Abstract
Helper-dependent adenoviral vectors (HD Ad) hold extreme promise for gene therapy of human diseases. All viral genes are deleted in HD Ad vectors, and therefore, the presence of a helper virus is required for their production. Current methods to minimize helper contamination in large-scale preparations rely on the use of the Cre/loxP system. The inclusion of loxP sites flanking the packaging signal results in its excision in the presence of Cre recombinase, preventing helper genome encapsidation. It is well established that the level of Cre recombinase activity is important in determining the degree of helper contamination. However, there is little information on other mechanisms that could also play an important role. We have generated several HD Ad vectors containing a rapalog-inducible system to regulate transgene expression, or LacZ under the control of the elongation factor 1 α promoter. Large-scale production of these vectors resulted in abundant helper contamination. Viral DNA analysis revealed the presence of rearrangements between vector and helper genomes. The rearrangements involved a helper DNA molecule with a fragment of the left arm of the HD Ad vector, including its ITR, packaging signal, and some stuffer sequence. Overall, our data suggest that helper DNA molecules that accumulate after Cre recombinase activity are prone to rearrangements, resulting in helper genomes that have incorporated a packaging signal from the vector. Helper particles with rearranged genomes have a growth advantage. This study identifies a novel mechanism leading to helper contamination during helper-dependent adenoviral vector production.
Ahn and colleagues demonstrate that extensive rearrangements occur between vector and helper genomes during helper dependent adenoviral (HD Ad) vector production. The rearrangements involve a helper DNA molecule with a fragment of the left arm of the HD Ad vector, including its inverted terminal repeat, packaging signal, and some stuffer sequences.
Introduction
Adenoviral vectors (Ad) are a useful delivery system for human gene therapy, and one of the most efficient viral vehicles for transferring genes into nondividing cells (Benihoud et al., 1999; Mountain, 2000). Ad vectors can easily be grown to high titers for use in large animals, and they lack risks associated with integration, as their genome is episomally maintained in the host cell (Mountain, 2000). Ad vectors have a natural tropism for the liver (Morral et al., 1999; Kim et al., 2001). Recent studies showed that hepatocyte transduction in vivo is largely mediated by the interaction of the capsid with blood-borne components such as factors II, VII, IX, and X, and anti-coagulant protein C (Shayakhmetov et al., 2005; Parker et al., 2006, 2007; Waddington et al., 2007, 2008).
Helper-dependent adenoviral vectors (HD Ad, also known as gutless or high-capacity vectors) are devoid of all viral-coding sequences and only retain the inverted terminal repeats (ITRs) as well as the packaging signal. HD Ad vectors can accommodate up to 37 kb of DNA, which allows for the delivery of multiple transgenes, and have shown to result in long-term transgene expression in the absence of adaptive immune responses or significant toxic effects (Parks and Graham, 1997; Morral et al., 1998; Schiedner et al., 1998). Several liver-directed gene therapies using HD Ad vectors in vivo have been reported, such as apolipoprotein E (ApoE) expression to treat atherosclerotic plaque formation, uridinediphosphoglucuronate glucuronosyltransferase (UGT1A1) gene replacement, and pancreatic duodenal homeobox-1 (Ipf1, also known as Pdx-1) expression to treat diabetes mellitus (Kim et al., 2001; Kojima et al., 2003; Strauss et al., 2006). In addition, recent advances in the RNAi field have multiplied the number of possible applications. Short hairpin RNA (shRNA)-mediated gene knockdown has become the most popular method for gene function studies in mammalian cells (Brummelkamp et al., 2002) and in vivo using viral vectors (Grimm and Kay, 2006; Ruiz et al., 2009). Currently, the Cre/loxP system is the most efficient method for generating HD Ad vectors (Parks et al., 1996; Sandig et al., 2000; Palmer and Ng, 2003). In order to obtain efficient packaging of vector genomes, the inclusion of stuffer DNA is required to maintain a genome size of at least 27 kb, which allows efficient and stable amplification during vector production. Genome sizes below 75% (<27 kb) or above 105% (>37.8 kb) tend to undergo DNA rearrangements to increase or decrease their size, respectively (Parks and Graham, 1997).
Several new tools for modulating gene expression in response to small molecule drugs have been developed (Clackson, 2000). In the dimerizer system (Pollock et al., 2000), expression relies on a DNA-binding domain and a transcription-activation domain fused to a drug-binding domain (Chen et al., 1995; Bayle et al., 2006). In the presence of a nonimmunosuppressive analog of rapamycin, rapalog AP21967, the two chimeric proteins dimerize, binding to a unique DNA target sequence in the promoter of the transgene (Rivera et al., 1996). This system has demonstrated tight, ligand-inducible control of gene expression in adeno-associated virus (AAV) and in first-generation adenovirus vectors (Rivera et al., 1999; Ye et al., 1999; Auricchio et al., 2002; Chong et al., 2002; Sanftner et al., 2006). A system that can be regulated by administration of a small molecule drug is also extremely useful in gene function studies, in particular for studies involving the liver. We have generated helper-dependent adenovirus-expressing E. coli LacZ under the control of the EF1α promoter, as well as vectors expressing carnitine palmitoyltransferase 1A (CPT1A) and short hairpin RNAs to target fatty acid-binding protein 5 (FABP5) under the control of the dimerizer system. CPT1A is the key enzyme in the carnitine-dependent fatty acid transport across the mitochondrial membrane and its deficiency results in a decreased rate of fatty acid β-oxidation (McGarry and Brown, 1997). FABP5 is a 15 kDa cytosolic protein that binds long chain fatty acids with high affinity and regulates fatty acid trafficking (Hoekstra et al., 2006). Here we report that vector and helper genome rearrangements can occur during production of HD Ad vectors, resulting in helper virus overgrowth.
Materials and Methods
Shuttle plasmids
The Cre/loxP system to rescue HD Ad adenoviral vectors uses the 30.9-kb shuttle plasmid pC4HSU (Sandig et al., 2000; Ruiz et al., 2009) (here named pSHL). When rescued into an adenoviral vector, it results in a viral genome of ∼29 kb. To accommodate different expression cassettes, pSHL was modified so that the final size of the virus genome would be 26 to 30 kb. Three different pSHL plasmids were generated: pSHL-23.8, pSHL-24.4, and pSHL-26.5 through removal of stuffer sequence. pSHL-23.8 was generated by pSHL digestion with SbfI and SwaI, which removed 7.1 kb of stuffer DNA. pSHL-26.5 was generated by pSHL digestion with SwaI and NotI, which removed 4.5 kb of stuffer DNA. pSHL-26.5 was digested with BaeI to remove 2 kb and generate pSHL-24.4. Table 1 shows the modified pSHL plasmid used to generate each of the HD Ad vectors.
Table 1.
Helper-Dependent Adenoviral Vectors Generated
| pSHL plasmid | pSHL-30.9 | pSHL-30.9 | pSHL-30.9 | pSHL-23.8 | pSHL-24.4 | pSHL-23.8 |
|---|---|---|---|---|---|---|
| Plasmid size (bp) | 30,989 | 30,989 | 30,989 | 23,889 | 24,429 | 23,889 |
| pBluescript (bp) | 2,890 | 2,890 | 2,890 | 2,890 | 2,890 | 2,890 |
| Expression cassette/s (bp) | 3,178 | 6,138 | 6,147 | 8,788 | 5,721 | 5,721 |
| Total size (bp) | 34,167 | 37,127 | 37,136 | 32,677 | 30,150 | 29,610 |
| HD Ad vector (genome size, bp) | gAd.EF1α FABP5 (31,277) |
gAd.AlbLacZ (34,237) |
gAd.EF1αLacZ (34,246) |
gAd.DBD-CPT1AWT; gAd.DBD-CPT1E3A (29,787) |
gAd.DBD-shFABP5 (27,260) |
gAd.DBD-shSCR; gAd.DBD-shFABP5 (26,720) |
Helper-dependent adenovirus rescue, propagation, and purification
Rescue and large-scale preparations of adenoviral vectors were done in 293Cre4 cells using H14 helper (Microbix Biosystems Inc.) (Chen et al., 1996; Parks et al., 1996), as previously described (Sandig et al., 2000; Ruiz et al., 2009). Plasmids pSHL-DBD/PL2-shRNA-mpA, pSHL-DBD/CPT1A, and pSHL-GFP (as a control) were digested overnight with PmeI to release pBluescript. 293Cre4 cells were transfected with 5 μg of PmeI-digested DNA in a 6-cm dish using Metafectene™ Pro (Biontex). The next day, helper H14 virus was added at multiplicity of infection (MOI) 3. Cells were incubated until 95% cytopathic effect (CPE) was observed. Cells were freeze-thawed and an aliquot was used to infect fresh cells and repeat the process. A total of six passages were conducted. 293Cre4 cells grown at 95% confluency in 15 triple flasks (500 cm2; 5–7×107 cells/flask) were infected with 15 ml of cell lysates and helper H14 at MOI 2. Cells were incubated at 37°C in a 5% CO2 atmosphere and were harvested upon presence of >90% cytopathic effect (CPE). The virus was purified by one CsCl step gradient centrifugation followed by one CsCl isopycnic separation. The helper-dependent adenovirus band was collected and dialyzed in TMN buffer (10 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 150 mM NaCl, 10% glycerol). Total virus particles were determined spectrophotometrically after particle disruption with 0.1% sodium dodecyl sulfate (SDS) (absorbance at 260 nm [A260]=1 corresponds to 1.1×1012 virus particle/mL). Stocks were stored at −80°C.
Plaque assay
Plaque assays were performed in low passage HEK 293 cells. Cells plated in 6-well plates were infected with 1 ml of 10−5 to 10−10 serial dilutions of virus. After overnight incubation, medium was removed and cells were covered with 4 mL of overlay solution (0.5% agarose, 1× MEM, 1× penicillin/streptomycin, 5% horse serum, 5 mM Hepes, and 0.05% yeast extract). Cells were incubated at 37°C. After 5 days, 2 mL of overlay solution was added to cells. Plaques were counted 10 days postinfection; the titer was estimated after adjusting for dilution.
Cell culture
HEK293 cells were cultured in MEMα supplemented with 10% fetal bovine serum (FBS). To evaluate the level of down-regulation with the shRNA-expressing constructs, plasmids expressing the mouse target gene under the control of the elongation factor 1 α (EF1α) promoter were used (Witting et al., 2008; Ruiz et al., 2009). HEK293 cells were cotransfected with plasmid pEF1αFABP5, pDBD and pBS-PL2-shRNA-mpA at 1:2:1 ratio using Metafectene™ Pro (Biontex) using a 1:3 plasmid to lipid ratio. Transfected cells were incubated in the absence or presence of rapalog AP21967 at concentrations of 0, 0.05, 0.25, 0.5, or 1.0 μM. Six hours later, fresh media with rapalog was added to cells. Cells were harvested 48 hr later and lysed using RIPA buffer (Thermo Scientific), containing protease inhibitors (Roche). All experiments were carried out in duplicates. Primary hepatocytes were isolated from C57BL/6 mice as previously described (Park et al., 2011). Wells were seeded with 4×105 cells, and 5 hours later cells were infected with gAd.DBD-CPT1AWT and rapalog AP21967 was added at concentrations of 0.25, 0.5, 1.0, 1.5, or 3.0 μM. After overnight infection, fresh media with rapalog was added to cells. Cells were harvested 48 hr later and lysed using RIPA buffer with protease inhibitors.
Western blotting
Proteins (20–30 μg) were separated in 4–15% Tris-HCl or 3–8% XT SDS PAGE Criterion gel (Bio-Rad) and transferred to 0.2-mm polyvinylidene fluoride (PVDF) membrane (Bio-Rad). Antibodies to FABP5, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-actin were purchased from R&D Scientific and Santa Cruz Biotechnology, Inc. The antibody against CPT1A was kindly provided by Dr. Carina Prip-Buus (INSERM, U1016, Institut Cochin). Blots were developed with Pierce ECL kit (Thermo Scientific) and exposed to enhanced chemiluminescence (ECL) film (GE Healthcare).
Viral DNA sequence analysis
Viral DNA was extracted from large-scale vector preparations. SDS (1%) and 100 μg/mL proteinase K (Roche) were added to 500 μl virus aliquots and incubated for 30 min at 37°C. DNA was extracted with phenol/chloroform and ethanol precipitated using 3 M sodium acetate (pH 5.2). DNA was resuspended in 20 μL dH2O. Viral DNAs were sequenced by the DNA Sequencing Core Facility at Indiana University School of Medicine using the primers shown in Table 2.
Table 2.
Primers Used for Sequencing/Polymerase Chain Reaction
| Section 1 | H14-loxP-R2 | 5′-ATGCGCGTTGTCAAATATGAG-3′ |
| H14-loxP-R | 5′-GCTCTAGACTCGAGGGATCTG-3′ | |
| Section 2 | H14-25582-F | 5′-AACTCTACGGGCTATTCTAAT-3′ |
| H14-28534-R | 5′-AAGACAGAGAATAACCCCAAC-3′ | |
| H14-27781R | 5′-ACTGTTTTCACTCTCTCAT-3′ | |
| Section 3 | H14-35356-F | 5′-CCTGAAAAACCCTCCTGCCTA-3′ |
| Section 4 | FKBP-13345-F | 5′-AACAATCAAGGTCCCCAAAC-3′ |
| FKBP-14432-R | 5′-GTGAATTCGTGTTTGCTTCTGT-3′ | |
| Section 5 | C4HSU-PS-F | 5′-TTGTAGTAAATTTGGGCGTAA- 3′ |
Polymerase chain reaction
To characterize the helper virus rearrangements, polymerase chain reaction (PCR) analysis to amplify the left and right arm of helper H14 viral genome was performed using 100 ng virus DNA isolated from large-scale HD Ad vector preparations. One primer was designed to specifically bind to the packaging signal of the HD-Ad vector (C4HSU-PS-F), and the second designed to bind in the E1 region of H14, in front of the loxP site (H14-loxP-R2, H14-loxP-R) or in front of the E4 promoter (H14-35356-F). The PCR reaction was conducted using a kit from Promega and 35 cycles of 30 sec at 95°C, 30 sec at 60°C, and 45 sec at 72°C, followed by 3 min at 72°C. PCR products were separated by agarose gel electrophoresis to confirm expected size. Bands were isolated using Qiaquick gel extraction kit (Qiagen) and were sequenced using primers C4HSU-PS-F, H14-loxP-R2, H14-loxP-R, or H14-35356-F (Table 2).
Results
Rapalog-regulated gene expression in HEK293 cells
The process to generate helper-dependent adenoviral vectors with the dimerizer system is depicted in Supplementary Figure 1 (Supplementary Data available online at www.liebertonline/hgtb.com), and details of the cloning steps are described in the Supplementary Materials and Methods section.
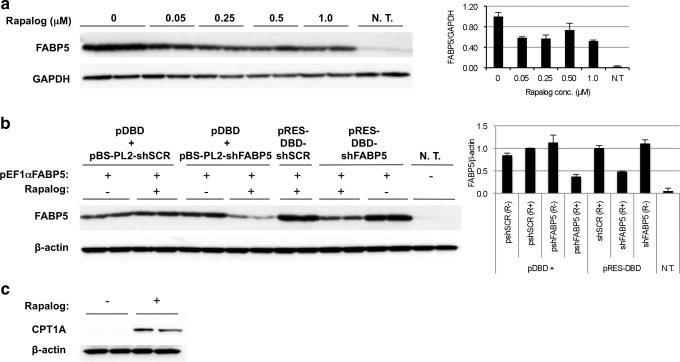
shRNA-expressing constructs were engineered to silence the fatty acid transporter FABP5 or to express a control scrambled sequence. To confirm that the shRNA-expressing constructs were tightly regulated by rapalog and to determine the optimal rapalog concentration needed to down-regulate target gene expression, HEK293 cells were cotransfected with pEF1α-FABP5, pDBD, and pBS-PL2-shFABP5-mpA (Supplementary Fig. 1). A plasmid-expressing mouse FABP5 was included in the transfection, as the shRNA sequence would not target human FABP5 expressed in HEK293 cells. Cells were cultured with 0, 0.05, 0.25, 0.5, and 1.0 μM rapalog AP21967 (Fig. 1a). FABP5 levels were reduced with 0.05 μM, reaching a plateau effect (Fig. 1a). This data showed that rapalog AP21967 tightly controls expression of shFABP5 in this construct and that gene silencing can be obtained with low concentrations of rapalog. Also, shSCR-treated cells cultured in the presence of 0.25 μM rapalog did not exhibit differences in levels of target genes relative to cells cultured without rapalog (Fig. 1b).
FIG. 1.
Rapalog-inducible transgene expression in vitro. (a) HEK293 cells were cotransfected with plasmids pEF1αFABP5 (expressing mouse FABP5), pDBD, and pBS-PL2-shFABP5-mpA in a ratio of 1:2:1 and cultured with rapalog AP21967 at the concentrations indicated above the wells or with ethanol as a negative control. After 48 hr, cells were harvested and expression of FABP5 was detected by Western blot. FABP5 knock-down is rapalog-concentration dependent. (b) HEK293 cells were cotransfected with plasmids pEF1αFABP5, pDBD, and pBS-PL2-shRNA-mpA (wells 1 through 8 from the left), or pEF1αFABP5 and pRES/DBD-shRNA-mpA at a 1:3 ratio. Cells were cultured in the presence of 0.25 μM rapalog AP21967 and 48 hr later were harvested. Expression of FABP5 was detected by Western blot. No transfection (N.T.) cells display endogenous levels of human FABP5 in HEK293 cells. FABP5 was significantly knocked-down only in the presence of rapalog. The shSCR-transfected cells do not show differences in FABP5 levels in the absence or presence of rapalog. (c) HEK293 cells were transfected with pRES/DBD-CPT1AWT and cultured with 0.25 μM rapalog (+) or ethanol (−). Carnitine palmitoyltransferase 1A (CPT1A) expression was detected only in the presence of rapalog.
The large cloning capacity of the helper-dependent adenoviral vector allows incorporating all the components of the dimerizer system into one construct. It was critical to determine that the amount of fusion proteins produced was sufficient to activate shRNA expression and silence the target gene, prior to rescuing a viral vector. To test this, HEK293 cells were transfected with pRES-DBD/PL2-shFABP5-mpA or pRES-DBD/PL2-shSCR-mpA and cultured in the presence or absence of 0.25 μM of rapalog AP21967 (Fig. 1b). FABP5 was significantly knocked-down only in the presence of rapalog (Fig. 1b). Thus, the level of transcription factor fusion proteins generated when all dimerizer components are in one molecule is sufficient to activate shRNA production and silence FABP5 expression.
In addition to the shRNA-expressing plasmids, bicistronic constructs were designed to express wild-type CPT1A or a mutant form of CPT1A (CPT1E3A) and green fluorescent protein (GFP). HEK293 cells transfected with pRES-DBD/CPT1AWT expressed CPT1A only in the presence of rapalog AP21967 (Fig. 1c). Altogether, these data indicate that when all components of the dimerizer system are incorporated in one molecule, tight regulation of shRNA or transgene expression is obtained.
Production of helper-dependent adenoviral vectors
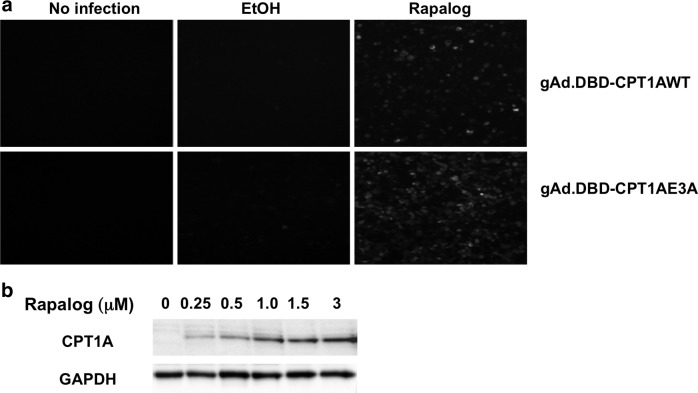
To generate helper-dependent adenoviral vectors, expression cassettes were excised from pRES3.2 and transferred to pSHL through recombination in E. coli BJ5183 cells. Three pSHL plasmids were generated in which stuffer sequence was deleted to accommodate the different sizes of the expression cassettes. The shFABP5 construct was cloned into two pSHL plasmids (pSHL-23.8 and pSHL-24.4) to generate vector genome sizes of 26.7 kb and 27.2 kb and to determine if small size differences affect the capacity to encapsidate the vector genome. Plasmids pSHL-DBD/PL2-shFABP5 (yielding a vector genome size of 26.7 kb), pSHL-DBD/PL2-shFABP5 (27.2 kb), pSHL-DBD/PL2-shSCR (26.7 kb), pSHL-DBD/CPT1AWT (29.7 kb), pSHL-DBD/CPT1E3A (29.7 kb), and pSHL-EF1αLacZ (34.2 kb) were digested with PmeI and helper-dependent adenoviral vectors were rescued in 293Cre4 cells. Given that the CPT1A vector was bicistronic, GFP was used to monitor virus production. An aliquot of gAd.DBD-CPT1AWT or gAd.DBD-CPT1E3A passage-4 lysate was inoculated into HEK293 cells in the absence/presence of 0.25 μM rapalog AP21967. GFP expression was observed only in the presence of rapalog (Fig. 2a). Thus, transgene expression in the virus was controlled by rapalog.
FIG. 2.
Helper-dependent adenoviral (HD Ad) vector expression of CPT1A. (a) Cell lysates from passage 4 were used to inoculate HEK293 cells. Ralapog (0.25 μM) was added to induce expression of CPT1 and green fluorescent protein (GFP). High induction of GFP expression was observed in the presence of rapalog. (b) CPT1AWT expression using the HD Ad vector produced from a large-scale preparation. Primary hepatocytes were infected at multiplicity of infection (MOI) 3 of gAd.DBD-CPT1AWT and cultured with 0.25, 0.5, 1.0, and 3.0 μM rapalog. In primary hepatocytes, highest expression of CPT1A was obtained in the presence of 1.0 μM rapalog.
Viral DNA rearrangements
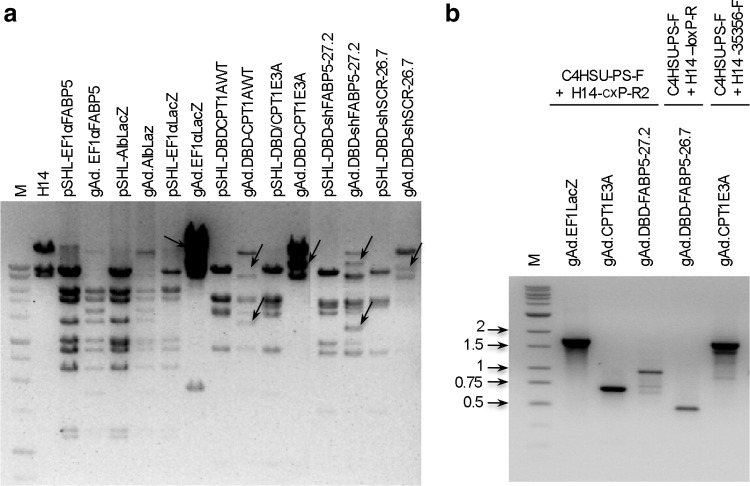
During CsCl gradient virus purification, vector and helper particles separate based on differences in their genome sizes (Parks et al., 1996). In large-scale preparations, we observed that the band corresponding to the helper was thicker than previously observed with other vectors rescued in the lab (data not shown). To determine whether this was due to insufficient Cre recombinase activity, the gAd.DBD-CPT1AWT vector was grown in 116 cells, a producer cell line expressing higher levels of Cre recombinase than 293Cre4 cells (Palmer and Ng, 2003). The vector DNA pattern again indicated the presence of very high amounts of helper (data not shown). Thus, Cre recombinase activity was not a limiting factor. In some of the preps, DNA restriction enzyme digestion showed an H14 and vector DNA pattern different from expected, suggesting genome rearrangements took place (Fig. 3a).
FIG. 3.
Restriction enzyme digestion of DNA isolated from HD Ad vectors. (a) Viral DNA was isolated and digested with BamHI. Constructs gAd.EF1αFABP5 and gAd.AlbLacZ showed the expected pattern of DNA fragments for vector and helper. However, gAd.DBD-shFABP5 (genome sizes 26.7 and 27.2 kb), gAd.DBD-shSCR, gAd.DBD-CPT1AWT, gAd.DBD-CPT1E3A, and gAd.EF1αLacZ showed unexpected DNA fragments (arrows). (b) PCR fragments obtained using primers described in Figure 4, to amplify specifically from: (i) packaging signal of the HD Ad and the E1 sequence, and (ii) H14 E4 sequence and packaging signal of the HD Ad.
An HD Ad vector with an expression cassette consisting of the elongation factor 1 α (EF1α) promoter driving LacZ gene expression also resulted in helper overgrowth (Fig. 3a). These results were puzzling because our lab has rescued multiple constructs expressing shRNA or transgenes and helper contamination has been consistently low—even after passaging large-scale preps to generate new lots of vector (Ruiz et al., 2009). To determine whether the sequence in the expression cassette had an impact on promoting rearrangements, two vectors were designed: the first one contained the EF1α promoter driving expression of a different transgene, fatty acid-binding protein 5 (FABP5), and the second contained the LacZ cDNA driven by the albumin promoter. Neither of these constructs resulted in rearrangements, even though the same H14 lot was used for rescue and large-scale production (Fig. 3a). Although there was a positive correlation between vector genome size and level of helper contamination, no rearrangements were observed, even after using the first large-scale preparations to generate additional lots (data not shown). Overall, our data suggests that the EF1α or LacZ sequence per se is not a determinant of rearrangements.
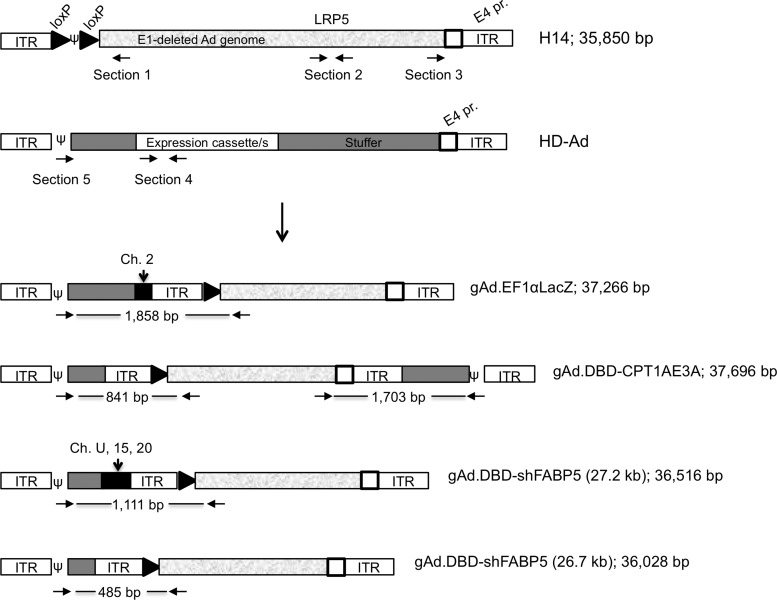
To understand the nature of the rearrangements, virus DNA was isolated from large-scale preparations of gAd.DBD-shFABP5 (genome sizes 26.7 and 27.2 kb), gAd.DBD-shSCR, gAd.DBD-CPT1AWT, gAd.DBD-CPT1E3A, and gAd.EF1αLacZ, and the helper DNA was sequenced using a primer that binds to the E1 sequence, only present in the H14 helper, and extends toward the ITR (Table 2; Fig. 4, Sec. 1). All constructs, except for gAd.DBD-CPT1AWT, showed the presence of one loxP site without the packaging signal, followed by the left ITR (as would be expected in H14 helper genomes that underwent Cre recombinase activity). However, 5′ upstream from the ITR, the helper of constructs gAd.EF1αLacZ, gAd.DBD-shFABP5 (genome size 26.7 kb), and gAd.DBD-CPT1E3A, had incorporated sequences from the HD Ad vector, including stuffer DNA, the entire packaging signal, and the left ITR. This sequence did not include the expression cassette/s (Fig. 4). Each construct had incorporated different amounts of the left arm of the HD Ad vector. In addition, the helper in gAd.EF1αLacZ had a fragment of 159 bp from human BAC clone RP11-324H23 (chromosome 2), which is not present in the stuffer sequence. Thus, a fragment of cellular DNA was incorporated. For constructs gAd.DBD-shFABP5 (genome size 27.2 kb) and gAd.DBD-shSCR, no conclusion could be drawn on whether or not a rearrangement was present, as the quality of the sequencing reaction was poor upstream from the ITR.
FIG. 4.
Helper H14 genome rearrangements. Top: Primers were designed to bind specific areas in helper (H14) and HD Ad vectors (see Table 2 for sequence). Bottom: Predicted structure of helper and vector rearrangements observed, based on DNA sequencing. The size of the expected PCR products is shown for each construct. The predicted size of the rearranged helper is shown after the name of the vector. DNA fragments of human origin are depicted on vectors gAd.EF1αLacZ and gAd.DBD-shFABP5 (27.2 kb). U, chromosome unknown.
To confirm the rearrangements, primers binding specifically to the packaging signal in the HD Ad vector (Fig. 4, Sec. 5) and to the E1 sequence in the H14 helper (Fig. 4, Sec. 1) were used to amplify the rearranged fragments by PCR in preps of gAd.EF1αLacZ, gAd.DBD-shFABP5 (genome size 27.2 kb), and gAd.DBD-CPT1E3A. Fragments of the expected size were obtained (Fig. 3b), and sequencing confirmed the results obtained from sequencing the helper genome. In addition, for construct gAd.DBD-shFABP5 (26.7 and 27.2 kb), sequencing revealed the presence of three fragments of human DNA (80-bp human PAC clone RP1-169K13, 275-bp chromosome 15, and 89-bp chromosome 20), not present in H14 or the stuffer of the HD Ad vector. Thus, like the helper in gAd.EF1αLacZ, the helper in gAd.DBD-shFABP5 also incorporated cellular DNA.
It has been shown that the packaging signal can function near either end of the adenovirus genome (Hearing et al., 1987). To investigate the possibility that the H14 helper had a rearrangement that incorporated the packaging signal at the right end, primers were designed specifically to bind to the H14 helper sequence, 5′ from the E4 promoter (Fig. 4, Sec. 3). No additional sequence was observed further downstream from the right-end ITR in the H14 helper of constructs gAd.DBD-shFABP5 (genome size 27.2 kb) and gAd.EF1αLacZ. However, a fragment consisting of the stuffer sequence, packaging signal, and ITR from the vector was found in H14 helper from the gAd.DBD-CPT1E3A construct. Sequencing of the PCR product confirmed the rearrangement (Fig. 3b). All rearranged helper molecules had a predicted size below 37.8 kb (Fig. 4), the limit for adenovirus genome packaging.
Plaque assays were performed on preparations gAd.EF1αLacZ and gAd.DBD-CPT1E3A to determine whether the rearranged H14 helper was able to provide all functions needed for adenovirus replication and encapsidation. High plaque forming units (PFU)/ml titers were observed (Table 3), confirming that the rearranged helpers have a functional packaging signal from the vector and were capable of growing in 293 cells. Particle:PFU ratios were 23:1 and 68:1 for gAd.EF1αLacZ and gAd.DBD-CPT1E3A, respectively. In contrast, the PFU titer was much lower in gAd.EF1αFABP5 and gAd.C4HSU (rescued from pSHL-30.9 plasmid; no expression cassette; genome size of 28.1 kb) (Ruiz et al., 2009). Particle:PFU ratios in these preparations were 357:1 and 3,758:1 respectively (Table 3). The ratio in gAd.C4HSU was similar to what we have previously described for six different shRNA-expressing vectors with genome sizes of ∼28.5 kb (Ruiz et al., 2009).
Table 3.
Particle and Plaque Titers
|
gAd.H1-shSREBP1 (Ruiz et al., 2009) |
gAd.C4HSU |
gAd.EF1α FABP5 |
gAd.EF1α LacZ |
gAd.DBD-CPT1E3A | |
|---|---|---|---|---|---|
| Particle/ml | 10.0×1011 | 12.4×1011 | 5×1011 | 14.2×1011 | 12.5×1011 |
| PFU/ml | 1.2×108 | 3.3×108 | 1.4×109 | 6.23×1010 | 1.83×1010 |
| Particle:PFU | 8,333:1 | 3,758:1 | 357:1 | 23:1 | 68:1 |
PFU, plaque forming units.
Finally, the H14 helper present in the gAd.DBD-CPT1AWT construct had lost one of the loxP sites, resulting in the inability of Cre recombinase to excise the packaging signal and leading to H14 helper outgrowth. Loss of one loxP site during vector production has been previously described (Hardy et al., 1997). In addition, one of the fragments observed in the pattern of the HD Ad vector suggested a deletion had occurred in that area during virus production (see different band sizes between plasmid and vector, Fig. 4). Sequencing using primers flanking the 3xFKBP drug-binding domains (Fig. 4, Sec. 4; Supplementary Fig. 1) indicated that two of the three tandemly repeated copies of FKBP were deleted. Nevertheless, CPT1A protein was expressed in primary hepatocytes transduced with vector gAd.DBD-CPT1AWT in a rapalog concentration-dependent manner (Fig. 2b). The presence of CPT1A protein suggests that even though two of the rapalog-binding sites were lost, the system was still able to activate transcription of CPT1A/GFP in the presence of the inducing drug. Neither the gAd.DBD-shSCR or the gAd.DBD-shFABP5 (27.2 kb) had a deletion in this region, suggesting this was an isolated event.
In E1-deleted adenovirus vector production, homologous recombination generates replication-competent adenovirus (RCA), which occurs during propagation in HEK293 cells due to the presence of common sequences in the vector genome and the 4.5 kb of adenovirus left arm inserted in the genome of the HEK293 cell line (Lochmuller et al., 1994; Louis et al., 1997). To prevent RCA formation during HD Ad vector propagation, the H14 helper has been modified to contain a 2,902-bp fragment of intronic sequence from chromosome 11 (low-density lipoprotein receptor-related protein 5 precursor gene) in the E3 region, so that the genome of the H14 helper would be unpackageable in case a recombination event took place during vector propagation. Given that this intronic sequence is from a human gene, we tested whether homologous recombination with the cellular genome could also have occurred. Constructs gAd.EF1αLacZ, gAd.DBD-shFABP5 (genome size 27.2 kb), and gAd.DBD-CPT1E3A were sequenced with primers flanking/within the 2,902-bp kb fragment (Fig. 4, Sec. 2). The correct sequence was observed in all constructs, suggesting the rearrangements affected only the genome ends.
Discussion
During production of helper-dependent adenoviral vectors, there is an inherent risk of recombination between the two virus genomes. Recombination events between homologous sequences in coreplicating adenoviruses can occur between the packaging signals, and this has been described when helper and vector have the same packaging signal sequence (Hardy et al., 1997; Sandig et al., 2000). To prevent recombination between the two DNA molecules, Sandig and colleagues modified the packaging signal of the H14 helper, which avoided rearrangements in more than 30 independent rescues (Sandig et al., 2000). Adenoviral vector DNA rearrangements have also been described when the viral genome size is <27 kb or >37.8 kb. Vector genome sizes less than 27 kb are inefficiently packaged and undergo rearrangements to produce a genome size of approximately 30 kb, comparable to the size of wild-type Ad5 (Parks and Graham, 1997). Similarly, the maximum capacity for genome encapsidation is 105% of wild-type genome size (36 kb). Vectors with larger genomes grow poorly and the genomes tend to rearrange, resulting in loss of the insert after a few passages (Bett et al., 1993).
We observed that rearrangements occur between HD Ad vector and the left or right arm of the helper, using the Cre/LoxP system described by Sandig and colleagues (Sandig et al., 2000). Interestingly, rearrangements always involved a H14 helper DNA molecule that had been subject to Cre recombinase activity, (i.e., the packaging signal and one loxP site were deleted). These molecules are in excess in the cell, as the genome of the helper is replicated without being packaged. In the gAd.EF1αLacZ and gAd.DBD-shFABP5 (27.2 kb) constructs, in addition to sequence from the HD Ad vector, the rearrangement in the helper included fragments from Homo sapiens DNA (gAd.EF1αLacZ, 159-bp chromosome 2; gAd.DBD-shFABP5, 80-bp PAC clone RP1-169K13, 275-bp chromosome 15 and 89-bp chromosome 20), which are not present in the stuffer DNA of the HD Ad vector or in H14 helper. Thus, the rearrangements involved helper, HD Ad vector, and in two occasions, cellular genome. The rearranged helper always had incorporated part of the left arm of the HD Ad vector, including ITR, packaging signal and stuffer sequence (Fig. 4), and was able to generate plaques (Table 3). Given that the left arm of the vector was incorporated upstream or downstream from the helper ITR, and in some instances it involved fragments of cellular DNA, it is possible that the rearrangements originated by nonhomologous end-joining (NHEJ). Nonhomologous end-joining occurs during adenovirus infection, as many linear single- and double-stranded molecules are generated, and the cellular proteins implicated in NHEJ ligate the DNA to generate concatemers (Weiden and Ginsberg, 1994). Furthermore, no homology exists between the sequence of the H14 helper and the vector, except for the ITRs and the E4 promoter, and no sequence homology was found between the cellular DNA and the HD vector (which also contains human DNA) around the junction areas. Thus, it is unlikely that the rearrangements occurred between homologous sequences between human DNA fragments and between human DNA and the ITR of the helper. Nevertheless, we cannot discount the possibility that microhomologies have facilitated recombination events. There is evidence that homologies of 8–9 nt between adenovirus type 12 and the cellular genome facilitate recombination (Wronka et al., 2002).
Sakhuja and colleagues described the presence of helper particles with genomes containing a loxP site and lacking the packaging signal, as would be expected from Cre recombinase activity during vector production in PERC6-Cre cells (Sakhuja et al., 2003). These molecules were characterized by PCR amplification of DNA isolated from HD Ad vector large-scale preparations. The investigators concluded that helper genomes without packaging signal could be encapsidated and hypothesized that production of HD Ad vector completely free from helper might not be possible (Sakhuja et al., 2003). It is possible that these particles represent rearranged helper molecules like the ones described in this study, and that the packaging signal from the vector was present upstream from the ITR. Hillgenberg and colleagues also observed the presence of mutated helper virus contaminating large-scale preparations of HD Ad vectors rescued using a different Cre/loxP system, although characterization of the mutants was not described (Hillgenberg et al., 2001). Therefore, in several Cre/loxP systems, rearranged molecules have been observed.
We have rescued 12 HD Ad vectors expressing a variety of transgenes and shRNAs, and none of these constructs showed evidence of rearrangements based on DNA restriction digestion (Witting et al., 2008; Ruiz et al., 2009). The level of helper contamination is low, even after passaging the large-scale preparations to generate new vector lots (Ruiz et al., 2009). The fact that four of five vectors containing the dimerizer system displayed rearrangements suggests the expression cassette may play a role at facilitating them. There is no reported evidence of DNA rearrangements using the rapalog-inducible system in first-generation adenovirus (Auricchio et al., 2002; Chong et al., 2002).
In addition, we have shown here that the sequence in the expression cassette is not sufficient to promote rearrangements and is unlikely to play a role. Despite the fact that a vector expressing LacZ under the control of the EF1α promoter resulted in a rearranged helper, two other vectors containing the EF1α promoter or the LacZ gene were rescued successfully and had the correct DNA pattern for both vector and helper. The fact that all cases of rearrangements involved the left arm of the HD vector suggests that fragments from this part of the genome accumulate during vector production. All expression cassettes in our constructs are placed within 4–5 kb from the left ITR. We believe that transcription of the transgene during DNA replication may interfere with the replication process, facilitating the accumulation of fragments that could then be used as substrates for rearrangements with the H14 helper. In the case of the dimerizer system, this may be further complicated by the fact that the DNA binding domain fusion protein (ZFHD1), which is under the control of the cytomegalovirus (CMV) promoter, is expressed at high levels during vector production. This fusion protein can bind to the 12xZFHD1 binding sites in the PL2 promoter, interfering with replication of the vector genome (even though the transgene downstream from it is not expressed in the absence of rapalog). Additional studies will be needed to determine whether transcription in the expression cassette interferes with replication.
In addition to rearrangements at the ends of the H14 helper genome, we observed that one of the HD Ad vectors, gAd.DBD-CPT1AWT, contained a deletion. The DNA binding domain (ZFHD1) is fused to three drug binding domains (3xFKBP), each of which is 321-bp long. Two of these domains were deleted during vector production, suggesting that repeated motifs generate secondary structures that can be deleted during vector DNA replication. Despite the decreased capacity for rapalog binding, the generated vector responded to the presence of rapalog, activating CPT1A expression.
In summary, during HD Ad adenoviral vector production, rearrangements between vector and helper genomes occur, likely by nonhomologous end-joining recombination. The rearrangements involve H14 helper DNA molecules that have been subject to Cre recombinase activity, which accumulates in 293Cre4 cells. By acquiring the packaging signal from the vector, these particles have a growth advantage, leading to large amounts of helper contamination in large-scale preparations. This process is distinct from helper contamination due to insufficient Cre recombinase activity (Ng et al., 2002) or from loss of one of the loxP sites in the helper genome during vector amplification (Hardy et al., 1997). Future research is needed to determine the role of the expression cassette or other factors that may promote the onset of rearrangements and helper overgrowth, so that appropriate modifications can be incorporated to HD Ad vectors prior to production for clinical applications.
Supplementary Material
Acknowledgments
We thank Ariad Pharmaceuticals, Inc., (Cambridge, MA) for kindly providing the rapalog system and rapalog AP21967; Dr. Philip Ng for providing the 116 cell line; and Dr. Carina Prip-Buus for providing the antibody against CPT1A. This research was supported by grants from the National Institutes of Health (DK069432, DK078595) and American Diabetes Association (1-08-RA-135). Miwon Ahn was supported by the T32-Indiana University Diabetes and Obesity Research Training Program [DK064466], Scott Witting by a postdoctoral fellowship from the American Heart Association, and Sneha Surendran by a DeVault fellowship.
Author Disclosure Statement
No competing financial interests exist.
References
- Auricchio A. Gao G.P. Yu Q.C., et al. Constitutive and regulated expression of processed insulin following in vivo hepatic gene transfer. Gene Ther. 2002;9:963–71. doi: 10.1038/sj.gt.3301746. [DOI] [PubMed] [Google Scholar]
- Bayle J.H. Grimley J.S. Stankunas K., et al. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem. Biol. 2006;13:99–107. doi: 10.1016/j.chembiol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Benihoud K. Yeh P. Perricaudet M. Adenovirus vectors for gene delivery. Curr. Opin. Biotechnol. 1999;10:440–7. doi: 10.1016/s0958-1669(99)00007-5. [DOI] [PubMed] [Google Scholar]
- Bett A.J. Prevec L. Graham F.L. Packaging capacity and stability of human adenovirus type 5 vectors. J. Virol. 1993;67:5911–21. doi: 10.1128/jvi.67.10.5911-5921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T.R. Bernards R. Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Chen J. Zheng X.F. Brown E.J. Schreiber S.L. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4947–51. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Anton M. Graham F. Production and characterization of human 293 cell lines expressing the site-specific recombinase Cre. Somat. Cell and Molec. Genet. 1996;22:477–488. doi: 10.1007/BF02369439. [DOI] [PubMed] [Google Scholar]
- Chong H. Ruchatz A. Clackson T., et al. A system for small-molecule control of conditionally replication-competent adenoviral vectors. Mol. Ther. 2002;5:195–203. doi: 10.1006/mthe.2002.0531. [DOI] [PubMed] [Google Scholar]
- Clackson T. Regulated gene expression systems. Gene Ther. 2000;7:120–5. doi: 10.1038/sj.gt.3301120. [DOI] [PubMed] [Google Scholar]
- Grimm D. Kay M.A. Therapeutic short hairpin RNA expression in the liver: viral targets and vectors. Gene Ther. 2006;13:563–75. doi: 10.1038/sj.gt.3302727. [DOI] [PubMed] [Google Scholar]
- Hardy S. Kitamura M. Harris-Stansil T., et al. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P. Samulski R.J. Wishart W.L. Shenk T. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J. Virol. 1987;61:2555–8. doi: 10.1128/jvi.61.8.2555-2558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillgenberg M. Schnieders F. Loser P. Strauss M. System for efficient helper-dependent minimal adenovirus construction and rescue. Hum. Gene Ther. 2001;12:643–57. doi: 10.1089/104303401300057342. [DOI] [PubMed] [Google Scholar]
- Hoekstra M. Stitzinger M. Van Wanrooij E.J., et al. Microarray analysis indicates an important role for FABP5 and putative novel FABPs on a Western-type diet. J. Lipid Res. 2006;47:2198–207. doi: 10.1194/jlr.M600095-JLR200. [DOI] [PubMed] [Google Scholar]
- Kim I.H. Jozkowicz A. Piedra P.A., et al. Lifetime correction of genetic deficiency in mice with a single injection of helper-dependent adenoviral vector. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13282–7. doi: 10.1073/pnas.241506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H. Fujimiya M. Matsumura K., et al. NeuroD-betacellulin gene therapy induces islet neogenesis in the liver and reverses diabetes in mice. Nat. Med. 2003;9:596–603. doi: 10.1038/nm867. [DOI] [PubMed] [Google Scholar]
- Lochmuller H. Jani A. Huard J., et al. Emergence of early region 1-containing replication-competent adenovirus in stocks of replication-defective adenovirus recombinants (delta E1+delta E3) during multiple passages in 293 cells. Hum. Gene Ther. 1994;5:1485–91. doi: 10.1089/hum.1994.5.12-1485. [DOI] [PubMed] [Google Scholar]
- Louis N. Evelegh C. Graham F.L. Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology. 1997;233:423–9. doi: 10.1006/viro.1997.8597. [DOI] [PubMed] [Google Scholar]
- McGarry J.D. Brown N.F. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur. J. Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- Morral N. Parks R. Zhou H., et al. High doses of a helper-dependent adenoviral vector yield supraphysiological levels of α1-antitrypsin with negligible toxicity. Hum. Gene Ther. 1998;9:2709–2716. doi: 10.1089/hum.1998.9.18-2709. [DOI] [PubMed] [Google Scholar]
- Morral N. O'Neal W. Rice K., et al. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12816–12821. doi: 10.1073/pnas.96.22.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain A. Gene therapy: the first decade. Trends Biotechnol. 2000;18:119–28. doi: 10.1016/s0167-7799(99)01416-x. [DOI] [PubMed] [Google Scholar]
- Ng P. Evelegh C. Cummings D. Graham F.L. Cre levels limit packaging signal excision efficiency in the Cre/loxP helper-dependent adenoviral vector system. J. Virol. 2002;76:4181–9. doi: 10.1128/JVI.76.9.4181-4189.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D. Ng P. Improved system for helper-dependent adenoviral vector production. Mol. Ther. 2003;8:846–52. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Park J.S. Surendran S. Kamendulis L.M. Morral N. Comparative nucleic acid transfection efficacy in primary hepatocytes for gene silencing and functional studies. BMC Res. Notes. 2011;4:8. doi: 10.1186/1756-0500-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A.L. Waddington S.N. Nicol C.G., et al. Multiple vitamin K-dependent coagulation zymogens promote adenovirus-mediated gene delivery to hepatocytes. Blood. 2006;108:2554–61. doi: 10.1182/blood-2006-04-008532. [DOI] [PubMed] [Google Scholar]
- Parker A.L. Mcvey J.H. Doctor J.H., et al. Influence of coagulation factor zymogens on the infectivity of adenoviruses pseudotyped with fibers from subgroup D. J. Virol. 2007;81:3627–31. doi: 10.1128/JVI.02786-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks R.J. Graham F.L. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J. Virol. 1997;71:3293–3298. doi: 10.1128/jvi.71.4.3293-3298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks R.J. Chen L. Anton M., et al. A helper-dependent adenovirus vector system: removal of helper virus by cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R. Issner R. Zoller K., et al. Delivery of a stringent dimerizer-regulated gene expression system in a single retroviral vector. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13221–6. doi: 10.1073/pnas.230446297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera V.M. Clackson T. Natesan S., et al. A humanized system for pharmacologic control of gene expression. Nat. Med. 1996;2:1028–32. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- Rivera V.M. Ye X. Courage N.L., et al. Long-term regulated expression of growth hormone in mice after intramuscular gene transfer. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8657–62. doi: 10.1073/pnas.96.15.8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz R. Witting S.R. Saxena R. Morral N. Robust hepatic gene silencing for functional studies using helper-dependent adenovirus vectors. Hum. Gene Ther. 2009;20:87–94. doi: 10.1089/hum.2008.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakhuja K. Reddy P.S. Ganesh S., et al. Optimization of the generation and propagation of gutless adenoviral vectors. Hum. Gene Ther. 2003;14:243–54. doi: 10.1089/10430340360535797. [DOI] [PubMed] [Google Scholar]
- Sandig V. Youil R. Bett A.J., et al. Optimization of the helper-dependent adenovirus system for production and potency in vivo. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1002–7. doi: 10.1073/pnas.97.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanftner L.M. Rivera V.M. Suzuki B.M., et al. Dimerizer regulation of AADC expression and behavioral response in AAV-transduced 6-OHDA lesioned rats. Mol. Ther. 2006;13:167–74. doi: 10.1016/j.ymthe.2005.06.480. [DOI] [PubMed] [Google Scholar]
- Schiedner G. Morral N. Parks R., et al. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat. Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- Shayakhmetov D.M. Gaggar A. Ni S., et al. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J. Virol. 2005;79:7478–91. doi: 10.1128/JVI.79.12.7478-7491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss K.A. Robinson D.L. Vreman H.J., et al. Management of hyperbilirubinemia and prevention of kernicterus in 20 patients with Crigler-Najjar disease. Eur. J. Pediatr. 2006;165:306–19. doi: 10.1007/s00431-005-0055-2. [DOI] [PubMed] [Google Scholar]
- Waddington S.N. Parker A.L. Havenga M., et al. Targeting of adenovirus serotype 5 (Ad5) and 5/47 pseudotyped vectors in vivo: fundamental involvement of coagulation factors and redundancy of CAR binding by Ad5. J. Virol. 2007;81:9568–71. doi: 10.1128/JVI.00663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington S.N. Mcvey J.H. Bhella D., et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Weiden M.D. Ginsberg H.S. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. U.S.A. 1994;91:153–7. doi: 10.1073/pnas.91.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting S.R. Brown M. Saxena R., et al. Helper-dependent adenovirus-mediated short hairpin rna expression in the liver activates the interferon response. J. Biol. Chem. 2008;283:2120–8. doi: 10.1074/jbc.M704178200. [DOI] [PubMed] [Google Scholar]
- Wronka G. Fechteler K. Schmitz B. Doerfler W. Integrative recombination between adenovirus type 12 DNA and mammalian DNA in a cell-free system: joining by short sequence homologies. Virus Res. 2002;90:225–42. doi: 10.1016/s0168-1702(02)00201-0. [DOI] [PubMed] [Google Scholar]
- Ye X. Rivera V.M. Zoltick P., et al. Regulated delivery of therapeutic proteins after in vivo somatic cell gene transfer. Science. 1999;283:88–91. doi: 10.1126/science.283.5398.88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.