Abstract
As HIV continues to be a global public health problem with no effective vaccine available, new and innovative therapies, including HIV gene therapies, need to be developed. Due to low transduction efficiencies that lead to low in vivo gene marking, therapeutically relevant efficacy of HIV gene therapy has been difficult to achieve in a clinical setting. Methods to improve the transplantation of enriched populations of anti-HIV vector-transduced cells may greatly increase the in vivo efficacy of HIV gene therapies. Here we describe the development of preselective anti-HIV lentiviral vectors that allow for the purification of vector-transduced cells to achieve an enriched population of HIV-resistant cells. A selectable protein, human CD25, not normally found on CD34+ hematopoietic progenitor cells (HPCs), was incorporated into a triple combination anti-HIV lentiviral vector. Upon purification of cells transduced with the preselective anti-HIV vector, safety was demonstrated in CD34+ HPCs and in HPC-derived macrophages in vitro. Upon challenge with HIV-1, improved efficacy was observed in purified preselective anti-HIV vector-transduced macrophages compared to unpurified cells. These proof-of-concept results highlight the potential use of this method to improve HIV stem cell gene therapy for future clinical applications.
Kalomoiris and colleagues develop and test an anti-HIV lentiviral vector based on the selectable marker CD25. The preselective vector allows for the purification of vector-transduced cells to achieve an enriched population of HIV-resistant cells. Upon purification of cells transduced with the preselective anti-HIV vector, safety was demonstrated in CD34+ hematopoietic progenitor cells (HPC) and in HPC-derived macrophages in vitro. Improved efficacy against HIV-1 challenge was observed in purified preselective anti-HIV vector-transduced macrophages.
Introduction
HIV infections continue to spread worldwide with no effective vaccine available in both developed and underdeveloped countries (Barouch et al., 2008; Edgeworth et al., 2002). Although antiretroviral therapy (ART) is effective in the majority of HIV-infected patients, challenges to therapeutic and curative success include the continuing emergence of drug-resistant HIV variants, drug toxicity, and incomplete viral suppression (Baldanti et al., 2010; Domingo et al., 2012; Johnson et al., 2010; Kuritzkes, 2011; Lewden et al., 2007; Macias et al., 2012; Tilton et al., 2010). ART also fails to eradicate viral reservoirs, which are established early in infection, leading to viral persistence and incomplete immune restoration (Gazzola et al., 2009; Mehandru et al., 2006). Interruption of ART results in rapid viral resurgence, the generation of escape mutants, and CD4+ T-cell loss in the peripheral blood of HIV-infected patients (Graham et al., 2012; Kalmar et al., 2012).
These challenges highlight the need for the further development of innovative HIV therapies with broad mechanisms of action. HIV gene therapy has potential as an alternative or complementary treatment strategy to ART, especially when hematopoietic stem cells (HSCs) are the cellular targets for genetic modification (Strayer et al., 2005). Advantages of HIV stem cell gene therapy include constitutive or controlled expression of anti-HIV genes, the generation of a durable and HIV-resistant immune system, and the possibility of a one-time treatment if enough anti-HIV vector-transduced cells can be transplanted into patients (Strayer et al., 2005). Many potent anti-HIV genes have been developed and tested both in preclinical and clinical settings, and the safety of some of these genes has been observed with engraftment of transduced stem cells and anti-HIV gene expression in transplanted patients (Bauer et al., 1997; DiGuisto et al., 2010; Kohn et al., 1999; Mitsuyasu et al., 2009; Podsakoff et al., 2005; Shimizu et al., 2010; ter Brake et al., 2009; Walker et al., 2012). However, efficacy in a clinical setting has been difficult to achieve due to low transduction efficiencies and low in vivo gene marking (DiGuisto et al., 2010; Mitsuyasu et al., 2009; Podsakoff et al., 2005).
In vitro HIV challenge experiments designed to evaluate the efficacy of anti-HIV genes in inhibiting HIV infection/replication rely on sorting or selection of the gene-transduced cells, resulting in a pure population of HIV-resistant cells prior to infection. However, this has not been feasible in a clinical setting because many reporter genes utilized for sorting may be immunoreactive. When unsorted/mixed populations of nontransduced and anti-HIV vector-transduced cells are infected with HIV, a selective survival advantage and an increase in the percentage of total immune cells of the anti-HIV gene-expressing cells has been observed (Anderson et al., 2009; Walker et al., 2012). These results highlight the ability of anti-HIV gene-modified cells to survive and possibly replenish the immune system with functioning HIV-resistant immune cells. However, when translated into a clinical setting, a large percentage of nontransduced HSCs are transplanted along with the anti-HIV vector-transduced cells due to low transduction efficiencies (DiGuisto et al., 2010; Mitsuyasu et al., 2009; Podsakoff et al., 2005). These nontransduced HSCs produce unprotected immune cells, which are targets for HIV infection and replication. These infected cells may also replenish viral reservoirs. By transplanting an enriched population of anti-HIV vector-transduced cells into patients where the majority of the cells express the anti-HIV genes, similar results observed with the Berlin patient may be achievable as this patient received a pure population of HIV-resistant HSCs from a donor who was homozygous for a CCR5 Δ32 bp allele (Hutter et al., 2009). Therefore, new methods need to be developed to increase the total percentage of anti-HIV vector-transduced cells transplanted into patients.
In our current proof-of-concept study, we have demonstrated in vitro safety and an improved efficacy of HIV stem cell gene therapy in the enriched population of HIV-resistant cells compared to unpurified cells. This was achieved by a triple combination anti-HIV vector that incorporated a selectable marker, human CD25, which is expressed on the surface of transduced cells. Human CD25, the low affinity IL-2 receptor alpha subunit, was chosen as the selectable marker because of its normal characteristics of not being expressed on the surface of HPCs or HSCs and its lack of intracellular signaling (Grant et al., 1992; Kuziel et al., 1990; Minami et al., 1993). Upon expressing CD25 on the surface of HPCs and purification of the transduced cells, safety of the enriched population of anti-HIV vector-transduced HPCs was observed along with potent HIV-1 inhibition. These proof-of-concept results highlight the potential use of this strategy to improve the efficacy of future HIV stem cell gene therapy clinical trials.
Materials and Methods
Lentiviral vector design and production
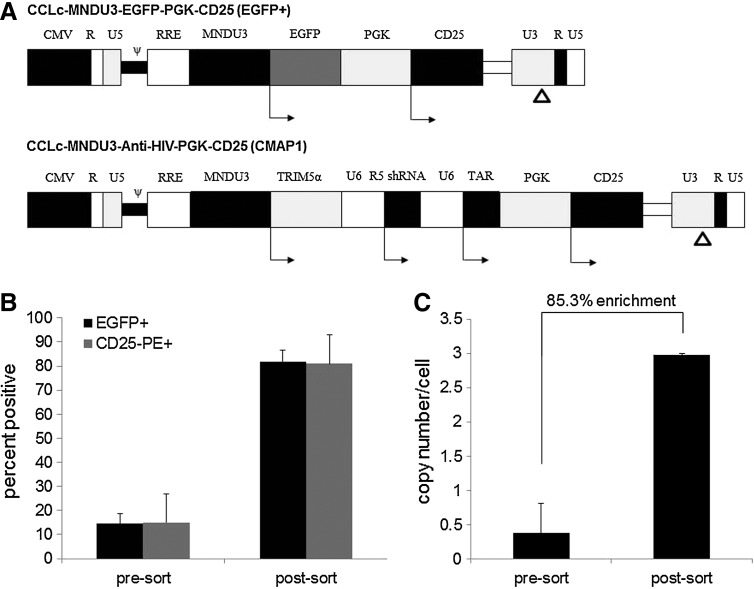
A third-generation self-inactivating lentiviral vector was utilized in this study, CCLc-MNDU3-X-PGK-X2. The U3 region of the 5' long terminal repeat (LTR) was replaced with the cytomegalovirus (CMV) promoter resulting in Tat-independent transcription but still maintaining high levels of expression. Self-inactivating (SIN): 400 bp in the U3 region of the 3' LTR were deleted, including the TATA box. The deletion is transferred to the 5' LTR after reverse transcription and integration in transduced cells, resulting in the transcriptional inactivation of the LTR in the provirus. To generate the control vector (named EGFP+), an enhanced green fluorescent protein (EGFP) reporter gene was inserted into position “X” under the control of the MNDU3 promoter, and a human CD25 coding region was inserted into position “X2” of this vector under the control of a phosphoglycerate kinase (PGK) promoter (Fig. 1A). This vector was only used to initially test the strategy of utilizing CD25 as a selective protein in purifying transduced cells. Therefore, we would be able to compare EGFP% positive cells to CD25% positive cells. To generate the preselective anti-HIV vector (named CMAP1 for Cclc-Mndu3-Antihiv-Protein-1), a triple combination of anti-HIV genes was inserted into position “X” and a human CD25 coding region was inserted into position “X2” of this vector under the control of a PGK promoter (Fig. 1B). The triple combination of anti-HIV genes includes a chimeric human/rhesus macaque TRIM5α gene under the control of the MNDU3 promoter, a polymerase-III U6 promoter-driven CCR5 shRNA expression cassette, and a polymerase-III U6 promoter-driven TAR decoy expression cassette (Fig. 1B). Sequencing of clones was confirmed by Laragen Inc. (Los Angeles, CA).
FIG. 1.
Preselective lentiviral vectors and purification of transduced HPCs. (A) A self-inactivating third generation lentiviral vector, CCLc-MNDU3-X-PGK-X2, was utilized to derive the preselective vectors. A 400 bp deletion in the 3’ LTR U3 region is denoted by Δ. The control EGFP+ vector contains an EGFP reporter gene under the control of the MNDU3 promoter and a human CD25 gene under the control of the PGK promoter. The CMAP1 anti-HIV vector contains a triple combination of anti-HIV genes, a human/rhesus macaque chimeric TRIM5α under the control of the MNDU3 promoter, a pol-III U6 promoter-driven CCR5 shRNA, a pol-III U6 promoter-driven TAR decoy, and a human CD25 gene under the control of the PGK promoter. (B) CD34+ HPCs were transduced with the control EGFP+ preselective vector, purified by CD25 immunomagnetic beads, and analyzed by flow cytometry for EGFP and CD25 expression. (C) CD34+ HPCs were transduced with the anti-HIV CMAP1 vector, purified by CD25 immunomagnetic beads, and analyzed by QPCR for vector copy number. All experiments were performed in triplicate. EGFP, enhanced green fluorescent protein; LTR, long terminal repeat; PGK, phosphoglycerate kinase; HPC, hematopoietic progenitor cells; QPCR, quantitative PCR; CMAP1, Cclc-mndu3-anti-hiv-protein-1.
Lentiviral vectors were generated in HEK-293T cells. Twenty-five micrograms of the packaging construct, Δ8.9 (packaging plasmid containing the gag and pol genes), 25 μg of EGFP+ or CMAP1, and 5 μg of VSVG (envelope) were transfected into cells in T225 flasks by lipofection. Vector supernatants were collected at 48 hr post-transfection and concentrated by ultrafiltration. Vector titers were calculated by transduction of HEK-293T cells. Forty-eight hr post-transduction, the HEK-293T cells were stained with a phycoerythrin (PE)-conjugated anti-human CD25 antibody (BD Biosciences, San Jose, CA) and analyzed by flow cytometry. All flow cytometry analyses were performed on a Beckman Coulter Cytomics FC500 using CXP software.
Transduction and purification of vector-transduced primary human CD34+ HPCs
CD34+ hematopoietic progenitor cells (HPCs) were isolated from human umbilical cord blood (NDRI, Philadelphia, PA) by Ficoll-Paque (GE Healthcare, Piscataway, NJ), and purified by CD34+ magnetic bead column separation (Miltenyi Biotec, Auburn, CA). CD34+ cell isolation purity (>90%) was routinely obtained. Total CD34+ cells were cultured in complete Iscove's modified Dulbecco's medium (IMDM) containing 10% fetal bovine serum (FBS) and supplemented with 50 ng/ml SCF, Flt-3 ligand, and TPO. Cells were transduced with the lentiviral vectors EGFP+ or CMAP1 (MOI 15) overnight at 37°C with 8 μg/ml protamine sulfate. Two days post-transduction, an aliquot of the EGFP+ vector-transduced cells was stained with a PE-conjugated anti-human CD25 antibody (BD Biosciences) and analyzed by flow cytometry for both EGFP and CD25 percentages. Total genomic DNA was extracted from an aliquot of the CMAP1 vector-transduced cells utilizing a Wizard Genomic DNA Isolation System (Promega, Madison, WI) and analyzed by quantitative PCR (QPCR) for vector copy number with a primer set specific for the chimeric TRIM5α gene: (forward) 5’-CTGGGTTGATGTGACAGTGG-3’ and (reverse) 5’-CGTGAGTGACGGAAACGTAA-3’. QPCR was performed using a SYBR Green PCR Master Mix Kit (Applied Biosystems, Foster City, CA). The rest of the transduced cells were labeled with CD25+ immunomagnetic beads (Miltenyi Biotec) according to the manufacturer's protocol and separated over a magnetic bead column. Purified cells were then used for subsequent experiments.
To evaluate the purity of the EGFP+ vector-transduced cells after CD25+ immunomagnetic bead selection, purified cells were stained with a PE-conjugated anti-human CD25 antibody (BD Biosciences) and analyzed for both EGFP and CD25 percentages. To evaluate the purity of the CMAP1 vector-transduced cells, total genomic DNA was extracted and analyzed by QPCR for vector copy number utilizing the chimeric TRIM5α primer set described above. QPCR was performed using a SYBR Green PCR Master Mix Kit (Applied Biosystems). GAPDH was used as an internal control.
Colony-forming unit assays
CD34+ HPCs, either nontransduced (NT) or CD25 immunomagnetic bead purified CMAP1 vector-transduced cells were cultured in semi-solid methylcellulose medium with growth factors (Stem Cell Technologies, Vancouver, Canada) for 10 days. After 10 days, total blood-forming colonies (BFU), granulocyte/erythrocyte/megakaryocyte/monocyte colonies (GEMM), and granulocyte/monocyte colonies (GM) were observed and counted.
To evaluate whether purified CMAP1 vector-transduced CD34+ HPCs (10,000 cells/3ml methylcellulose) had an increased proliferation potential in the presence of IL-2, 1 μg/ml of IL-2 was added to the methylcellulose medium. After 10 days, total cell numbers were counted.
To derive macrophages from the nontransduced and the purified CMAP1 vector-transduced CD34+ HPCs, cells were removed from the methylcellulose and plated in 6-well plates in complete DMEM with 10% FBS supplemented with 10 ng/ml of GM-CSF and M-CSF (R&D Systems, Minneapolis, MN). Media was changed every two days for four days to derive mature macrophages. Both nontransduced (NT) and purified CMAP1 vector-transduced CD34+ cell-derived macrophages were used for subsequent experiments.
Phenotypic and genotypic analysis of CD34+ cell-derived macrophages
To determine if CMAP1 vector-transduced CD34+ cells were able to mature into phenotypically normal macrophages, cells were visualized by microscopy and analyzed by flow cytometry. Macrophages were stained with antibodies to detect normal macrophage cell surface markers, including PE-conjugated CD14, allophycocyanin (APC)-conjugated HLADR, PE-conjugated CD4, PECY7-conjugated CD80, PE-conjugated CCR5, or APC-conjugated CD25 (BD Biosciences).
To determine if the addition of IL-2 induced the expression of proto-oncogenes in CMAP1 vector-transduced macrophages, IL-2 (1 μg/ml) was added to the macrophage cultures. On day three post-IL2 addition, total cellular RNA was extracted and QPCR was performed. Total cellular RNA was extracted from cells using RNA-STAT-60 (Tel-Test Inc., Friendswood, TX). First strand cDNA synthesis was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). QPCR was then performed using the SYBR Green PCR Master Mix Kit (Applied Biosystems) with primers: myc (forward) 5’-TCCATTCCGAGGCCACAGCAAG-3’ and (reverse) 5’-TCAGCTCGTTCCTCCTCTGACG-3’; myb (forward) 5’-AAGACCCTGAGAAGGAAAAGCG-3’ and (reverse) 5’-GTGTTGGTAATGCCTGCTGTCC-3’; fos (forward) 5’-ACTACCACTCACCCGCAGAC-3’ and (reverse) 5’-GACGGGAAGCCAGCCTTAC-3’; and jun (forward) 5’-CCCCAAGATCCTGAAACAGA-3’ and (reverse) 5’-CCGTTGCTGGACTGGATTAT-3’. Glyceraldehyde-6-phosphate was used as an internal control. Peripheral blood mononuclear cells (PBMCs) were cultured in complete Roswell Park Memorial Institute (RPMI) media supplemented with 10% FBS and used as a positive control for proto-oncogene up-regulation upon IL-2 (1 μg/ml) stimulation.
Stability of CD25 preselective anti-HIV vectors in transduced cells
To determine if there were any deletions or rearrangements of the vector transgenes in CD25 preselective anti-HIV vector-transduced cells, PCR was performed. Total genomic DNA was extracted from nontransduced and vector-transduced CD34+ HPC-derived macrophages using the Wizard Genomic DNA Isolation System (Promega, Madison, WI). PCR was performed using high fidelity Taq and primers corresponding to vector-specific sequences: MNDU3 (forward) 5’-CGCCCTCAGCAGTTTCTAGAG-3’, TRIM5α (forward) 5’-CATTATTTACGTTTCCGTCACTCACG-3’, CCR5 shRNA (forward) 5’-GGACGAAACACCGAGCATGACTGACATC-3’, CCR5 shRNA (reverse) 5’-GATGTCAGTCATGCTCGGTGTTTCGTCC-3’, and CD25 (reverse) 5’-AACTCATCAGATTGTTCTTCTACTCTTCC-3’ (IDT DNA Technologies, Coralville, IA). Albumin was used as an internal control: (forward) 5’-TGAAACATACGTTCCCAAAGAGTTT-3’ and (reverse) 5’-CTCTCCTTCTCAGAAAGTGTGCATAT-3’. The CD25 preselective anti-HIV DNA plasmid used for vector production was utilized as a positive control. PCR products were visualized on an agarose gel.
Detection of anti-HIV gene expression
To detect expression of the three anti-HIV genes in transduced cells, QPCR was performed. Total RNA was extracted from NT and CMAP1 vector-transduced CD34+ HPC-derived macrophages using RNA-STAT-60 (Tel-Test Inc., Friendswood, TX). cDNA was synthesized using a QuantiMir RT kit (System Biosciences, Mountain View, CA) according to the manufacturer's protocol. QPCR was performed using a SYBR Green PCR Master Mix kit (Applied Biosystems, Foster City, CA). The primers used for detection of the anti-HIV genes are as follows: TRIM5α (forward) 5’-CTGGGTTGATGTGACAGTGG-3’ and (reverse) 5’-CGTGAGTGACGGAAACGTAA-3’, CCR5 shRNA 5’-GTAGATGTCAGTCATGCTC-3’, TAR decoy 5’-TGACGAAAATTCTTACTGAGCAAG, and control U6 snRNA 5’-CGCAAGGATGACACGCAAATTC-3’. These experiments were performed in triplicate.
HIV-1 challenge of vector-transduced macrophages
To determine whether the purified CMAP1 vector-transduced macrophages were capable of improved inhibition of HIV-1 infection compared to unpurified CMAP1 vector-transduced macrophages, cells were challenged with an R5-tropic BaL-1 strain of HIV-1 (MOI 0.05). On various days post-infection, supernatants were collected and analyzed for levels of HIV-1 replication by p24 antigen ELISA (Zeptometrix Corp., Buffalo, NY). On day 28 post-infection, the HIV-1-challenged cells were also visualized by microscopy.
Statistical analysis
Two sample t-tests were used for statistical analyses. They were conducted in R (version 2.10.1) for Windows. A significance level of 0.05 was used for testing hypotheses.
Results
Enrichment of cells transduced with the preselective vectors
A third-generation self-inactivating lentiviral vector, CCLc-MNDU3-X-PGK-X2, was used to derive the control (EGFP+) and the anti-HIV (CMAP1) preselective vectors. A selectable marker, human CD25, which is not normally found on the surface of CD34+ HPCs, was incorporated into the control EGFP+ and CMAP1 vectors under the control of the PGK promoter and used to purify vector-transduced cells (Fig. 1A). CMAP1 vector titers ranged between 2.3×108 to 3.8×108 transducing units/ml. The EGFP+ control vector was only used to initially test the strategy of utilizing CD25 as a selective protein in purifying transduced cells and to compare the purification levels to CMAP1 vector-transduced cells. By using EGFP in the same vector as CD25, we would be able to compare EGFP% positive cells to CD25% positive cells. To evaluate the levels of purification of vector-transduced cells, flow cytometry and quantitative PCR (QPCR) were performed on EGFP+ and CMAP1 vector-transduced CD34+ HPCs, respectively. As displayed in Figure 1B, the percentage of control EGFP+ vector-transduced cells in the total cell population increased on average from 14.5% EGFP+ unpurified to 81.9% EGFP+ after purification. This correlated with expression of CD25 on the surface of the transduced HPCs, which increased on average from 15.1% CD25+ before purification to 81.0% CD25+ post-purification. As displayed in Figure 1C, the levels of vector copy number per cell in the total cell population of CMAP1 vector-transduced cells increased on average from 0.38 vector copies per cell unpurified to 2.97 vector copies per cell after purification. This amounted to an average enrichment percentage of 85.3%. These data successfully demonstrate that upon expression of CD25 on the surface of cells transduced with the preselective vectors, a high level of enrichment can be achieved.
Vector stability and expression of anti-HIV genes
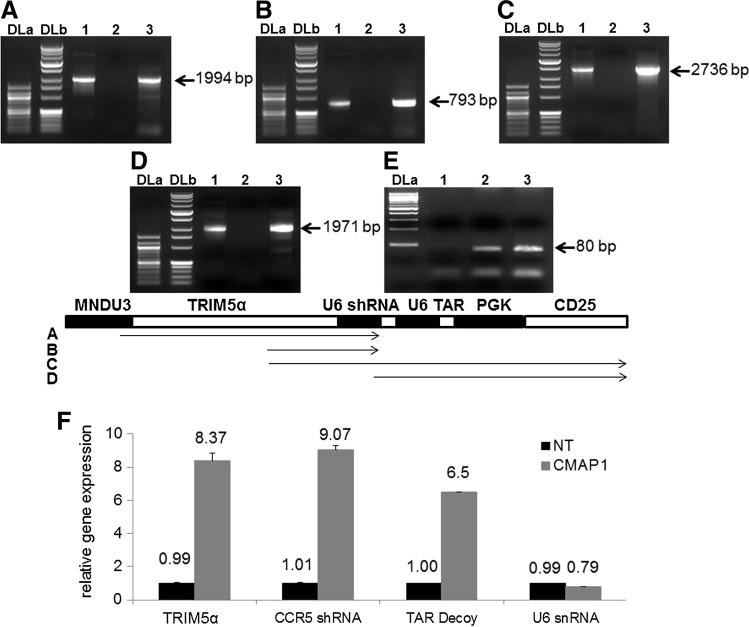
The anti-HIV preselective vector, CMAP1, is complex due to the incorporation of four different expression cassettes. Therefore, to evaluate the stability of the vector in transduced cells, PCR on genomic DNA was performed to detect vector transgene deletions or rearrangements using CMAP1 vector-specific primers: Figure 2A, MNDU3 forward and CCR5 shRNA reverse (1994 bp); Figure 2B, TRIM5α forward and CCR5 shRNA reverse (793 bp); Figure 2C, TRIM5α forward and CD25 reverse (2736 bp); Figure 2D, CCR5 shRNA forward and CD25 reverse (1971 bp); and Figure 2E, albumin forward and reverse (80 bp). As displayed in Figure 2, no deletions or rearrangements were detected as PCR bands corresponded to the appropriate sizes for both the positive control plasmid DNA (lane 1) and the CMAP1 vector-transduced cell-genomic DNA (lane 3). This was in contrast to NT cells (lane 2), which did not amplify the correct PCR products. A schematic of the proposed PCR products is displayed below the panels. Smaller bands were observed below the expected PCR size, however, these bands were faint and the major bands observed were those of the correct sizes. These results were similar to those obtained in our previous experiments (Anderson et al., 2009).
FIG. 2.
Stability of the CMAP1 vector and expression of the anti-HIV genes. Total genomic DNA was extracted from nontransduced (lane 2) and CMAP1 vector-transduced (lane 3) HPC-derived macrophages and analyzed by PCR with vector-specific primers. (A) MNDU3 (forward) and CCR5 shRNA (reverse); (B) TRIM5α (forward) and CCR5 shRNA (reverse); (C) TRIM5α (forward) and CD25 (reverse); and (D) CCR5 shRNA (forward) and CD25 (reverse). The CMAP1 plasmid used in vector production was utilized as a positive control (lane 1). DLa, DNA ladder a (100 bp), DLb, DNA ladder b (1 kb). A schematic of the PCR products is depicted below the gel pictures. (F) Total cellular RNA was extracted from either nontransduced (NT) or CMAP1 vector-transduced HPC-derived macrophages and analyzed by QPCR for the expression of the chimeric TRIM5α, CCR5 shRNA, and TAR decoy. U6 snRNA was used as an internal control.
To detect expression of the three anti-HIV genes, QPCR was performed on CMAP1 vector-transduced CD34+ HPC-derived macrophages. Expression of all three anti-HIV genes was detected as displayed in Figure 2F. The expression levels of the chimeric TRIM5α, CCR5 shRNA, and TAR decoy were on average 8.37 (standard deviation of 0.46), 9.07 (standard deviation of 0.23), and 6.50-fold (standard deviation of 0.00) higher, respectively, compared to control NT cells, which do not express any of the anti-HIV genes. The levels of the internal control U6 snRNA were similar among NT and CMAP1 cells.
Safety of CMAP1-transduced HPCs in CFU assays
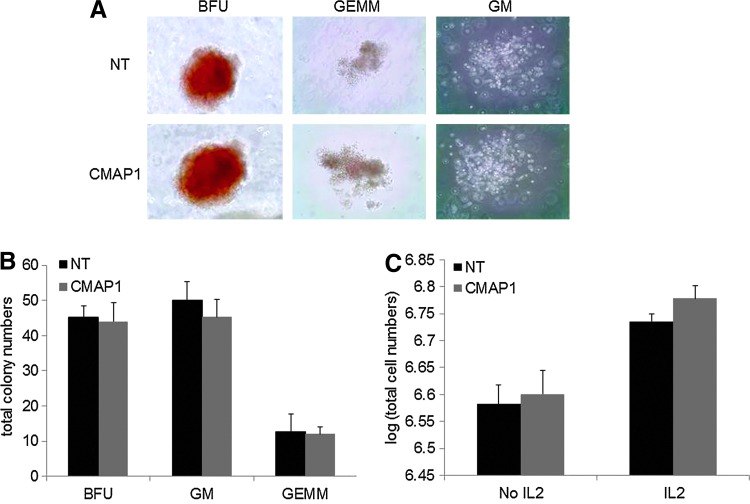
CD25 is not normally found on the surface of human HPCs. Therefore, to evaluate the effect of CD25 expression on HPCs and their ability to form normal quantities and types of hematopoietic colonies, CFU assays were performed on purified CMAP1 vector-transduced HPCs. As displayed in Figure 3A, normal colony phenotypes of BFUs, GEMMs, and GMs formed in the purified CMAP1 cultures compared to NT cultures. The representative pictures displayed were taken with a 10× objective. Total numbers of each type of colony were also counted. As displayed in Figure 3B, no significant differences (p>0.05) in the numbers of each colony, BFUs, GMs, and GEMMs had formed, on average, in the purified CMAP1 cultures, 44.0 BFUs, 45.3 GMs, and 12.0 GEMMs with standard deviations of 5.29, 5.03, and 2.00, respectively, compared to the NT cultures, 45.3 BFUs, 50.0 GMs, and 12.7 GEMMs with standard deviations of 3.06, 5.29 and 5.03, respectively.
FIG. 3.
Safety analysis of purified CMAP1-transduced HPCs. (A) HPCs, either nontransduced (NT) or purified CMAP1 vector-transduced, were cultured in semi-solid methylcellulose medium for 10 days and CFUs (BFUs, GEMMS, and GMs) were visualized by microscopy under 10× magnification. (B) Total BFU, GM, and GEMM colonies were counted on day 10 for the NT and purified CMAP1 cultures. (C) Total cells were counted in HPC methylcellulose NT and purified CMAP1 vector-transduced cultures in the absence or presence of IL-2. All experiments were performed in triplicate. Representative cell pictures are displayed. Color images available online at www.liebertpub.com/hgtb
As CD25 is part of the IL-2 receptor complex, which is involved in the immune response and immune cell proliferation, we wanted to investigate whether the addition of IL-2 to the purified CMAP1 vector-transduced HPC methylcellulose cultures resulted in an increase in HPC proliferation and in total cell numbers. As displayed in Figure 3C, no significant differences (p>0.05) in cell numbers were observed, on average, in the NT and CMAP1 cultures with the addition of IL-2 (106.74 and 106.78 total cells, respectively). An increase in total cell numbers was observed in the CMAP1 cultures with IL-2 compared to the cultures without IL-2 (an increase of 0.18 log); however, a similar increase was observed in the NT cultures (an increase of 0.16 log). This was likely due to the presence of cells that normally respond to IL-2 since during the course of the 10-day culture, the majority of the cells have differentiated.
These results highlight the initial safety of expressing CD25 on the surface of vector-transduced HPCs and demonstrated that no apparent aberrations in HPC differentiation or proliferation had occurred with overexpression of CD25.
Derivation of phenotypically normal macrophages from CMAP1-transduced HPCs
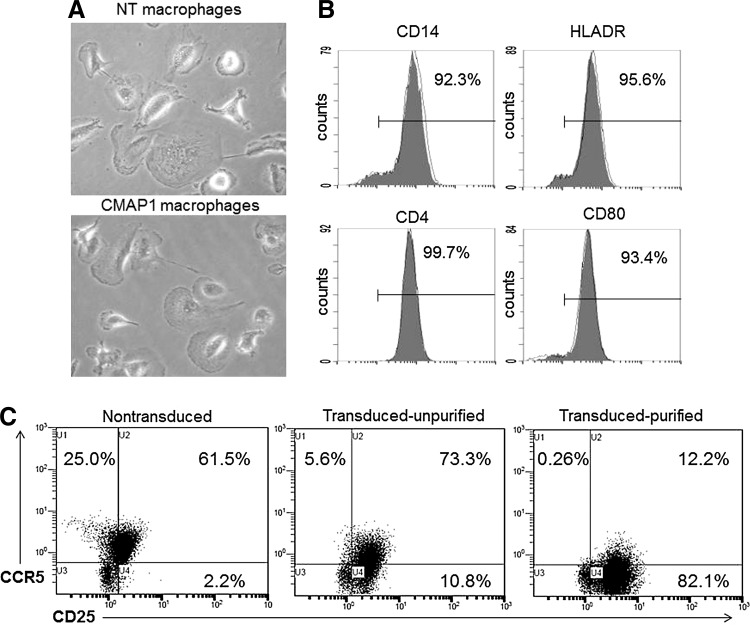
To determine whether normal immune cells could be derived from the purified CMAP1-transduced cells, macrophages were differentiated from the HPCs. Macrophage cultures were visualized by microscopy under a 10× objective. As displayed in Figure 4A, normal macrophage phenotypes were observed with the appearance of attached cells having a “fried-egg” appearance in both the NT and purified CMAP1 vector-transduced cultures. These macrophages were then phenotypically analyzed by flow cytometry for normal macrophage cell-surface markers. As displayed in Figure 4B, a normal phenotype of purified CMAP1 macrophages (shaded histograms) was demonstrated as compared to NT macrophages (unshaded histograms). Representative overlay histograms are displayed (Fig. 4B). CMAP1 cells displayed 92.3% of CD14, 95.6% of HLADR, 99.7% of CD4, and 93.4% of CD80.
FIG. 4.
Derivation of phenotypically normal macrophages. (A) Macrophages derived from the nontransduced (NT) and purified CMAP1 HPC cultures were visualized by microscopy under 10× magnification. (B) The NT (unshaded) and purified CMAP1 (shaded) macrophages were analyzed by flow cytometry for the cell-surface markers CD14, HLADR, CD4, and CD80. (C) Macrophages, NT, unpurified CMAP1 transduced, and purified CMAP1 transduced were also analyzed by flow cytometry for the expression of CD25 and CCR5. All experiments were performed in triplicate. Representative cell pictures and flow cytometry histogram overlays and cell plots are displayed.
Macrophages normally express a measureable level of CD25 on their cell surface. To evaluate any increase in cell surface expression of CD25 on CMAP1-transduced macrophages, flow cytometry was performed. As displayed in Figure 4C, a gradient increase was observed, on average, in CD25 expression on the surface of macrophages from NT macrophages (63.7%) to unpurified CMAP1-transduced macrophages (84.1%) to purified CMAP1-transduced macrophages (94.3%). This increase in CD25 expression, which correlates to CMAP1 vector transduction, resulted in the downregulation of CCR5 due to the expression of the CCR5 shRNA. In purified CMAP1 macrophages, CCR5 expression was 12.5%, which compared to NT macrophages (86.5%) and unpurified CMAP1 macrophages (78.9%).
These results demonstrate that phenotypically normal macrophages can be derived from purified CMAP1 vector-transduced HPCs and that overexpression of CD25 did not disrupt normal macrophage differentiation from HPCs. These results also demonstrate that the CMAP1 vector worked as hypothesized by displaying an increase in CD25 expression on purified populations of transduced cells while decreasing expression of CCR5 due to the expression of the CCR5 shRNA.
Proto-oncogene expression in purified CMAP1 macrophages
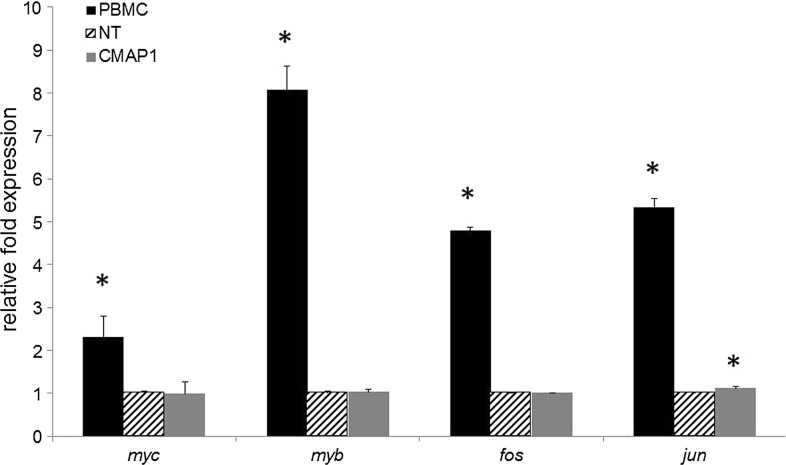
As mentioned above, CD25 is part of the IL-2 receptor complex, which is involved in the cell proliferation of immune cells. After IL-2 binds to the IL-2 receptor, expression of proto-oncogenes, including myc, myb, fos, and jun, are upregulated to promote cell division. To evaluate whether overexpression of CD25 on the surface of purified CMAP1 macrophages upregulated the expression of the mentioned proto-oncogenes, QPCR was performed on total RNA from macrophage cultures with the addition of IL-2. PBMCs were used as a positive control for proto-oncogene upregulation in the presence of IL-2. As displayed in Figure 5, similar levels of proto-oncogene expression were measured in NT and purified CMAP1 macrophage cultures. No significant difference (p>0.05) was observed with the expression of myc, myb, and fos in CMAP1 macrophages compared to NT macrophages in the presence of IL-2. However, an average relative expression level of 1.1 (standard deviation of 0.04) for jun (p=0.0121, statistically significant) was observed with CMAP1 macrophages compared to NT macrophages, which were used as the reference cells with an average relative expression level of 1.0 (standard deviation of 0.04). As a positive control for proto-oncogene upregulation, PBMCs were cultured in the presence of IL-2. A significant upregulation in the expression of myc (2.3-fold) (p=0.0418), myb (8.1-fold) (p=0.0018), fos (4.8-fold) (p=0.0001), and jun (5.3-fold) (p=0.0004) was observed in the IL-2 stimulated PBMC cultures relative to NT macrophages.
FIG. 5.
Expression of proto-oncogenes in purified CMAP1 macrophages. Cell cultures containing either peripheral blood mononuclear cells (PBMCs), nontransduced (NT) HPC-derived macrophages, or purified CMAP1 vector-transduced HPC-derived macrophages were evaluated by QPCR for their expression of the proto-oncogenes myc, myb, fos, and jun in the presence of IL-2. Experiments were performed in triplicate. Statistical significance (p<0.05) is represented by an asterisk.
These results demonstrate that even though the purified CMAP1 macrophages express an increased level of CD25, proto-oncogene expression levels remained similar to nontransduced cells.
HIV-1 inhibition of CMAP1 HPC-derived macrophages
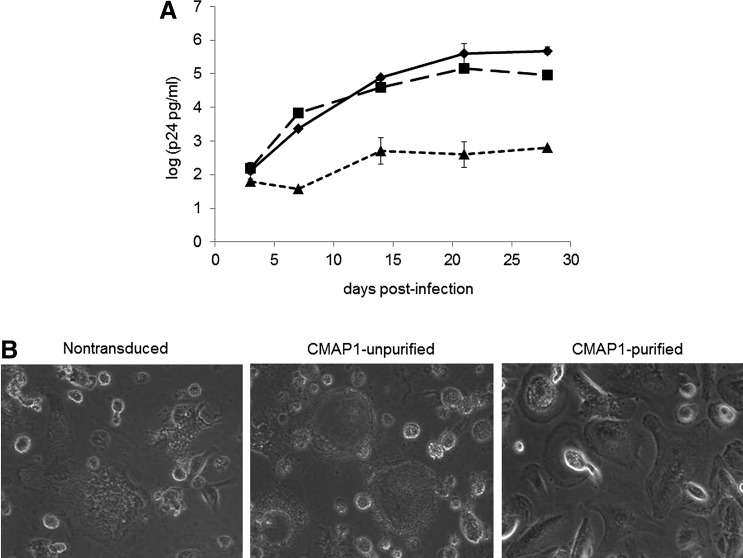
Previous challenge experiments performed with this triple combination of anti-HIV genes, both in vitro and in vivo, demonstrated strong viral inhibition to both CCR5 and CXCR4-tropic strains of HIV-1 (Anderson et al., 2009; Kohn et al., 1999). These experiments, however, relied on sorting the cells based on a nonclinically acceptable reporter gene, EGFP, prior to viral challenge. To evaluate whether purified CMAP1 macrophages displayed an increased efficacy of HIV-1 inhibition compared to NT and unpurified CMAP1 macrophages, cells were challenged with an R5-tropic BaL-1 strain of HIV-1. As displayed in Figure 6A at the end of the challenge experiments, potent inhibition of HIV-1 infection was observed in the purified CMAP1 cultures compared to NT macrophages (2.9 log difference) and unpurified CMAP1 macrophages (2.2 log difference). A slight inhibition of HIV-1 infection was observed in the unpurified CMAP1 macrophage cultures (0.7 log difference) compared to NT macrophages due to the presence of anti-HIV gene-expressing cells. On day 28 post infection, HIV-1 infected macrophages were also visualized by microscopy under 10× magnification. As displayed in Figure 6B with representative pictures, cell death from HIV infection was observed throughout the cultures of infected NT and unpurified CMAP1 macrophages. This was in comparison to purified CMAP1 macrophages, which displayed a small amount of cell death but where the majority of macrophages still appeared healthy.
FIG. 6.
HIV-1 challenge of CMAP1 HPC-derived macrophages. (A) HPC-derived macrophages, either nontransduced (NT) (♦), unpurified CMAP1-transduced (CMAP1-UP) (▪), or purified CMAP1 (CMAP1-P) (▲)-transduced, were challenged with an R5-tropic BaL-1 strain of HIV-1 at an MOI of 0.05. On various days postinfection, culture supernatants were analyzed for HIV-1 replication by p24 ELISA. (B) On day 28 post-infection, infected cultures were visualized by microscopy under 10× magnification. All experiments were performed in triplicate. Representative cell pictures are displayed.
These results highlight the increased efficacy of anti-HIV vector-transduced cells when they are purified to a cell population in which the majority of the cells express the anti-HIV genes and demonstrate the utility of this novel CMAP1 vector.
Discussion
HIV gene therapy holds considerable promise as an alternative treatment strategy for HIV-infected patients. As observed with the Berlin patient who received a pure population of HIV-resistant hematopoietic stem cells in a bone marrow transplant from a donor homozygous for a CCR5 Δ32 bp allele, HIV-resistant stem cells are capable of repopulating the immune system with cells, which can inhibit HIV infection in the absence of ART for a prolonged period of time (Hutter et al., 2009). The safety of numerous anti-HIV genes has been demonstrated in previous HIV stem cell gene therapy clinical trials (DiGuisto et al., 2010; Mitsuyasu et al., 2009; Podsakoff et al., 2005). However, patients were given a mixed population of cells, with the vast majority not being transduced with anti-HIV genes. This led to the derivation of an immune system where the majority of the immune cells were still susceptible to HIV infection. A low level of efficacy was observed with a measureable increase in the levels of anti-HIV gene-expressing cells in the presence of a viral load; however, initial transduction efficiencies and in vivo gene marking were too low for patients to remain off ART. If a pure population of anti-HIV gene-transduced cells could be transplanted into patients, similar results observed with the Berlin patient may be achievable.
We have recently demonstrated strong ex vivo resistance of infection to multiple strains of HIV-1 and a selective survival advantage of cells transduced with a triple combination anti-HIV lentiviral vector expressing a human/rhesus macaque TRIM5α, a CCR5 shRNA, and a TAR decoy in vivo in a humanized mouse model (Walker et al., 2012). However, plasma viremia levels in HIV-1 infected mice transplanted with anti-HIV vector-transduced cells did not decrease over time due to the transplantation of a majority of nontransduced HSCs that continually produced HIV susceptible target cells (Walker et al., 2012). If anti-HIV vector-transduced cells could be enriched to a pure population or at least to a population in which the majority of the cells express the anti-HIV genes, improved in vivo efficacy may be demonstrated.
Current reporter genes for cell sorting are not clinically relevant due to their foreign nature, which would invoke an immune response against transduced cells. Another approach similar to the one presented here utilizes a P140K mutant methylguanine methyltransferase (MGMT) transgene to select for transduced cells in vivo after transplantation into patients. This, however, would require another patient infusion with agents that would select for vector-transduced cells (Trobridge et al., 2009). Therefore, as a first step to improve on the efficacy of HIV gene therapy, we have developed preselective anti-HIV lentiviral vectors that express a normal human cell surface protein as to avoid rejection of transduced cells and allow for the purification of vector-transduced cells prior to transplantation. Human CD25 was chosen as a selectable marker based on its characteristics of not being expressed on the surface of HPCs or HSCs, it is a normal immune cell surface protein, and it has been previously shown to have no direct intracellular signaling (Grant et al., 1992; Kuziel et al., 1990; Minami et al., 1993).
When overexpressing a protein on the surface of cells, especially cells that do not normally express the protein, the safety and function of the engineered cells is a concern. Upon transduction and high-level purification (>85%) of CMAP1 vector-transduced HPCs, phenotypically normal CFUs and the number of CFUs formed in methylcellulose medium were similar to nontransduced cells (Fig. 3). Safety was also observed with macrophages derived from the purified CMAP1 vector-transduced HPCs as they appeared phenotypically normal compared to nontransduced macrophages (Fig. 4).
As CD25 is part of the IL-2 receptor complex, we wanted to investigate whether the over expression of CD25 in purified vector-transduced cells had any effect on cell proliferation or the upregulation of proto-oncogenes in the addition of IL-2. In the presence of IL-2, no adverse effects were observed in purified CMAP1 vector-transduced HPCs or macrophages. No increased cell proliferation of HPCs (Fig. 3) and no increase in the expression of the proto-oncogenes myc, myb, or fos (Fig. 5) was observed compared to nontransduced cells. A small increase in the expression of jun in CMAP1 vector-transduced macrophages was observed (1.1) compared to NT cells (1.0). This difference was, however, significantly different to the positive control PBMCs that displayed a 5.3 relative increase in expression of jun. No other adverse effects were observed with purified CMAP1 vector-transduced cells. Our findings of a lack of cell proliferation and proto-oncogene upregulation are likely due to CD25's normal function of being involved with the assembly of the IL-2 receptor but having no mitotic signaling capabilities (Grant et al., 1992; Kuziel et al., 1990; Minami et al., 1993).
The ultimate goal of HIV stem cell gene therapy is to provide an alternative therapeutic intervention for HIV-infected patients and to provide a possibility for them to withdraw ART medications. For this to be achievable, an enriched population of anti-HIV gene-transduced cells with little to no nontransduced cells needs to be transplanted. Upon HIV-1 challenge of purified CMAP1 vector-transduced HPC-derived macrophages, strong inhibition of HIV-1 infection was observed. CMAP1 vector-transduced cells that were not purified displayed high levels of HIV-1 replication similar to nontransduced macrophage cultures. Even though unpurified CMAP1 cultures contained anti-HIV gene-expressing cells, the majority of the cells were nontransduced and, thus, were capable of being infected and replicating HIV-1 at a high level. This highlights the improved efficacy of this preselective anti-HIV vector and allows for the purification of anti-HIV gene-expressing cells to an enriched population that could potentially be used in future clinical trials for HIV stem cell gene therapy.
In previous studies of graft-versus-host-disease (GVHD), conflicting results have been observed over the reactivity of CD25+ regulatory T-cells. One study explained that chronic GVHD was associated with an increased number of CD4+/CD25+ T-cells (Clark et al., 2004). Contrastingly, QPCR studies demonstrated decreased expression levels of FOXP3 in both acute and chronic GVHD patients (Miura et al., 2004). Yet another study observed decreased levels of CD4+/CD25+ T-cells and decreased levels of FOXP3 expression in patients with chronic GVHD compared to patients without chronic GVHD (Zorn et al., 2005). However, when tested in functional assays, the regulator T-cells from both patients and from healthy donors acted similarly (Zorn et al., 2005). Therefore, the use of CD25 as a preselective molecule may have limitations. However, these previous reports have focused on allogeneic transplants where we are planning on using autologous HSCs from patients. Also, in future experiments, truncated and inactive forms of CD25 may be used as preselective molecules. As these experiments were proof-of-concept to demonstrate increased efficacy with the preselective lentiviral vector method, our group is currently evaluating other cell surface proteins for their use as purification molecules.
Acknowledgments
This work was supported by the University of California Davis Health System start-up funds from the dean's office for the Stem Cell Program and by the James B. Pendleton Charitable Trust. This work was also supported in part by the Gin and Imy Mar stem cell research fund. The NIH AIDS Research and Reference Reagent Program provided the HIV-1 viral strains. Biostatistics support for this publication was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through grant number TR 000002.
Disclosure Statement
No competing financial interests exist.
References
- Anderson J.S. Javien J. Nolta J.A., et al. Preintegration HIV-1 inhibition by a combination lentiviral vector containing a chimeric TRIM5alpha protein, a CCR5 shRNA, and a TAR decoy. Mol. Ther. 2009;17:2103–2114. doi: 10.1038/mt.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldanti F. Paolucci S. Gulminetti R., et al. Early emergence of raltegravir resistance mutations in patients receiving HAART salvage regimens. J. Med. Virol. 2010;82:116–122. doi: 10.1002/jmv.21651. [DOI] [PubMed] [Google Scholar]
- Barouch D.H. Challenges in the development of an HIV-1 vaccine. Nature. 2008;455:613–619. doi: 10.1038/nature07352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer G. Valdez P. Kearns K., et al. Inhibition of human immunodeficiency virus-1 (HIV-1) replication after transduction of granulocyte colony-stimulating factor-mobilized CD34+ cells from HIV-1-infected donors using retroviral vectors containing anti-HIV-1 genes. Blood. 1997;89:2259–2267. [PubMed] [Google Scholar]
- Clark F.J. Gregg R. Piper K., et al. Chronic graft-versus-host-disease is associated with increased numbers of peripheral blood CD4+CD25high regulatory T cells. Blood. 2004;103:2410–2416. doi: 10.1182/blood-2003-06-2073. [DOI] [PubMed] [Google Scholar]
- DiGiusto D. Krishnan A. Li L., et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010;36:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo P. Estrada V. Lopez-Aldeguer J., et al. Fat redistribution syndromes associated with HIV-1 infection and combination antiretroviral therapy. AIDS Rev. 2012;14:112–123. [PubMed] [Google Scholar]
- Edgeworth R.L. San J.H. Rosenweig J.A., et al. Vaccine development against HIV-1: current perspectives and future directions. Immunol. Res. 2002;25:53–74. doi: 10.1385/ir:25:1:53. [DOI] [PubMed] [Google Scholar]
- Gazzola L. Tincati C. Bellistri G.M., et al. The absence of CD4+ T cell count recovery despite receipt of virologically suppressive highly active antiretroviral therapy: clinical risk, immunological gaps, and therapeutic options. Clin. Infect. Dis. 2009;48:328–337. doi: 10.1086/595851. [DOI] [PubMed] [Google Scholar]
- Graham SM. Jalalian-Lechak Z. Shafi J, et al. Antiretroviral treatment interruptions predict female genital shedding of genotypically resistant HIV-1 RNA. J. Acquir. Immune. Defic. Syndr. 2012;60:511–518. doi: 10.1097/QAI.0b013e31825bd703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A.J. Roessler E. Ju G., et al. The interleukin 2 receptor (IL-2R) the IL-2R alpha subunit alters the function of the IL-2R beta subunit to enhance IL-2 binding and signaling by mechanisms that do not require binding of IL-2 to IL-2R alpha subunit. Proc. Natl. Acad. Sci. USA. 1992;89:2165–2169. doi: 10.1073/pnas.89.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter G. Nowak D. Mossner M., et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- Johnson V.A. Brun-Vezinet F. Clotet B., et al. Update of the drug resistance mutations in HIV-1: December 2010. Top. HIV Med. 2010;18:156–63. [PubMed] [Google Scholar]
- Kalmar E.M. Sanabani S.S. Charlys da Costa A, et al. Evaluation of HIV-1 resistance to antiretroviral drugs among 150 patients after six months of therapeutic interruption. Int. J. STD. AIDS. 2012;23:120–125. doi: 10.1258/ijsa.2011.011124. [DOI] [PubMed] [Google Scholar]
- Kohn D.B. Bauer G. Rice C.R., et al. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood. 1999;94:368–371. [PubMed] [Google Scholar]
- Kuritzkes D.R. Drug Resistance in HIV-1. Curr. Opin. Virol. 2011;1:582–589. doi: 10.1016/j.coviro.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuziel W.A. Greene W.C. Interleukin-2 and IL-2 receptor: new insights intostructure and function. J. Invest. Dermatol. 1990;94:27S–32S. doi: 10.1111/1523-1747.ep12875017. [DOI] [PubMed] [Google Scholar]
- Lewden C. Chene G. Morlat P., et al. HIV-infected adults with a CD4 cell count greater than 500 cells/mm3 on long-term combination antiretroviral therapy reach same mortality rates as the general population. J. Acquir. Immune. Defic. Syndr. 2007;46:72–77. doi: 10.1097/QAI.0b013e318134257a. [DOI] [PubMed] [Google Scholar]
- Macias J. Neukam K. Mallolas J., et al. Liver toxicity of initial antiretroviral drug regimens including two nucleoside analogs plus one non-nucleoside analog or one ritonavir-boosted protease inhibitor in HIV/HCV-coinfected patients. HIV Clin. Trials. 2012;13:61–69. doi: 10.1310/hct1302-61. [DOI] [PubMed] [Google Scholar]
- Mehandru S. Poles M.A. Tenner-Racz K., et al. Lack of mucosal immune reconstitution during prolonged treatment of acute and early HIV-1 infection. PLoS Med. 2006;3:e484. doi: 10.1371/journal.pmed.0030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y. Kono T. Miyazaki T., et al. The IL-2 receptor complex: Its structure, function, and target genes. Annu. Rev. Immunol. 1993;11:245–267. doi: 10.1146/annurev.iy.11.040193.001333. [DOI] [PubMed] [Google Scholar]
- Mitsuyasu R.T. Merigan T.C. Carr A., et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat. Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsakoff G.M. Engel B.C. Carbonaro D.A., et al. Selective survival of peripheral blood lymphocytes in children with HIV-1 following delivery of an anti-HIV gene to bone marrow CD34(+) cells. Mol. Ther. 2005;12:77–86. doi: 10.1016/j.ymthe.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Shimizu S. Hong P. Arumugam B., et al. A highly efficient short hairpin RNA potently down-regulates CCR5 expression in systemic lymphoid organs in the hu-BLT mouse model. Blood. 2010;115:1534–1544. doi: 10.1182/blood-2009-04-215855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer D.S. Akkina R. Bunnel B.A., et al. Current Status of Gene Therapy Strategies to Treat HIV/AIDS. Mol. Ther. 2005;11:823–841. doi: 10.1016/j.ymthe.2005.01.020. [DOI] [PubMed] [Google Scholar]
- ter Brake O. Legrand N. von Eije K.J., et al. Evaluation of safety and efficacy of RNAi against HIV-1 in the human immune system (Rag-2(-/-)gammac(-/-)) mouse model. Gene Ther. 2009;16:148–153. doi: 10.1038/gt.2008.124. [DOI] [PubMed] [Google Scholar]
- Tilton J.C. Wilen C.B. Didigu C.A., et al. A maraviroc resistant HIV-1 with narrow cross-resistance to other CCR5 antagonists depends on both N-terminal and extracellular loop domains of drug-bound CCR5. J. Virol. 2010;84:10863–10876. doi: 10.1128/JVI.01109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobridge G.D. Wu R.A. Beard B.C., et al. Protection of stem cell-derived lymphocytes in a primate AIDS gene therapy model after in vivo selection. PloS One. 2009;4:e7693. doi: 10.1371/journal.pone.0007693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.E. Chen R.X. McGee J., et al. Generation of an HIV-1-resistant immune system with CD34(+) hematopoietic stem cells transduced with a triple-combination anti-HIV lentiviral vector. J. Virol. 2012;86:5719–5729. doi: 10.1128/JVI.06300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn E. Kim H.T. Lee S.J., et al. Reduced frequency of FOXP3+ CD4+CD25+ regulatory T cells in patients with chronic graft-versus-host disease. Blood. 2005;106:2903–2911. doi: 10.1182/blood-2005-03-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]