Abstract
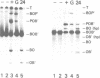
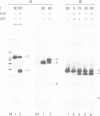
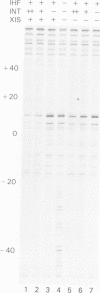
Lambda's Int protein acts as a specific topoisomerase at attachment sites, the DNA segments that are required for site-specific recombination. Int cleaves each strand of an attachment site at a unique place and creates strand exchanges by joining broken ends from two different parents. To study the action of Int topoisomerase in more detail, heteroduplex attachment sites were made by annealing strands that are complementary except for a few base pairs that lie in the region between the points of top and bottom strand exchange in the attachment site core. These heteroduplexes appear to interact normally with Int and its accessory proteins IHF and Xis. Although the heteroduplex sites are specifically cleaved by Int topoisomerase, rejoining of the broken DNA is hindered by the lack of Watson--Crick complementarity adjacent to the break. Because of this, heteroduplexes accumulate broken intermediates which are then processed in novel ways. We have used this feature to provide new information about functional differences between attachment sites, to investigate the way Xis protein controls directionality of site-specific recombination, and to demonstrate that Int protein can join strands indiscriminately and can therefore generate recombinants with either of two genetic polarities.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K., Gottesman S. Purification of the bacteriophage lambda xis gene product required for lambda excisive recombination. J Biol Chem. 1982 Aug 25;257(16):9658–9662. [PubMed] [Google Scholar]
- Abremski K., Hoess R. Escherichia coli plasmid vectors for high-level regulated expression of the bacteriophage lambda xis gene product. Gene. 1983 Nov;25(1):49–58. doi: 10.1016/0378-1119(83)90166-x. [DOI] [PubMed] [Google Scholar]
- Alberts B. M., Doty P. Characterization of a naturally occurring, cross-linked fraction of DNA. 1. Nature of the cross-linkage. J Mol Biol. 1968 Mar 14;32(2):379–403. doi: 10.1016/0022-2836(68)90017-x. [DOI] [PubMed] [Google Scholar]
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour A. G., Garon C. F. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987 Jul 24;237(4813):409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- Bauer C. E., Gardner J. F., Gumport R. I. Extent of sequence homology required for bacteriophage lambda site-specific recombination. J Mol Biol. 1985 Jan 20;181(2):187–197. doi: 10.1016/0022-2836(85)90084-1. [DOI] [PubMed] [Google Scholar]
- Bauer C. E., Hesse S. D., Gardner J. F., Gumport R. I. DNA interactions during bacteriophage lambda site-specific recombination. Cold Spring Harb Symp Quant Biol. 1984;49:699–705. doi: 10.1101/sqb.1984.049.01.079. [DOI] [PubMed] [Google Scholar]
- Better M., Wickner S., Auerbach J., Echols H. Role of the Xis protein of bacteriophage lambda in a specific reactive complex at the attR prophage attachment site. Cell. 1983 Jan;32(1):161–168. doi: 10.1016/0092-8674(83)90506-8. [DOI] [PubMed] [Google Scholar]
- Bushman W., Thompson J. F., Vargas L., Landy A. Control of directionality in lambda site specific recombination. Science. 1985 Nov 22;230(4728):906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983 Dec;35(3 Pt 2):795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- Craig N. L. The mechanism of conservative site-specific recombination. Annu Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- Greenstein D., Horiuchi K. Double-strand cleavage and strand joining by the replication initiator protein of filamentous phage f1. J Biol Chem. 1989 Jul 25;264(21):12627–12632. [PubMed] [Google Scholar]
- Hsu P. L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984 Oct 25;311(5988):721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Kneale G., Anand N. N., Rabinovich D., Kennard O. The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution. J Biol Chem. 1987 Jul 25;262(21):9962–9970. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J Mol Biol. 1988 Nov 5;204(1):95–107. doi: 10.1016/0022-2836(88)90602-x. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987 Sep 24;329(6137):346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- Lewis S. M., Hesse J. E., Mizuuchi K., Gellert M. Novel strand exchanges in V(D)J recombination. Cell. 1988 Dec 23;55(6):1099–1107. doi: 10.1016/0092-8674(88)90254-1. [DOI] [PubMed] [Google Scholar]
- Meyer-Leon L., Huang L. C., Umlauf S. W., Cox M. M., Inman R. B. Holliday intermediates and reaction by-products in FLP protein-promoted site-specific recombination. Mol Cell Biol. 1988 Sep;8(9):3784–3796. doi: 10.1128/mcb.8.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Pargellis C. A., Hasan N. M., Bushman E. W., Landy A. Autonomous DNA binding domains of lambda integrase recognize two different sequence families. Cell. 1988 Sep 23;54(7):923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Bauer C. E., Gardner J. F. Role of homology in site-specific recombination of bacteriophage lambda: evidence against joining of cohesive ends. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4049–4053. doi: 10.1073/pnas.84.12.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A. Integrative recombination of bacteriophage lambda DNA in vitro. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1072–1076. doi: 10.1073/pnas.72.3.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A., Flamm E., Weisberg R. A., Miller H. I. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J Bacteriol. 1987 Sep;169(9):4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987 Aug 28;50(5):779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Reddy M. K., Bauer W. R. Activation of the vaccinia virus nicking-joining enzyme by trypsinization. J Biol Chem. 1989 Jan 5;264(1):443–449. [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein-DNA complex. Cell. 1988 Jan 15;52(1):9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Ross W., Shulman M., Landy A. Biochemical analysis of att-defective mutants of the phage lambda site-specific recombination system. J Mol Biol. 1982 Apr 15;156(3):505–522. doi: 10.1016/0022-2836(82)90263-7. [DOI] [PubMed] [Google Scholar]
- Shulman M., Gottesman M. Attachment site mutants of bacteriophage lambda. J Mol Biol. 1973 Dec 25;81(4):461–482. doi: 10.1016/0022-2836(73)90517-2. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., de Vargas L. M., Skinner S. E., Landy A. Protein-protein interactions in a higher-order structure direct lambda site-specific recombination. J Mol Biol. 1987 Jun 5;195(3):481–493. doi: 10.1016/0022-2836(87)90177-x. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Weisberg R. A., Enquist L. W., Foeller C., Landy A. Role for DNA homology in site-specific recombination. The isolation and characterization of a site affinity mutant of coliphage lambda. J Mol Biol. 1983 Oct 25;170(2):319–342. doi: 10.1016/s0022-2836(83)80151-x. [DOI] [PubMed] [Google Scholar]
- de Massy B., Dorgai L., Weisberg R. A. Mutations of the phage lambda attachment site alter the directionality of resolution of Holliday structures. EMBO J. 1989 May;8(5):1591–1599. doi: 10.1002/j.1460-2075.1989.tb03543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ende A., Langeveld S. A., Teertstra R., van Arkel G. A., Weisbeek P. J. Enzymatic properties of the bacteriophage phi X174 A protein on superhelical phi X174 DNA: a model for the termination of the rolling circle DNA replication. Nucleic Acids Res. 1981 May 11;9(9):2037–2053. doi: 10.1093/nar/9.9.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]