Abstract
T cell-sorting technologies with peptide–MHC multimers or antibodies against gene markers enable enrichment of antigen-specific T cells and are expected to enhance the therapeutic efficacy of clinical T cell therapy. However, a direct comparison between sorting reagents for their ability to enrich T cells is lacking. Here, we compared the in vitro properties of primary human T cells gene-engineered with gp100280–288/HLA-A2-specific T cell receptor-αβ (TCRαβ) on magnetic-activated cell sorting (MACS) with various peptide–MHC multimers or an antibody against truncated CD34 (tCD34). With respect to peptide–MHC multimers, we observed that Streptamer®, when compared with pentamers and tetramers, improved T cell yield as well as level and stability of enrichment, of TCR-engineered T cells (>65% of peptide–MHC-binding T cells, stable for at least 6 weeks). In agreement with these findings, Streptamer, the only detachable reagent, revealed significant T cell expansion in the first week after MACS. Sorting TCR and tCD34 gene-engineered T cells with CD34 monoclonal antibody (mAb) resulted in the most significant T cell yield and enrichment of T cells (>95% of tCD34 T cells, stable for at least 6 weeks). Notably, T cells sorted with CD34 mAb, when compared with Streptamer, bound about 2- to 3-fold less peptide–MHC but showed superior antigen-specific upregulated expression of CD107a and production of interferon (IFN)-γ. Multiparametric flow cytometry revealed that CD4+ T cells, uniquely present in CD34 mAb-sorted T cells, contributed to enhanced IFN-γ production. Taken together, we postulate that CD34 mAb-based sorting of gene-marked T cells has benefits toward applications of T cell therapy, especially those that require CD4+ T cells.
Govers and colleagues perform direct comparisons between T cell sorting reagents for their ability to enrich T cells. They compare in vitro properties of primary human T cells gene-engineered with gp100280-288 Human Leukocyte Antigen A2-specific T-cell receptorαβ undergoing Magnetic-Activated Cell Sorting either with different peptide-MHC multimers or with an antibody against truncated CD34 (tCD34). Among the multimers, streptamers resulted in the best T cell yield, stability, and expansion ability after sorting, while tCD34-sorted cells resulted in the most significant T cell yield and enrichment.
Introduction
Adoptive therapy with tumor-infiltrating T cells, preceded by lymphodepletion, shows significant clinical responses in patients with melanoma (Dudley et al., 2005; Besser et al., 2010). In an effort to make T cell therapy a more universally applicable treatment, T cells have been gene-engineered to express virus and tumor-specific T cell receptors (TCRs). T cells transduced with TCRs directed against the HLA-A2-restricted antigens MART-1 (melanoma antigen recognized by T cells-1), gp100, CEA (carcinoembryonic antigen), and NY-ESO-1 have been tested in clinical trials, and clinical responses have been observed in patients with metastatic melanoma, colorectal carcinoma, and synovial carcinoma (Morgan et al., 2006; Johnson et al., 2009; Parkhurst et al., 2011; Robbins et al., 2011). Clinical responses, although variable and based on a relatively small number of patients, are promising but generally lag behind those observed with tumor-infiltrating T cells (Dudley et al., 2005; Besser et al., 2010). The clinical use of high-affinity TCRs enhances response rates and may provide a means to improve clinical responses (Morgan et al., 2006; Johnson et al., 2009). However, high-affinity TCRs, when directed against differentiation antigens that are overexpressed on tumors but also present, albeit to a small extent, on normal tissues, result in on-target toxicities (Johnson et al., 2009; Parkhurst et al., 2011). An alternative strategy to enhance functional responses of T cells is to enhance the frequency of TCR-transduced T cells. The threshold antigen concentration for T cell activation correlates inversely with the level of TCR expression in T cell populations (Cooper et al., 2000; Weijtens et al., 2000), providing a rationale for the enrichment of TCR-transduced T cells before their clinical use. T cell populations used in TCR gene therapy studies generally demonstrated a peptide–MHC binding of about 62%, being in some cases as low as 3% and in some cases as high as 97%, which presents a window for further improvement (Morgan et al., 2006; Johnson et al., 2009; Robbins et al., 2011).
Magnetic-activated cell sorting (MACS) was developed from the late 1970s onward (Molday et al., 1977; Miltenyi et al., 1990) and now represents an established technology to enrich cells that is compatible with conditions of Good Medical Practice and can be applied in clinical cellular therapy trials (Rauser et al., 2004; Hammer et al., 2007; Feuchtinger et al., 2008). When MACS is used in combination with peptide–MHC multimers, this technology provides a versatile and potent platform to enrich T cells with a defined antigen specificity. For example, T cells specific for multiple antigens, such as cytomegalovirus (CMV), Epstein–Barr virus epitopes 1–3 (EBV1–3), influenza epitope 1 (Flu1), or MART-1 antigens from a single sample can be simultaneously enriched with peptide-MHC tetramers (Newell et al., 2009). Also, MART-1/HLA-A2-specific T cells have been enriched up to a 1000-fold from peripheral blood mononuclear cells (PBMCs) and tumor-infiltrating lymphocytes from patients with metastatic melanoma without loss of in vitro reactivity (Labarriere et al., 2008). At present, various forms of peptide–MHC multimers exist, of which tetramers (Altman et al., 1996), pentamers (Sebestyen et al., 2008), and Streptamers (Knabel et al., 2002; Neudorfer et al., 2007) represent the ones most extensively characterized. These peptide–MHC multimers differ in avidity and reversibility of binding to T cells (as detailed in Table 1), and it is currently not known how these reagents compare with one another with respect to MACSorting of T cells. In addition to peptide–MHC multimers, a surrogate gene marker such as truncated CD34 (tCD34) (Fehse et al., 2000) can be applied to enrich T cells by CD34 monoclonal antibody (mAb)-based MACS (Stull et al., 2000). Cotransduction of tCD34 and a herpes simplex virus thymidine kinase (HSVtk) suicide gene into T cells, to allow ganciclovir-mediated elimination of alloreactive gene-modified T cells, and subsequent MACS resulted in T cell populations that were highly enriched for both tCD34 and HSVtk (Preuss et al., 2010).
Table 1.
Reagents Used in Magnetic-Activated Cell Sorting of T Cell Receptor-Engineered T Cellsa
| Sort reagent | Label | Second step sort reagent | TCR transgenes | Concentrationb(ng/107 T cells) | pMHC valency | Detachable binding | Key reference(s) |
|---|---|---|---|---|---|---|---|
| gp 100/A2 tetramerc | R-Phycoerythrin | Anti-PE microbeads | TCRαβ | 700 | 4 | No | Altman et al., 1996 |
| gp 100/A2 pentamerd | R-Phycoerythrin | Anti-PE microbeads | TCRαβ | 500 | 5 | No | Sebestyén et al., 2008 |
| gp 100/A2 Streptamere | R-Phycoerythrin | Anti-PE microbeads | TCRαβ | 200 | 8–12 | Yesf | Knabel et al., 2002 |
| Neudorfer et al., 2007 | |||||||
| CD34 mAb microbeadsg | — | — | TCR-tCD34 | ND | None | No | Stull et al., 2000 |
An overview of specific characteristics of various sort reagents to enrich TCR-engineered T cells is shown.
Concentrations of peptide–MHC multimers are based on molecular weight provided by manufacturers.
Tetramers contain four biotinylated peptide–MHC monomers, which are multimerized with Streptavidin–PE to form a tetrahedral complex. A maximum of three peptide–MHC molecules is available per focal plane (Yao et al., 2008).
Pentamers contain five peptide–MHC–PE monomers, which are multimerized through a coiled-coil structure. All five peptide–MHC molecules are available per focal plane (Yao et al., 2008).
Streptamers contain between 8 and 12 Strep-tagged peptide–MHC monomers, which are multimerized with Streptactin–PE.
Release of peptide–MHC monomers is established by addition of an excess of d-biotin that has a high affinity for streptactin (Kd, ∼10–13 M) (Neudorfer et al., 2007).
CD34 mAb is directly coupled to magnetic microbeads.
mAb, monoclonal antibody; ND, not determined; PE, phycoerythrin; pMHC, peptide–MHC; tCD34, truncated CD34; TCR, T cell receptor.
In this paper, we have compared tetramers, pentamers, Streptamers, and CD34 mAb for their ability to enrich TCR-engineered T cells (Fig. 1 provides a schematic representation of sort reagents used in our study). To this end, we gene-transduced human T cells with a gp100280–288/HLA-A2 (gp100/A2) TCR and tested the various sort reagents for T cell output numbers, T cell yield at later time points, T cell expansion, enrichment for peptide–MHC binding, and gp100/A2-specific functions. With respect to peptide–MHC multimers, Streptamers significantly enhanced T cell yield, expansion, and enrichment of peptide–MHC-binding T cells. MACS with CD34 mAb resulted in significant T cell yield, expansion and enrichment of tCD34-positive T cells, but not in significant enrichment of peptide–MHC-binding T cells. Notably, CD34 mAb-sorted T cells demonstrated enhanced antigen-specific T cell functions, which were related to CD4+ T cells that were uniquely present in CD34 mAb-sorted T cells.
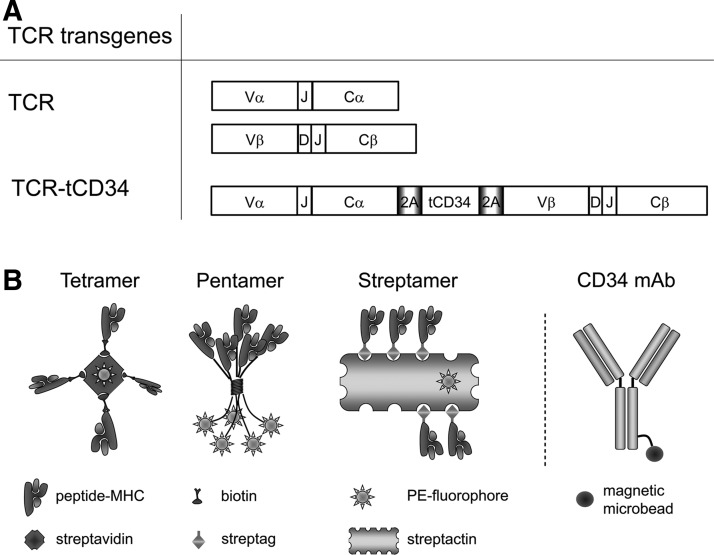
FIG. 1.
Gene constructs and sorting reagents to MACSort TCR-engineered human T cells. (A) Schematic representation of TCRα, TCRβ, and TCRα-2A-tCD34-2A-TCRβ transgenes used to gene-engineer primary human T cells. TCR specific for gp100/A2 comprised TRAV13-1*02/J52*01/CA and TRBV27*01/J2-7*01/D2*02/CB2 (Schaft et al., 2003). tCD34 represents a truncated and functionally inert variant of CD34. T cells were transduced either with pBullet:TCRα+pBullet:TCRβ and termed “TCR T cells” or with pBullet:TCRα-2A-tCD34-2A-TCRβ and termed “TCR-tCD34 T cells.” Abbreviations: V, TCRαβ-variable domain; C, TCRαβ-constant domain; D, TCRβ-diversity domain; J, TCRαβ-joining domain; 2A, 2A sequence encoding a self-cleaving peptide; tCD34, truncated CD34 molecule. (B) Schematic illustration of reagents used to MACSort TCR-engineered human T cells. From left to right: Tetramer (Altman et al., 1996), pentamer (Sebestyen et al., 2008), Streptamer (Knabel et al., 2002; Neudorfer et al., 2007), and CD34 mAb (Stull et al., 2000). Tetramers consist of 4 gp100 peptide (YLEPGPVTA)/HLA-A2 (gp100/A2) monomers that were biotinylated and multimerized with streptavidin–phycoerythrin; pentamers consist of 5 gp100/A2 monomers that were linked to phycoerythrin and multimerized by a self-assembling, coiled-coil structure; and Streptamers consist of 8–12 gp100/A2 monomers that were Strep-tagged and multimerized with Strep-Tactin–phycoerythrin (depicted with 5 monomers). Anti-CD34 mAbs consist of heavy and light chains providing two antigen-binding sites and are directly coupled to magnetic microbeads.
Materials and Methods
Cells and reagents
T lymphocytes derived from healthy donors were isolated and expanded as described elsewhere (Van de Griend et al., 1984) and cultured in HEPES-buffered RPMI 1640 medium (BioWhittaker, Verviers, Belgium) supplemented with 10% human serum, 2 mM l-glutamine, and the antibiotics streptomycin and penicillin. The human embryonic kidney cell line 293T, the packaging cell line Phoenix-A, and the melanoma cell lines BLM and FM3 were cultured in Dulbecco's modified Eagle's medium (DMEM; BioWhittaker) supplemented with 10% fetal bovine serum (FBS; Stonehouse, Gloucestershire, UK), 2 mM l-glutamine, nonessential amino acids, and antibiotics. Monoclonal antibodies used in this study were as follows: anti-CD34 microbeads (clone QBEND/10; Miltenyi Biotec, Bergisch Gladbach, Germany), anti-phycoerythrin (PE) microbeads (Miltenyi Biotec), fluorescein isothiocyanate (FITC)- and PE-conjugated anti-TCR-Vβ27 (Beckman Coulter, Marseille, France), allophycocyanin (APC)- and nonconjugated anti-CD3ɛ (OKT3; BD Biosciences [San Jose, CA] and Beckman Coulter, respectively), APC-conjugated anti-CD8α (RPA-T8; BD Biosciences), FITC-conjugated anti-CD34 (AC136; Miltenyi Biotec), PE-conjugated anti-CD107a (H4A3; BD Biosciences), FITC-conjugated anti-interferon (IFN)-γ (B27; BD Biosciences), FITC-conjugated anti-interleukin (IL)-2 (MQ1-17H12; BD Biosciences), and FITC-conjugated anti-IL-2q receptor-α (IL2Rα) (M-A251; BD Biosciences). Other reagents included RetroNectin (human fibronectin fragments CH-296; Takara Shuzo, Otsu, Japan); phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich, St. Louis, MO), phytohemagglutinin (PHA; Remel Europe, Dartford, UK), gp100 peptide (YLEPGPVTA) (ProImmune, Oxford, UK); GolgiStop (BD Biosciences), GolgiPlug (BD Biosciences), and paraformaldehyde (PFA; Brunschwig, Amsterdam, The Netherlands).
TCR and tCD34 transgenes
TCRαβ specific for gp100/A2 was derived from cytotoxic T lymphocyte (CTL) clone 296 and uses the TCR-V genes TRAV13-1*02/J52*01/C and TRBV27*01/J2-7*01/D2*02/C2 (nomenclature according to http://imgt.org) (Schaft et al., 2003). TCRα and TCRβ chains were cloned in pBullet vectors either as single TCR chains (i.e., pBullet:TCRα+pBullet:TCRβ; Schaft et al., 2003) or as combined TCRα and TCRβ chains linked by 2A sequences and truncated CD34 (i.e., pBullet:TCRα-2A-tCD34-2A-TCRβ). See Fig. 1A for a schematic representation of TCR transgenes. Truncated CD34, a naturally occurring splice variant of human CD34 that lacks protein kinase C-binding sites in its cytoplasmic tail (Fehse et al., 2000), was flanked by 2A sequences (kindly provided by D. Gilham, University of Manchester, Manchester, UK), and introduced into the pBullet vector via PCR and an NcoI ligation. Primers used to amplify 2A-tCD34-2A were as follows: tCD34 5′ primer, attcggccatggcggggcgcgcccgctcgagcgagtgaaacagactttgaat; and tCD34 3′ primer, acgcgtccaagcttggcaattgattccctggcccggggttggactc and introduced additional restriction sites. These allowed subsequent introduction of TCRα and TCRβ chains into pBullet:tCD34. TCRα and TCRβ were subcloned via NcoI/XhoI and MfeI/MluI, respectively, into pBullet:tCD34.
Retroviral gene transfer into T cells
Moloney murine leukemia retroviruses, positive for TCR and TCR-tCD34, were produced by cocultures of the packaging cell lines 293T and Phoenix-A. Packaging cells were calcium phosphate-transfected with pBullet:TCRα+pBullet:TCRβ or with pBullet:TCRα-2A-tCD34-2A-TCRβ, and the helper vectors pHIT60 MLV GAG/POL and pCOLT-GALV ENV. Transduction of human T cells was performed as described previously (Lamers et al., 2006).
Peptide–MHC complexes
Complexes of gp100280–288/HLA-A*0201 (gp100/A2) peptide–MHC used in this study were tetramers, pentamers and Streptamers. Tetramers were generated by multimerization of 5 μg of biotinylated monomers (Sanquin, Amsterdam, The Netherlands) with 12.5 μl of Streptavidin–PE (BD Biosciences), which were mixed and incubated for 40 min at 4°C. Streptavidin–PE was added and mixed in steps of 2.5 μl that were followed by incubations of 8 min; pentamers were purchased as PE-conjugated multimers (ProImmune); Streptamers were generated by multimerization of 1 μg of Strep-tagged monomers (IBA, Göttingen, Germany) with 0.75 μg of Strep-Tactin–PE (IBA) in 50 μl of buffer (phosphate-buffered saline [PBS] with 0.5% bovine serum albumin [BSA], 2 mM EDTA; pH 7.4), and were mixed and incubated for 45 min. Sort reagents, during preparations and once ready, were kept at 4°C and protected from light. Concentrations of tetramers, pentamers, and Streptamers for use in flow cytometry were determined per batch by serial dilutions and set at 1:100, 1:20, and undiluted, respectively. See Fig. 1 for a schematic representation of peptide–MHC reagents, and Table 1 for properties and use of peptide–MHC multimers in MACS.
MACS to enrich gene-engineered T cells
Human T cells were labeled either with PE-conjugated peptide–MHC multimers and microbead-conjugated PE mAb or microbead-conjugated CD34 mAb according to the manufacturer's instructions (Miltenyi Biotec). In the case of peptide–MHC multimer stainings, reagents were added to cell pellets (see Table 1 for final concentrations) and incubated for 45 min (all solutions were ice-cold and all incubations were performed at 4°C and protected from light). T cells were washed twice with PBS–0.5% BSA (pH 7.4), resuspended in PBS–0.5% BSA with PE mAb microbeads (volume ratio, 4:1), and incubated for 15 min. In the case of CD34 mAb stainings, T cells were washed with PBS, resuspended in PBS–0.5% BSA with CD34 mAb microbeads and Fc receptor (FcR)-blocking reagent (volume ratio, 5:1:1), and incubated for 30 min. Microbead-labeled T cells were washed, resuspended in PBS-0.5% BSA, passed over a MACS preseparation filter, and separated in MACS separation columns that were exposed to a magnetic field. Sorted T cells were washed and subsequently flushed from the column with PBS–0.5% BSA. In the case of Streptamers, sorted T cells were treated twice with 1 mM d-biotin (Invitrogen, Carlsbad, CA) for 20 min and washed repeatedly. Last, sorted T cells were counted and cultured on feeder cells (1×104 T cells/200 μl of feeder medium; Van de Griend et al., 1984).
T cell counts
T cells were counted directly after MACS, 1 week after MACS, and weekly up to 6 weeks after MACS. T cell viability was determined by trypan blue exclusion and viable T cells were counted microscopically with Bürker chambers and a Leitz Laborlux 12 microscope (Leica Geosystems, Rijswijk, The Netherlands). T cell yields and expansions are represented as absolute numbers and fold increases in cell numbers, respectively.
Flow cytometry
All T cell stainings were performed according to standard protocols. T cells (1×104 cells) were incubated with peptide–MHC multimers for 15 min at room temperature or with mAbs for 30 min on ice, and fixed with 1% PFA. To measure antigen-specific T cell responses, T cells were stimulated with target cells and assessed for surface expression of CD107a and intracellular expression of IFN-γ and IL-2. Target cells used were BLM cells (without or with 10 μM gp100 peptide) and FM3 cells. CD107a expression was detected as described previously (Govers et al., 2011). Briefly, T cells were resuspended in a mixture of T cell medium, GolgiStop, and CD107a mAb–PE. Next, target cells and T cells were mixed at a 1:1 ratio and incubated for 2 hr at 37°C and 5% CO2. T cells were subsequently stained with CD3ɛ mAb to allow distinction of T cells from target cells at the time of analysis. Intracellular cytokine levels were detected in T cells (2×105) that were stimulated with target cells (6×104) in the presence of GolgiPlug for 6 hr at 37°C and 5% CO2. Next, T cells were washed and stained with CD8α mAb–APC, after which T cells were washed again, permeabilized (permeabilization solution 2; BD Biosciences) for 10 min at room temperature, and stained with IFN-γ mAb–FITC or IL-2 mAb–FITC. Samples were measured on a FACSCalibur dual-laser flow cytometer (Becton Dickinson, Alphen aan den Rijn, The Netherlands). Data analysis was performed on viable and in some cases CD3-positive T cells, using CellQuest software (BD Biosciences), and data are displayed either as dot plots or histograms.
IFN-γ production
T cells were assayed for their IFN-γ production as described previously (Schaft et al., 2006). BLM melanoma cells, without or with titrated amounts of gp100 peptide (between 10–4 and 101 μM), and FM3 melanoma cells were used to stimulate T cells overnight, and supernatants were tested in triplicate. T cell IFN-γ levels were determined by ELISA (Sanquin) and a TiterTek Plus reader (Merlin, Breda, The Netherlands) according to the manufacturer's instructions.
Statistical analyses
Student t tests (unpaired, two-tailed) and GraphPad (San Diego, CA) Prism 4 software were used to test the various sort reagents with respect to in vitro properties of T cells. Differences with p values less than 0.05 were considered significant.
Results
MACS with Streptamers or CD34 mAb results in enhanced T cell yield and expansion
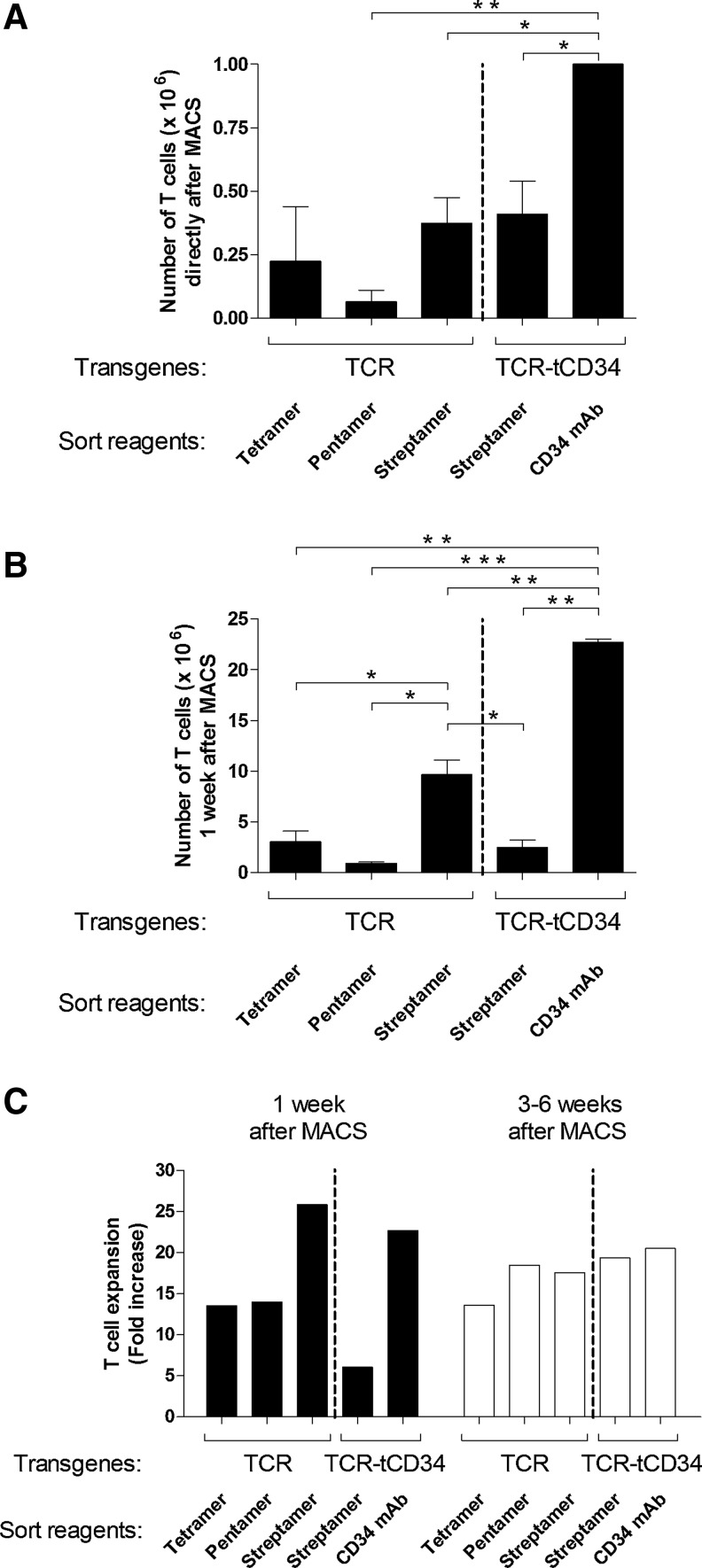
Primary human T cells were transduced with gp100/A2-specific TCR and TCR-tCD34 genes and MACSorted with tetramers, pentamers, Streptamers, or CD34 mAb. Flow cytometric analyses showed that presort TCR T cells labeled similarly with the various peptide–MHC multimers, which extends an earlier report by Yao and colleagues (2008). In addition, TCR and TCR-tCD34 T cells showed comparable binding of peptide–MHC multimers (data not shown). MACS of TCR T cells or TCR-tCD34 T cells with peptide–MHC multimers (input for all labeling conditions, 10×106 T cells) resulted in comparable numbers of T cells directly after MACS (output, 0.07–0.41×106 T cells), whereas MACS of TCR-tCD34 T cells with CD34 mAb (input again, 10×106 T cells) resulted in significantly enhanced numbers of T cells (output, 1.0×106 T cells; Fig. 2A). MACS of TCR-tCD34 T cells with CD34 mAb also resulted in the highest yield of T cells 1 week after MACS (22.7×106 T cells), which was significantly higher when compared with MACS of TCR-tCD34 T cells with Streptamers (2.5×106 T cells) (Fig. 2B). The increased yield of CD34 mAb-sorted T cells was due to both enhanced T cell numbers directly after MACS and enhanced T cell expansion in the first week after MACS (Fig. 2A and C). MACS of TCR T cells with Streptamers resulted in a significantly enhanced yield of T cells 1 week after MACS (9.7×106 T cells) when compared with tetramers and pentamers (3.1 and 0.9×106 T cells, respectively) (Fig. 2B). The increased yield of Streptamer-sorted T cells was due primarily to enhanced T cell expansion in the first week after MACS (Fig. 2C) rather than enhanced T cell numbers directly after MACS (Fig. 2A). Notably, Streptamer-sorted TCR-tCD34 T cells, when compared with Streptamer-sorted TCR T cells, yielded lower T cell numbers, which appeared to be related to reduced T cell expansion in the first week after MACS (Fig. 2B and C). When analyzing T cell yields at later time points, that is, between 3 and 6 weeks after MACS, we observed clear T cell expansion rates (weekly T cell expansion>10-fold) with no differences observed between the sort reagents (Fig. 2C).
FIG. 2.
MACS with Streptamer or CD34 mAb improves T cell yield. Primary human T cells were transduced either with TCR or TCR-tCD34 transgenes as depicted in Fig. 1A. TCR T cells were MACSorted with tetramers, pentamers, and Streptamers, whereas TCR-tCD34 T cells were MACSorted with Streptamers and CD34 mAbs. After MACSorting, T cell numbers were counted microscopically and T cells were expanded with feeder cultures (as described in Materials and Methods). Mean T cell numbers and SEM (A) directly after MACS or (B) 1 week after MACS are from two to six repeat experiments with T cells from two to four healthy donors. (C) T cell expansion 1 week (solid columns) and 3–6 week(s) (open columns) after MACS is expressed as the fold increase in T cell numbers when comparing 1 week and directly after MACS and 3–6 weeks and 1 week after MACS, respectively. T cell expansions are from representative T cell cultures out of two to four repeat experiments with T cells from two to four healthy donors with similar results. Statistically significant differences were calculated with Student t tests: *p<0.05; **p<0.005; ***p<0.0005.
MACS with Streptamers, but not CD34 mAb, results in T cell populations significantly enriched for peptide–MHC binding
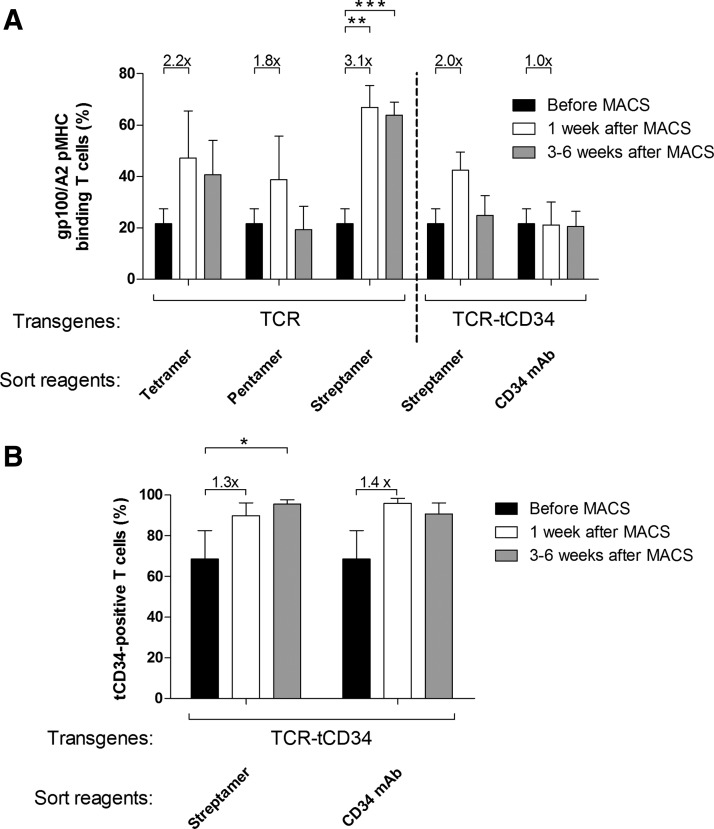
MACS of TCR T cells with tetramers, pentamers, or Streptamers resulted in clear enrichments of peptide–MHC-binding T cells (2.2-, 1.8-, and 3.1-fold increase, respectively; Fig. 3A). Streptamers, however, were the only peptide–MHC multimers that resulted in percentages of peptide–MHC-binding T cells (>65%) that were significantly enhanced when compared with presort T cells (20%). Please note that limited output of T cell numbers directly after MACSort with tetramers and pentamers (in some cases<50,000 cells) did not allow for flow cytometry with peptide–MHC multimers because the total number of T cells was expanded to provide the yield 1 week after MACSort necessary for downstream assays. In general, in our experience, peptide–MHC enrichment 1 week after MACS or FACS correlates well with the actual sorting efficiency. Percentages of Streptamer-sorted T cells that bind peptide–MHC proved stable over a culture period of up to 6 weeks (Fig. 3A). In contrast, percentages of pentamer-sorted T cells that bind peptide–MHC decreased to presort values in a 6-week time period.
FIG. 3.
MACS of TCR T cells with Streptamers results in enhanced peptide–MHC binding that is stable over time. Primary human T cells were transduced and MACSorted as described in the legend to Fig. 2. Sorted T cells were analyzed by flow cytometry for (A) peptide–MHC binding (using gp100/A2 pentamer) and (B) surface expression of tCD34 (using CD34 mAb) at various time points: before MACS (solid columns), 1 week after MACS (open columns), and 3–6 weeks after MACS (shaded columns). Columns and error bars represent the mean percentage and SEM of two to four repeat experiments with T cells from two to four healthy donors. Binding of peptide–MHC or CD34 mAb before MACS was 20 and 70%, respectively. Enrichment of T cells that bind peptide–MHC or CD34 mAb is presented as the fold difference between T cells 1 week after and before MACS, and is indicated above the corresponding bars. Statistically significant differences were calculated with Student t tests: *p<0.05; **p<0.005; ***p<0.0005.
MACS of TCR-tCD34 T cells with either CD34 mAb or Streptamers resulted in a maximal enrichment of tCD34-expressing T cells, nearly 100%, which was stable over time (Fig. 3B). Unexpectedly, MACS of TCR-tCD34 T cells with CD34 mAb did not result in an enrichment of peptide–MHC-binding T cells (Fig. 3A). Also, Streptamer MACS of TCR-tCD34 T cells resulted in an enrichment of peptide–MHC-binding T cells that was 2-fold less and not stable over time when compared with TCR T cells (Fig. 3A).
MACS with CD34 mAb yields T cells with enhanced CD107a mobilization and IFN-γ production in response to antigen-positive melanoma cells
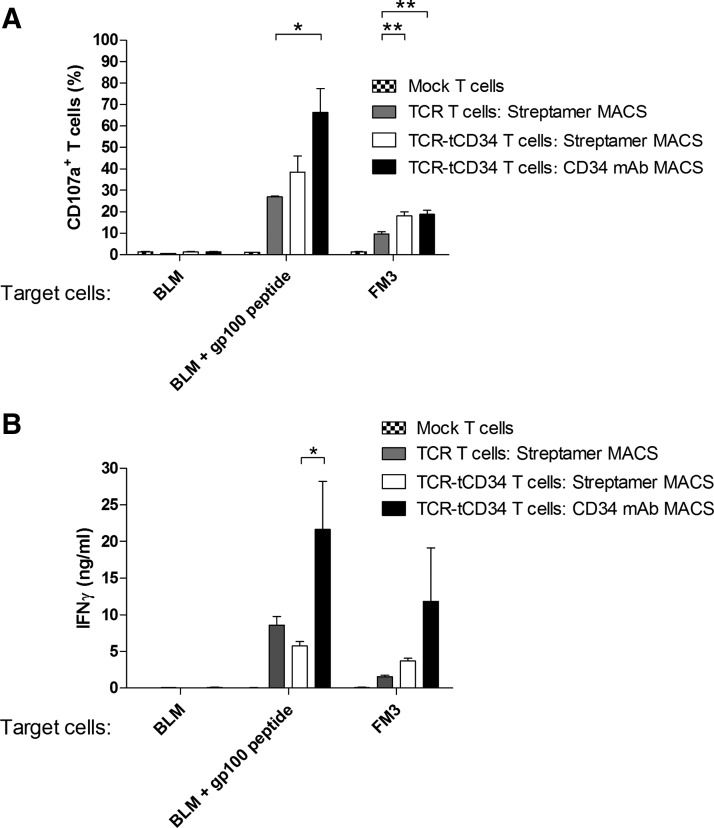
To analyze T cell functions, we decided to compare Streptamers, which among the peptide–MHC multimers tested showed the best T cell yield and enrichment for peptide–MHC-binding T cells, with CD34 mAb as sort reagents. Three to 6 weeks after MACS, TCR and TCR-tCD34 T cells were stimulated with peptide-loaded BLM or native FM3 melanoma cells and analyzed for surface expression of CD107a and IFN-γ production. To better compare T cell functions, values were standardized for peptide–MHC binding (see Supplementary Fig. S1A; supplementary data are available online at www.liebertpub.com/hum). Streptamer-sorted TCR-tCD34 T cells, when compared with similarly sorted TCR T cells, revealed a higher level of CD107a expression and similar levels of IFN-γ production in response to FM3 melanoma cells (Fig. 4A and B). Strikingly, CD34 mAb-sorted T cells showed the highest level of CD107a expression and IFN-γ production in response to melanoma cells (Fig. 4A and B). To assess T cell sensitivity, we have tested IFN-γ responses toward titrated amounts of gp100 peptide. CD34 mAb-sorted T cells generally produced about 2-fold more IFN-γ in response to 10–3 to 101 μM peptide; however, equal EC50 values of approximately 0.04 μM peptide were observed (Supplementary Fig. S1B).
FIG. 4.
Human T cells sorted with CD34 mAb demonstrate increased antigen-specific responses. Primary human T cells were transduced with empty virus particles (mock T cells) or with TCR (TCR T cells) or TCR-tCD34 transgenes (TCR-tCD34 T cells) and were either not MACSorted (mock T cells, patterned columns) or MACSorted with Streptamers (TCR T cells, shaded columns; TCR-tCD34 T cells, open columns) or anti-CD34 mAbs (TCR-tCD34 T cells, solid columns). T cells sorted with CD34 mAb show upregulated (A) surface expression of T cell CD107a and (B) IFN-γ production on stimulation with antigen-positive melanoma cells. T cells were stimulated with the following target cells: BLM cells (gp100–/A2+) loaded without or with 10 μM gp100 peptide, and FM3 cells (gp100+/A2+). CD107a expression (percent) was measured by flow cytometry, gating on viable and CD3-positive T cells, after a 2-hr stimulation, and IFN-γ production (ng/ml) was measured after overnight stimulation with conditioned supernatants by ELISA. Columns and error bars represent mean values and SEM of two independent measurements from two healthy donors. CD107a and IFN-γ data were corrected for differences in peptide–MHC binding, with peptide–MHC binding of CD34 mAb-sorted T cells set to 100% (see Supplementary Fig. S1A for details). Statistically significant differences were calculated by Student t test: *p<0.05; **p<0.005.
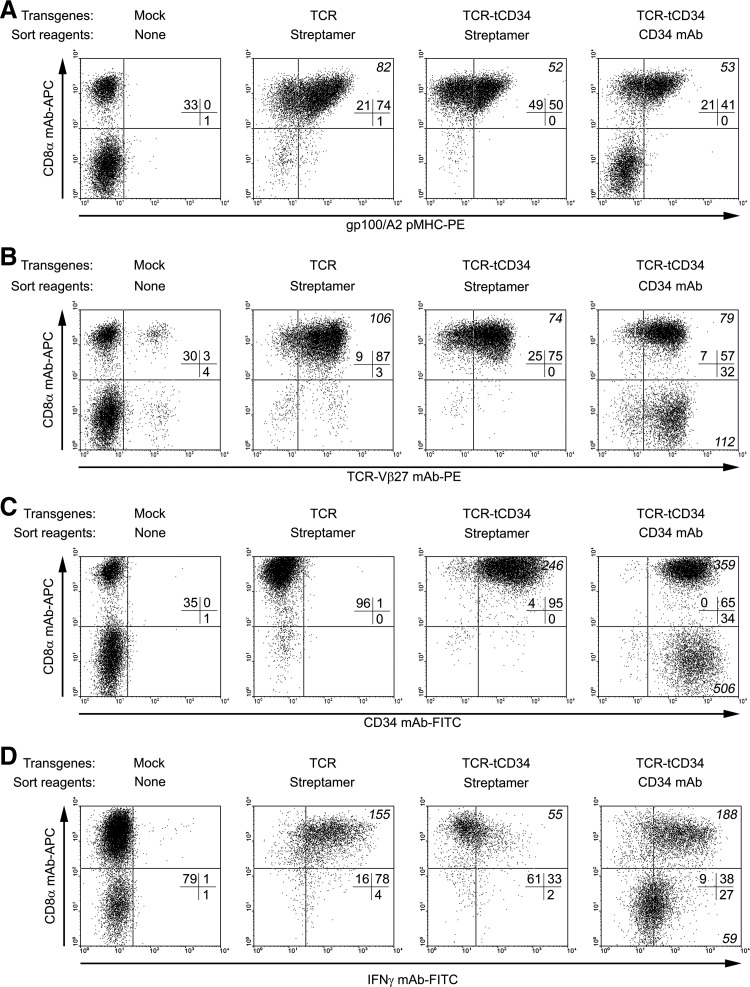
MACS with CD34 mAb, but not Streptamers, results in CD4+ and CD8+ T cells that both contribute to antigen-specific IFN-γ production
To follow up on the enhanced function of CD34 mAb-sorted T cells, we performed multiparametric flow cytometry to study the contribution of CD4+ and CD8+ T cell subsets to IFN-γ production. Besides intracellular IFN-γ, sorted T cells were analyzed for peptide–MHC binding, and surface expression of TCR-Vβ27, tCD34, and CD8α. Percentages of CD4+ T cells in presorted T cells were 67% (Fig. 5A, nonsorted mock T cells), whereas CD4+ T cell percentages in TCR T cells sorted with Streptamers or TCR-tCD34 T cells sorted with either Streptamers or CD34 mAb were 5, 1, and 38%, respectively (Fig. 5A). As expected, MACS with Streptamers yielded a T cell population that was biased for CD8+ T cells. In contrast, MACS with CD34 mAb yielded T cells that contained significant populations of both CD4+ and CD8+ T cells. CD4+ T cells within CD34 mAb-sorted T cells expressed TCR-Vβ27 and tCD34 at levels equal to those observed in CD8+ T cells, yet CD4+ T cells did not bind peptide–MHC (Fig. 5A–C). Stimulation of T cells with peptide-loaded BLM cells resulted in enhanced intracellular levels of IFN-γ in CD8+ T cells irrespective of the MACS reagent used. Notably, in CD34 mAb-sorted T cells both CD4+ and CD8+ T cell subsets demonstrated antigen-specific IFN-γ production (Fig. 5D). Similarly, although to a lesser extent, CD4+ T cells contributed to antigen-specific IL-2 production of CD34 mAb-sorted T cells (Supplementary Fig. S2A and B). Table 2 provides a summary of our data with respect to yield, expansion, enrichment for peptide–MHC binding and tCD34, and antigen-specific functions of T cells MACSorted with tetramers, pentamers, Streptamers, or CD34 mAb.
FIG. 5.
IFN-γ production of CD34 mAb-sorted T cells, but not peptide–MHC-sorted T cells, depends on both CD8+ and CD4+ T cells. Primary human T cells were transduced and MACSorted as described in the legend to Fig. 4. T cells were analyzed by flow cytometry after staining with APC-conjugated anti-CD8α mAb in combination with one of the following reagents: (A) PE-conjugated gp100/A2 tetramer (n=12); (B) PE-conjugated anti-TCR-Vβ27 mAb (n=6); (C) FITC-conjugated anti-CD34 mAb (n=14); and (D) FITC-conjugated anti-IFN-γ mAb (n=2). In the case of IFN-γ stainings, T cells were stimulated for 6 hr with BLM (gp100–/A2+) cells loaded with 10 μM gp100 peptide, permeabilized, and stained with anti-IFN-γ mAb, as described in more detail in Materials and Methods. Shown are representative dot plots, indicating percentages (all quadrants) and MFIs (upper and lower right quadrants when percent ≥5, in italic) of stained T cells.
Table 2.
Results of Magnetic-Activated Cell Sorting of T Cells with Peptide–MHC Multimers or CD34 Monoclonal Antibody: In Vitro Evaluationa
| TCR T cells | TCR-tCD34 T cells | ||||
|---|---|---|---|---|---|
| T | P | S | S | CD34 | |
| T cell output numbers (directly after MACS)b | ± | – | + | + | ++ |
| T cell yield (1 week after MACS)c | ± | – | + | ± | ++ |
| T cell expansion (1 week after MACS)d | ± | ± | ++ | – | ++ |
| T cell expansion (3–6 weeks after MACS)d | ± | + | + | + | ++ |
| pMHC binding (1 week after MACS)e | + | + | ++ | + | – |
| pMHC binding (3–6 weeks after MACS)e | + | – | ++ | ± | – |
| tCD34 expression (1 week after MACS)f | NA | NA | NA | ++ | ++ |
| tCD34 expression (3–6 weeks after MACS)f | NA | NA | NA | ++ | ++ |
| Antigen-specific CD107a mobilizationg | ND | ND | ± | ± | ++ |
| Antigen-specific IFN-γ secretionh | ND | ND | ± | ± | ++ |
Primary human T cell populations were transduced with TCR or TCR-tCD34 genes, MACSorted with gp100/A2 tetramers (T), pentamers (P), or Streptamers (S), or with CD34 mAb (CD34), and evaluated for various in vitro parameters. See Materials and Methods for details.
T cell numbers directly after MACS (counted microscopically): –, 0–0.10×106:±, 0.10–0.25×106;+, 0.26–0.75×106; ++, >0.75×106.
T cell numbers 1 week after MACS (counted microscopically): –, 0–1×106;±, 2–5×106; +, 6–15×106; ++, >15×106.
T cell expansion 1 or 3–6 week(s) after MACS (fold increase in T cell numbers at 1 week vs. directly after MACS and 3–6 weeks vs. 1 week after MACS, respectively): –, 0–10;±, 11–15;+, 16–20; ++, >20.
Peptide–MHC binding 1 and 3–6 week(s) after MACS (measured by flow cytometry with gp100/A2 pentamer): –, ≤20% (value before MACS);±, 21–30; +, 31–50; ++, >50%.
tCD34 expression 1 and 3–6 week(s) after MACS (measured by flow cytometry with CD34 mAb):–, ≤70% (value before MACS);±, 71–80; +, 81–90; ++, >90%.
T cell CD107a mobilization 3–6 weeks after MACS (measured by flow cytometry with CD107a mAb on peptide-specific stimulation): –, 0–20;±, 21–40; +, 41–60; ++, >60%.
T cell IFN-γ production 3–6 weeks after MACS (measured by ELISA on peptide-specific stimulation): –, 0–5;±, 6–10; +, 11–20; ++, >20 ng/ml.
MACS, magnetic-activated cell sorting; NA, not applicable; ND, not determined; pMHC, peptide–MHC; tCD34, truncated CD34.
Discussion
In this study, we have performed a head-to-head comparison of tetramers, pentamers, Streptamers, and CD34 mAb as sort reagents to obtain a population of MACS-enriched TCR-engineered T cells. First, we compared the various peptide–MHC multimers and noted that Streptamers provided significantly improved output of T cell numbers, which was due to primarily enhanced T cell expansion in the first week after MACS. This observation is in line with a previous report showing that dissociation of Streptamers into monomers, as we have done after MACS, reduces activation-induced cell death (AICD) and improves T cell proliferation when compared with nondissociated multimers (Neudorfer et al., 2007). In fact, multimeric peptide–MHC is reported to result in upregulated expression of Fas ligand (FasL), which in turn results in FasL/Fas-mediated T cell death (Xu et al., 2001). In contrast, peptide–MHC monomers do not lead to full and enduring TCR-mediated T cell activation, most likely due to relatively low-affinity interactions with TCRs and high off-kinetics of monomeric peptide–MHC (Matsui et al., 1991; Minguet et al., 2007). Besides Streptamers, the use of an mAb directed against a truncated CD34 molecule represents another means of circumventing AICD. When comparing peptide–MHC multimers with CD34 mAb, we observed that CD34 mAb sorting of gene-marked T cells provided the highest output of T cell numbers, which was due to both enhanced T cell numbers directly after MACS and enhanced T cell expansion in the first week after MACS. T cell populations all expanded at similar rates up to 6 weeks after MACS, implying that T cells bound to sort reagents such as tetramers and pentamers are ultimately “diluted out” or lost (Fig. 2).
With respect to enrichment for peptide–MHC binding, Streptamer-based MACS of TCR T cells resulted in the most effective enrichment, which proved stable over time. The reversibility of Streptamer binding, which, as discussed previously, potentially aids T cell expansion of sorted T cells, may have contributed to the observed T cell enrichment. MACS of TCR-tCD34 T cells with CD34 mAb resulted in a T cell population that was nearly completely positive for tCD34 for at least 6 weeks but did not show improved binding of peptide–MHC. Similar results were obtained with Streptamer MACS of TCR-tCD34 T cells, showing enrichment of tCD34 but only a weak and transient enrichment for peptide–MHC-binding T cells. Findings with TCR-tCD34 T cells suggest that CD34 mAb sorting does not result in concomitant enrichment of peptide–MHC-binding T cells, whereas peptide–MHC multimer sorting does result in enrichment of tCD34-positive T cells. This observation may be related to the fact that surface expression of TCRαβ chains, but not tCD34, is prone to competition for endogenous proteins and TCR mispairing (Govers et al., 2010).
When comparing TCR-tCD34 and TCR T cells with respect to peptide–MHC binding, we postulate that the TCRα-2A-tCD34-2A-TCRβ cassette may compromise gene expression of TCRαβ. Although we cannot exclude that the products of TCR genes intervened by a tCD34 are more prone to mispair with endogenous TCR chains, TCR genes expressed from a single construct are generally less prone to mispair when compared with TCR genes expressed from separate constructs (Bendle et al., 2010). Sizes of transgenes in pBullet retroviral vectors may adversely affect gene expression (TCR-tCD34, 2867 nucleotides; TCRα or TCRβ, 827 and 923 nucleotides) (data not shown). More specifically, surface expression of the TCRα gene, positioned 5′ of 2A sequences, may be reduced as a consequence of the 2A amino acids added to the 3′ end of the TCRα protein (Garcia et al., 1996; Rudolph et al., 2006). Addition of 2A amino acids to the intracellular domain of TCRα potentially decreases the stability of immunoglobulin constant region-containing proteins. Because TCR-Vα13 mAbs are commercially not available, we cannot formally prove the decreased surface expression of TCRα. However, two lines of evidence support decreased expression of TCRα by TCR-tCD34 T cells. First, peptide–MHC binding by TCR-tCD34 T cells is decreased compared with TCR T cells whereas TCRβ surface expression is not (Fig. 3A and Supplementary Fig. S3, and also Fig. 5A and B). Second, mean fluorescence intensity of peptide–MHC binding by Streptamer-sorted TCR-tCD34 T cells was 1.6-fold lower when compared with similarly sorted TCR T cells (Fig. 5A). Along these same lines, binding of peptide–MHC monomers (on reversal of Streptamers into peptide–MHC monomers) is expected to be less persistent in the case of TCR-tCD34 T cells, potentially providing a decreased T cell proliferation signal in the setting of feeder cultures (with T cell costimulation) (Fig. 2C). TCRβ expression appears generally less sensitive for additional 3′ amino acids (Hart et al., 2008; Leisegang et al., 2008; Yang et al., 2008), suggesting that a TCRβ-2A-tCD34-2A-TCRα or tCD34-2A-TCRβ-2A-TCRα configuration, in particular in those vectors that can carry large-sized transgenes, may be more favorable for stable TCRα expression and peptide–MHC multimer binding by TCR-tCD34 T cells.
When analyzing T cell functions, CD34 mAb-sorted T cells revealed enhanced CD107a mobilization and IFN-γ production in response to melanoma target cells. The enhanced functional avidity of these T cells did not correlate with the surface expression of TCRβ, which is in line with the aforementioned discrepancy between TCRβ expression and peptide–MHC binding. Supplementary Fig. S3 demonstrates that MACSorting enhanced the percentages of TCRβ in a manner that appeared independent of T cell population and sort reagent used. Within our panel of MACSorted T cells, CD34 mAb-sorted T cells are unique with respect to the presence of CD4+ T cells (Fig. 5A), and these CD4+ T cells contributed to higher antigen-specific T cell IFN-γ responses (Fig. 5D). It is noteworthy that besides T cell function, it is not expected that CD4+ T cells contributed to differences observed with respect to either T cell output numbers, T cell yield, or enrichment for peptide–MHC binding (Figs. 2 and 3). First, the relative presence of CD4+ T cells was identical in presort T cells from individual donors (as in nonsorted mock T cells; Fig. 5A). Second, T cell expansion rates did not differ among all sorted T cell populations tested (Fig. 2C) suggesting that differences in T cell yield and enrichment for peptide–MHC binding 1 week after MACS (Figs. 2 and 3) were not due to the presence of CD4+ T cells. Somewhat unexpectedly, CD4+ T cells did not contribute to peptide–MHC binding of CD34 mAb-sorted T cells. We hypothesize that MACS procedures, in contrast to, for instance, FACS procedures, may apply additional forces to the TCR:peptide–MHC interactions, such as those caused by downward flows of washing buffers (Miltenyi et al., 1990). Consequently, MACS of T cells may select for T cells with the highest avidity for peptide–MHC, in which case CD8+ T cells generally have an advantage over CD4+ T cells. Downward flow may not affect MACS with CD34 mAb because antibodies generally have a 10,000-fold higher ligand-binding affinity than TCRs (Matsui et al., 1991). The absence of peptide–MHC binding by TCR-positive CD4+ T cells is an observation that seemingly contradicts a previous report, which showed that the same gp100/A2 TCR acts independently of the CD8α coreceptor (Willemsen et al., 2006). In fact, in the latter study we have shown that CD4+ and CD8+ T cells revealed comparable peptide–MHC binding and nearly comparable T cell cytotoxicity, but T cells were transduced with TCR genes and obtained via peptide–MHC/CD8α mAb-based FACSorting (Willemsen et al., 2006). The lowered and unstable TCRα expression in TCR-tCD34 T cells may reduce T cell avidity and hamper peptide–MHC binding, resulting, as discussed, in lowered peptide–MHC binding mean fluorescence intensity of CD8+ T cells and potentially, in this study, in absent peptide–MHC binding by CD4+ T cells (Fig. 5A).
Despite the absence of peptide–MHC binding, the CD4+ T cell population in CD34 mAb-sorted TCR-tCD34 T cells contributed to antigen-specific responses as shown by intracellular IFN-γ and, to a lesser extent, IL-2 production (Fig. 5D and Supplementary Fig. S2A). This observation is in agreement with the notion that T cell function requires only a few peptide–MHC molecules, which may be well below the detection limit of T cell peptide–MHC binding by standard flow cytometric methods (Irvine et al., 2002). IFN-γ and IL-2 production by CD4+ T cells is important for recruitment and mobilization of CD8+ T cells to the tumor site (Nakanishi et al., 2009; Bos and Sherman, 2010). In addition, CD4+ T cells aid the induction of CD8+ T cell-mediated immune responses, targeting of tumor stroma, and formation of T cell memory (Keene and Forman, 1982; Jennings et al., 1991; Bennett et al., 1997; Hung et al., 1998; Gao et al., 2002; Janssen et al., 2003; Smith et al., 2004; Schietinger et al., 2010). Findings on IFN-γ and IL-2 production by CD34 mAb-sorted CD4+ T cells extend a previous study, where we observed antigen-specific production of high levels of IFN-γ and low levels of IL-2 in gp100/A2 TCR-positive CD4+ T cells (Willemsen et al., 2006). In another study, CD8+ T cells transduced with an ERBB2-specific chimeric antigen receptor (CAR) revealed the best T cell expansion and protection against metastases when cocultured with helper T cell type 1 (Th1) rather than Th2 T cells (Moeller et al., 2007). Thus, the antitumor activity of CD34 mAb-sorted T cells may be further exploited by skewing CD4+ T cells toward a Th1 phenotype.
Our study has focused on three types of peptide–MHC multimers to enrich TCR-transduced T cells by MACS. To date, other peptide–MHC multimers have been designed that contain up to 200 peptide–MHC monomers (Lebowitz et al., 1999; Mallet-Designe et al., 2003; Chattopadhyay et al., 2006; Scholler et al., 2010; Davis et al., 2011). Although the use of these novel peptide–MHC reagents needs to be tested for T cell sorting, the enhanced valencies of these peptide–MHC multimers may allow enrichment for CD4+ T cells, yet the accompanying lower “off-rates” and inability to revert binding are likely to result in AICD.
In short, MACSorting of T cells with detachable Streptamers improves T cell yield and enrichment for peptide–MHC-binding T cells when compared with tetramers and pentamers. Truncated CD34 proves a valid surrogate marker to MACSort T cells, resulting in further enhancement of T cell yield and maximal CD34 enrichment, yet does not enrich T cells for peptide–MHC binding. CD34 mAb sorting yields CD4+ T cells that significantly contribute to antigen-specific T cell functions. Therefore, we conclude that CD34 mAb-based MACSorting of T cells has benefits toward applications of T cell therapy, especially those that require CD4+ T cells.
Supplementary Material
Acknowledgments
This work was supported by the European Union 6th framework grant (018914), entitled: Adoptive Engineered T Cell Targeting to Activate Cancer Killing (ATTACK).
Author Disclosure Statement
The authors declare no conflict of interest.
References
- Altman J.D. Moss P.A. Goulder P.J., et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [Google Scholar]
- Bendle G.M. Linnemann C. Hooijkaas A.I., et al. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat. Med. 2010;16:565–570. doi: 10.1038/nm.2128. 561p following 570. [DOI] [PubMed] [Google Scholar]
- Bennett S.R. Carbone F.R. Karamalis F., et al. Induction of a CD8+ cytotoxic T lymphocyte response by cross-priming requires cognate CD4+ T cell help. J. Exp. Med. 1997;186:65–70. doi: 10.1084/jem.186.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser M.J. Shapira-Frommer R. Treves A.J., et al. Clinical responses in a phase II study using adoptive transfer of short-term cultured tumor infiltration lymphocytes in metastatic melanoma patients. Clin. Cancer Res. 2010;16:2646–2655. doi: 10.1158/1078-0432.CCR-10-0041. [DOI] [PubMed] [Google Scholar]
- Bos R. Sherman L.A. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res. 2010;70:8368–8377. doi: 10.1158/0008-5472.CAN-10-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay P.K. Price D.A. Harper T.F., et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat. Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- Cooper L.J. Kalos M. Lewinsohn D.A., et al. Transfer of specificity for human immunodeficiency virus type 1 into primary human T lymphocytes by introduction of T-cell receptor genes. J. Virol. 2000;74:8207–8212. doi: 10.1128/jvi.74.17.8207-8212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.M. Altman J.D. Newell E.W. Interrogating the repertoire: Broadening the scope of peptide–MHC multimer analysis. Nat. Rev. Immunol. 2011;11:551–558. doi: 10.1038/nri3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley M.E. Wunderlich J.R. Yang J.C., et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J. Clin. Oncol. 2005;23:2346–2357. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehse B. Richters A. Putimtseva-Scharf K., et al. CD34 splice variant: An attractive marker for selection of gene-modified cells. Mol. Ther. 2000;1:448–456. doi: 10.1006/mthe.2000.0068. [DOI] [PubMed] [Google Scholar]
- Feuchtinger T. Richard C. Joachim S., et al. Clinical grade generation of hexon-specific T cells for adoptive T-cell transfer as a treatment of adenovirus infection after allogeneic stem cell transplantation. J. Immunother. 2008;31:199–206. doi: 10.1097/CJI.0b013e31815ef862. [DOI] [PubMed] [Google Scholar]
- Gao F.G. Khammanivong V. Liu W.J., et al. Antigen-specific CD4+ T-cell help is required to activate a memory CD8+ T cell to a fully functional tumor killer cell. Cancer Res. 2002;62:6438–6441. [PubMed] [Google Scholar]
- Garcia K.C. Degano M. Stanfield R.L., et al. An αβ T cell receptor structure at 2.5 Å and its orientation in the TCR–MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- Govers C. Sebestyen Z. Coccoris M., et al. T cell receptor gene therapy: Strategies for optimizing transgenic TCR pairing. Trends Mol. Med. 2010;16:77–87. doi: 10.1016/j.molmed.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Govers C. Sebestyen Z. Berrevoets C., et al. T cell receptor fused to CD3ζ: Transmembrane domain of CD3ζ prevents TCR mis-pairing, whereas complete CD3ζ directs functional TCR expression. Open Gene Ther. J. 2011;4:11–22. [Google Scholar]
- Hammer M.H. Brestrich G. Mittenzweig A., et al. Generation of EBV-specific T cells for adoptive immunotherapy: A novel protocol using formalin-fixed stimulator cells to increase biosafety. J. Immunother. 2007;30:817–824. doi: 10.1097/CJI.0b013e318155a11c. [DOI] [PubMed] [Google Scholar]
- Hart D.P. Xue S.A. Thomas S., et al. Retroviral transfer of a dominant TCR prevents surface expression of a large proportion of the endogenous TCR repertoire in human T cells. Gene Ther. 2008;15:625–631. doi: 10.1038/sj.gt.3303078. [DOI] [PubMed] [Google Scholar]
- Hung K. Hayashi R. Lafond-Walker A., et al. The central role of CD4+ T cells in the antitumor immune response. J. Exp. Med. 1998;188:2357–2368. doi: 10.1084/jem.188.12.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine D.J. Purbhoo M.A. Krogsgaard M., et al. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- Janssen E.M. Lemmens E.E. Wolfe T., et al. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- Jennings S.R. Bonneau R.H. Smith P.M., et al. CD4-positive T lymphocytes are required for the generation of the primary but not the secondary CD8-positive cytolytic T lymphocyte response to herpes simplex virus in C57BL/6 mice. Cell. Immunol. 1991;133:234–252. doi: 10.1016/0008-8749(91)90194-g. [DOI] [PubMed] [Google Scholar]
- Johnson L.A. Morgan R.A. Dudley M.E., et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.A. Forman J. Helper activity is required for the in vivo generation of cytotoxic T lymphocytes. J. Exp. Med. 1982;155:768–782. doi: 10.1084/jem.155.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knabel M. Franz T.J. Schiemann M., et al. Reversible MHC multimer staining for functional isolation of T-cell populations and effective adoptive transfer. Nat. Med. 2002;8:631–637. doi: 10.1038/nm0602-631. [DOI] [PubMed] [Google Scholar]
- Labarriere N. Gervois N. Bonnin A., et al. PBMC are as good a source of tumor-reactive T lymphocytes as TIL after selection by Melan-A/A2 multimer immunomagnetic sorting. Cancer Immunol. Immunother. 2008;57:185–195. doi: 10.1007/s00262-007-0361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers C.H. Willemsen R.A. van Elzakker P., et al. Phoenix-ampho outperforms PG13 as retroviral packaging cells to transduce human T cells with tumor-specific receptors: Implications for clinical immunogene therapy of cancer. Cancer Gene Ther. 2006;13:503–509. doi: 10.1038/sj.cgt.7700916. [DOI] [PubMed] [Google Scholar]
- Lebowitz M.S. O'Herrin S.M. Hamad A.R., et al. Soluble, high-affinity dimers of T-cell receptors and class II major histocompatibility complexes: Biochemical probes for analysis and modulation of immune responses. Cell. Immunol. 1999;192:175–184. doi: 10.1006/cimm.1999.1441. [DOI] [PubMed] [Google Scholar]
- Leisegang M. Engels B. Meyerhuber P., et al. Enhanced functionality of T cell receptor-redirected T cells is defined by the transgene cassette. J. Mol. Med. (Berl) 2008;86:573–583. doi: 10.1007/s00109-008-0317-3. [DOI] [PubMed] [Google Scholar]
- Mallet-Designe V.I. Stratmann T. Homann D., et al. Detection of low-avidity CD4+ T cells using recombinant artificial APC: Following the antiovalbumin immune response. J. Immunol. 2003;170:123–131. doi: 10.4049/jimmunol.170.1.123. [DOI] [PubMed] [Google Scholar]
- Matsui K. Boniface J.J. Reay P.A., et al. Low affinity interaction of peptide–MHC complexes with T cell receptors. Science. 1991;254:1788–1791. doi: 10.1126/science.1763329. [DOI] [PubMed] [Google Scholar]
- Miltenyi S. Muller W. Weichel W., et al. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- Minguet S. Swamy M. Alarcon B., et al. Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity. 2007;26:43–54. doi: 10.1016/j.immuni.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Moeller M. Kershaw M.H. Cameron R., et al. Sustained antigen-specific antitumor recall response mediated by gene-modified CD4+ T helper-1 and CD8+ T cells. Cancer Res. 2007;67:11428–11437. doi: 10.1158/0008-5472.CAN-07-1141. [DOI] [PubMed] [Google Scholar]
- Molday R.S. Yen S.P. Rembaum A. Application of magnetic microspheres in labelling and separation of cells. Nature. 1977;268:437–438. doi: 10.1038/268437a0. [DOI] [PubMed] [Google Scholar]
- Morgan R.A. Dudley M.E. Wunderlich J.R., et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi Y. Lu B. Gerard C., et al. CD8+ T lymphocyte mobilization to virus-infected tissue requires CD4+ T-cell help. Nature. 2009;462:510–513. doi: 10.1038/nature08511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudorfer J. Schmidt B. Huster K.M., et al. Reversible HLA multimers (Streptamers) for the isolation of human cytotoxic T lymphocytes functionally active against tumor- and virus-derived antigens. J. Immunol. Methods. 2007;320:119–131. doi: 10.1016/j.jim.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Newell E.W. Klein L.O. Yu W., et al. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat. Methods. 2009;6:497–499. doi: 10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst M.R. Yang J.C. Langan R.C., et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol. Ther. 2011;19:620–626. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss E. Treschow A. Newrzela S., et al. TK.007: A novel, codon-optimized HSVtk(A168H) mutant for suicide gene therapy. Hum. Gene Ther. 2010;21:929–941. doi: 10.1089/hum.2009.042. [DOI] [PubMed] [Google Scholar]
- Rauser G. Einsele H. Sinzger C., et al. Rapid generation of combined CMV-specific CD4+ and CD8+ T-cell lines for adoptive transfer into recipients of allogeneic stem cell transplants. Blood. 2004;103:3565–3572. doi: 10.1182/blood-2003-09-3056. [DOI] [PubMed] [Google Scholar]
- Robbins P.F. Morgan R.A. Feldman S.A., et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph M.G. Stanfield R.L. Wilson I.A. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Schaft N. Willemsen R.A. de Vries J., et al. Peptide fine specificity of anti-glycoprotein 100 CTL is preserved following transfer of engineered TCR αβ genes into primary human T lymphocytes. J. Immunol. 2003;170:2186–2194. doi: 10.4049/jimmunol.170.4.2186. [DOI] [PubMed] [Google Scholar]
- Schaft N. Lankiewicz B. Drexhage J., et al. T cell re-targeting to EBV antigens following TCR gene transfer: CD28-containing receptors mediate enhanced antigen-specific IFNγ production. Int. Immunol. 2006;18:591–601. doi: 10.1093/intimm/dxh401. [DOI] [PubMed] [Google Scholar]
- Schietinger A. Philip M. Liu R.B., et al. Bystander killing of cancer requires the cooperation of CD4+ and CD8+ T cells during the effector phase. J. Exp. Med. 2010;207:2469–2477. doi: 10.1084/jem.20092450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholler J. Singh M. Bergmeier L., et al. A recombinant human HLA-class I antigen linked to dextran elicits innate and adaptive immune responses. J. Immunol. Methods. 2010;360:1–9. doi: 10.1016/j.jim.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Sebestyen Z. Schooten E. Sals T., et al. Human TCR that incorporate CD3ζ induce highly preferred pairing between TCRα and β chains following gene transfer. J. Immunol. 2008;180:7736–7746. doi: 10.4049/jimmunol.180.11.7736. [DOI] [PubMed] [Google Scholar]
- Smith C.M. Wilson N.S. Waithman J., et al. Cognate CD4+ T cell licensing of dendritic cells in CD8+ T cell immunity. Nat. Immunol. 2004;5:1143–1148. doi: 10.1038/ni1129. [DOI] [PubMed] [Google Scholar]
- Stull R.A. Hyun W.C. Pallavicini M.G. Simultaneous flow cytometric analyses of enhanced green and yellow fluorescent proteins and cell surface antigens in doubly transduced immature hematopoietic cell populations. Cytometry. 2000;40:126–134. [PubMed] [Google Scholar]
- Van de Griend R.J. Van Krimpen B.A. Bol S.J., et al. Rapid expansion of human cytotoxic T cell clones: Growth promotion by a heat-labile serum component and by various types of feeder cells. J. Immunol. Methods. 1984;66:285–298. doi: 10.1016/0022-1759(84)90340-5. [DOI] [PubMed] [Google Scholar]
- Weijtens M.E. Hart E.H. Bolhuis R.L. Functional balance between T cell chimeric receptor density and tumor associated antigen density: CTL mediated cytolysis and lymphokine production. Gene Ther. 2000;7:35–42. doi: 10.1038/sj.gt.3301051. [DOI] [PubMed] [Google Scholar]
- Willemsen R.A. Sebestyen Z. Ronteltap C., et al. CD8 α coreceptor to improve TCR gene transfer to treat melanoma: Down-regulation of tumor-specific production of IL-4, IL-5, and IL-10. J. Immunol. 2006;177:991–998. doi: 10.4049/jimmunol.177.2.991. [DOI] [PubMed] [Google Scholar]
- Xu X.N. Purbhoo M.A. Chen N., et al. A novel approach to antigen-specific deletion of CTL with minimal cellular activation using α3 domain mutants of MHC class I/peptide complex. Immunity. 2001;14:591–602. doi: 10.1016/s1074-7613(01)00133-9. [DOI] [PubMed] [Google Scholar]
- Yang S. Cohen C.J. Peng P.D., et al. Development of optimal bicistronic lentiviral vectors facilitates high-level TCR gene expression and robust tumor cell recognition. Gene Ther. 2008;15:1411–1423. doi: 10.1038/gt.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J. Bechter C. Wiesneth M., et al. Multimer staining of cytomegalovirus phosphoprotein 65-specific T cells for diagnosis and therapeutic purposes: A comparative study. Clin. Infect. Dis. 2008;46:e96–e105. doi: 10.1086/587749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.