Levine R.L. Quantitation of protein on gels and blots by infrared fluorescence of Coomassie blue and fast green. Anal. Biochem. 2006;350:233–238. doi: 10.1016/j.ab.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Maguire A.M. High K.A. Auricchio A., et al. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: A phase 1 dose-escalation trial. Lancet. 2009;374:1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshri N. Koerber J.T. Kaspar B.K. Schaffer D.V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- Nathwani A.C. Tuddenham E.G. Rangarajan S., et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapti K. Louis-Jeune V. Kohlbrenner E., et al. Neutralizing antibodies against AAV serotypes 1, 2, 6, and 9 in sera of commonly used animal models. Mol. Ther. 2012;20:73–83. doi: 10.1038/mt.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr U.P. Wulf M.A. Stahn S., et al. Fast and reliable titration of recombinant adenoassociated virus type-2 using quantitative real-time PCR. J. Virol. Methods. 2002;106:81–88. doi: 10.1016/s0166-0934(02)00138-6. [DOI] [PubMed] [Google Scholar]
- Sommer J.M. Smith P.H. Parthasarathy S., et al. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol. Ther. 2003;7:122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- Wright J.F. Manufacturing and characterizing AAV-based vectors for use in clinical studies. Gene Ther. 2008;15:840–848. doi: 10.1038/gt.2008.65. [DOI] [PubMed] [Google Scholar]
- Zeltner N. Kohlbrenner E. Clement N., et al. Near-perfect infectivity of wild-type AAV as benchmark for infectivity of recombinant AAV vectors. Gene Ther. 2010;17:872–879. doi: 10.1038/gt.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin S. Byrne B.J. Mason E., et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.