Abstract
Integral to the characterization of radiation-induced tissue damage is the identification of unique biomarkers. Biomarker discovery is a challenging and complex endeavor requiring both sophisticated experimental design and accessible technology. The resources within the National Institute of Allergy and Infectious Diseases (NIAID)-sponsored Consortium, Medical Countermeasures Against Radiological Threats (MCART), allow for leveraging robust animal models with novel molecular imaging techniques. One such imaging technique, MALDI (matrix-assisted laser desorption ionization) mass spectrometry imaging (MSI), allows for the direct spatial visualization of lipids, proteins, small molecules, and drugs/drug metabolites—or biomarkers—in an unbiased manner. MALDI-MSI acquires mass spectra directly from an intact tissue slice in discrete locations across an x, y grid that are then rendered into a spatial distribution map composed of ion mass and intensity. The unique mass signals can be plotted to generate a spatial map of biomarkers that reflects pathology and molecular events. The crucial unanswered questions that can be addressed with MALDI-MSI include identification of biomarkers for radiation damage that reflect the response to radiation dose over time and the efficacy of therapeutic interventions. Techniques in MALDI-MSI also enable integration of biomarker identification among diverse animal models. Analysis of early, sublethally irradiated tissue injury samples from diverse mouse tissues (lung and ileum) shows membrane phospholipid signatures correlated with histological features of these unique tissues. This paper will discuss the application of MALDI-MSI for use in a larger biomarker discovery pipeline.
Keywords: bioassay, imaging, laboratory animals, radiation effects
Introduction
The spectrum of techniques in molecular histology is expanding rapidly from more traditional antibody- or probe-based approaches to include novel mass spectrometry imaging approaches, allowing for discovery of biomarkers or description of targets lacking specific reagents (Schwamborn and Caprioli 2010; Touboul et al. 2011). Mass spectrometry imaging is a developing technique that couples more traditional molecular histological approaches with advanced mass spectrometry resulting in mass-based tissue imaging. Front-end sample ionization techniques include matrix assisted laser desorption-ionization (MALDI), desorption electroscopy ionization (DESI), and secondary ion mass spectrometry (SIMS), each having intrinsic power and utility (vanHove et al. 2010). Matrix assisted laser desorption ionization mass spectrometry imaging (MALDI-MSI) is currently the most widely published technique in molecular histology with reports of its use in identifying an array of molecular classes, including lipids, proteins, peptides, and metabolites (Fig. 1) (Angel and Caprioli 2013; Cazares et al. 2011). At its core, a MALDI-MSI experiment consists of mass spectra acquisition directly from an intact tissue section in discrete locations across a grid pattern (Fig. 2). Collected spectra are then rendered into a spatial distribution map to identify the location of individual molecules of interest on a tissue slice. The spatial resolution of these MSI techniques ranges from micron scale (MALDI) to nanometer scale (SIMS) and is dependent on instrument setup. Unbiased data acquisition is a powerful feature of MSI experiments, particularly with respect to biomarker detection and characterization.
Fig. 1.
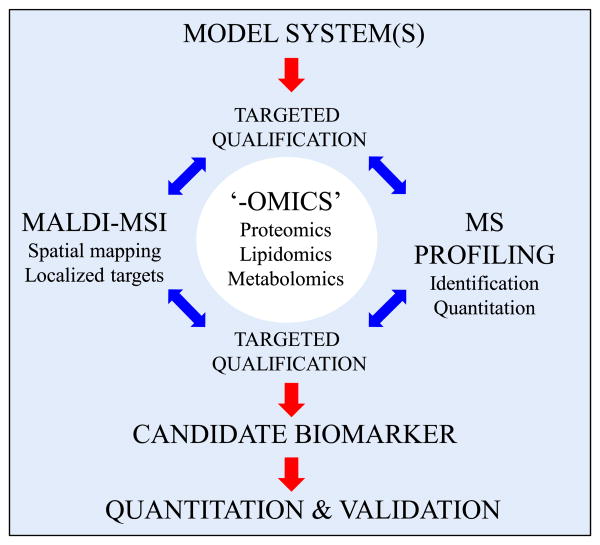
Discovery power of paired biomarker characterization techniques. The biomarker discovery process begins with a straightforward goal of defining measurable targets that correlate with an outcome. Within the goals of the consortium on medical countermeasures against radiological threats (MCART), the authors seek to define molecular correlates of radiation exposure, dosage experience, countermeasure efficacy, and cross-species utility, to name only a few. To achieve these goals, MCART has established a cyclical biomarker discovery and target validation approach centered around two complimentary methods: matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI-MSI) and mass spectrometry (MS) profiling. Both techniques have specific advantages, but their commonalities for use in unbiased characterization (lipidomics, proteomics, metabolomics, etc.) make them particularly strong when used together. For example, where MS profiling can quantify changes in abundance of a target-of-interest, MALDI-MSI can aid mechanistic work by providing histologically significant spatial information. Similarly, MALDI-MSI may detect robust differential abundance of a target-of-interest within a localized tissue feature, information that may be lost to dilution or extraction during sample preparation for MS profiling.
Fig. 2.
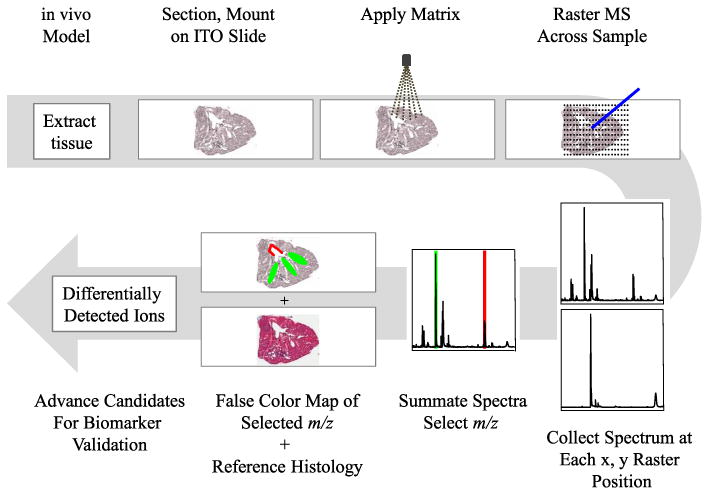
MALDI-MSI workflow. A MALDI-MSI experiment begins with tissue extraction from an in vivo model system, and the sample is snap-frozen (unfixed) and cryosectioned. Sections are lifted onto an indium tin oxide (ITO) coated slide, fixed into place, and prepared for use. Matrix, dissolved in a solvent, is applied to the slide with a vibrational nebulizer to achieve uniform matrix application. Following matrix application, an x,y coordinate grid is assigned, the MALDI laser is rastered across the tissue capturing data at each point on the grid, and finally, the entire series of mass spectra (m/z, mass over charge) are summated. From the summated spectra, peaks are selected, and the intensity of the peak is mapped by individual spectrum back to the x,y coordinate and displayed color intensity (green and red selections). Post-MALDI-MSI, the tissue is stripped of matrix and stained by traditional methods and scanned at high resolution to highlight useful histological features and facilitate co-registration of the tissue image and mass signals.
Discovery experiments, however, are not the only types of experiments suited for MALDI-MSI. In the context of a known target molecule, MALDI-MSI studies can provide additional insight from the sub-histological localization of a target molecule and the localized concentration of a target of interest. Following identification and spatial mapping of candidate biomarkers in one model system, this untargeted approach can be used to interrogate the same or similar molecules in diverse model systems, allowing for crossstrain and cross-species biomarker validation. Additionally, by using alternative combinations of MALDI matrices, solvent systems, tissue preparation, and ionization modes, a variety of targets can be queried without bias.
Biomarker studies, in particular, can benefit from including a targeted or untargeted MALDI-MSI experiment. Traditional biomarker discovery studies have included elements of broad techniques such as genomics, transcriptomics, proteomics, and metabolomics to identify quantifiable correlates of a biological outcome. Generally, these techniques revolve around the extraction of potential biomarkers from a biological fluid or tissue sample, which results in the destruction of important histological information and loss of significant differences observable only as a localized target concentration. By including an untargeted MALDI-MSI biomarker study, it is possible to identify target molecules that may be locally concentrated and represent a significant correlate; a correlate that may lose significance on dilution during tissue extraction steps. Targeted MALDI-MSI studies can add power to a traditionally identified biomarker by adding histological information, which could lead to the starting point for future mechanistic studies. Combining MALDI-MSI to existing biomarker discovery techniques, including mass spectrometry (MS) profiling, makes a powerful pairing that enriches both the discovery and validation phases (Fig. 1).
Recent developments have resulted in the application of MALDI-MSI to better understand a wide variety of biological problems. MALDI-MSI has been used to identify differential lipid distributions in mouse brain, map peptides in leeches, localize drug metabolites in rats, and characterize metabolic signals between pathogenic bacteria (Bruand et al. 2011; Norris and Caprioli 2013; Yang et al. 2009). In addition, retrospective studies of human cancers have identified molecular correlates of disease or pathology, a molecular-level cancer signature (Schone et al. 2013; Schwamborn and Caprioli 2010). MSI is not, however, a standalone discovery technique. In the context of hypothesis-driven research, MSI experiments may be included to develop new leads, but more traditional techniques would necessarily follow to confirm and validate targets, including immunohistochemistry (IHC), in situ hybridization, traditional histological stains, component extracts, and quantitative mass spectrometry (Cazares et al. 2011). Evident throughout MSI literature is the discovery power, regardless of disease, target molecular class, or model species (Gruner et al. 2012). These advantages make MSI a natural addition to the Medical Countermeasures Against Radiological Threats (MCART) research program.
Early molecular correlates, or biomarkers, of radiation exposure are particularly valuable as they can be used to identify individuals early in the progression of the acute- and delayed-onset syndromes for which interventional therapy may be recommended. Identification of biomarkers begins with understanding a measurable sentinel event, usually downstream components of molecular cascades that may be traced back not only to direct initiation but also to concomitant events that may lead to alternative biomarker candidates. An ideal biomarker is easily sampled (circulating in blood or collected in a minimally-intrusive manner), readily and reliably measured, and is unambiguously correlated with an outcome (radiation exposure, presence of tissue damage, response to mitigator).
The MCART Consortium is uniquely equipped to address the lack of biomarkers available for various endpoints in radiation research. MCART has established a series of appropriate animal models, murine and primate, to study the physiology of both acute radiation syndrome (ARS) and delayed effects of acute radiation exposure (DEARE), organ-specific response to radiation exposure, and efficacy of medical countermeasures (MCM) (MacVittie 2012; Booth et al. 2012a and b; Chua et al. 2012; Jackson et al. 2012; MacVittie et al. 2012a and b; Plett et al. 2012). Two hallmark organs of survivable, radiation-induced damage are the gastrointestinal tract (GI) and lung in mouse models. GI remodeling and lung injury are two long-term consequences of exposure to sublethal irradiation with crypt remodeling and collagen deposition noted in the ileum and fibrosis and vascular lymphocytic cuffing observed in the lung. (Booth et al. 2012a; Jackson et al. 2012) In this preliminary study, the authors sought to assess the utility of MALDI-MSI in these specific tissues, mouse lung and GI, to address its use in a broader biomarker discovery and validation pipeline.
Materials and Methods
Mice
Specific pathogen free (SPF) C57BL/6 mice (50/50 male/female; Jackson Laboratory, Bar Harbor, ME) were received at 10 wk of age and housed up to five mice per age in microisolator cages on sterilized direct contact bedding. Mice were provided certified commercial extruded lab rodent chow (Harlan 2018SXC; Harlan, Indianapolis, IN) ad libitum in cage hoppers and acidified water (pH 2.0–3.0) in sipper tube bottles. Animal rooms on a 12-h light/dark cycle were maintained at 21 ± 3ºC with 30–80% relative humidity and at least 10 air changes per hour of 100% conditioned fresh air. All studies were approved by the Indiana University School of Medicine Institutional Animal Care and Use Committee.
Irradiation and dosimetry
Mice (16–17 wk old) were placed in single chambers of a Plexiglass irradiation apparatus and exposed to 0.65 Gy of gamma radiation delivered as a single uniform total body dose from a 137Cs radiation source (GammaCell 40; Nordion International, Kanata, Ontario, Canada) at an exposure rate of 0.57–0.61 Gy min−1. Exposure was confirmed using Inlight Dot dosimeters (Landauer Inc., Glenwood, IL) placed inside of a parafilm mouse phantom and irradiated along with the mice. Dosimeters were read using a validated Landauer microStar reader calibrated with standard Dot dosimeters exposed with a NIS-traceable 137Cs source (Battelle Memorial Institute, Seattle, WA). Reproducibility of individual dots was 3 ± 1% with accuracy of 4 ± 2%, well within the 10% industry standard for experimental radiation dosimetry.
Tissue collection
Forty-eight hours after irradiation, mice were euthanized by CO2 inhalation followed by exsanguination. Lungs, small intestine, colon, sternum, liver, and heart were harvested and snap-frozen (unfixed) by floating on foil in liquid nitrogen. Intestine samples were not cleared of lumenal contents prior to freezing but were staged in inch-long, straight sections for ease of handling. Tissue samples were stored frozen (−80°C) prior to sectioning or extraction.
Tissue preparation and matrix application
Frozen, unfixed tissues were cut into 12-μm sections on a Leica AM1925 cryomicrotome, mounted and heat-fixed onto an Indium Tin Oxide (ITO)-coated MALDI glass slide (Bruker Daltonics, Billerica, MA) by holding a finger on the slide directly underneath the section. All slides were stored in a vacuum desiccator for at least 20 min prior to preparation for MALDI-MSI (or storage at heat−20°C). For lipid-targeting experiments a 12.5 mg mL−1 solution of norharmane (NH), 9H-Pyrido[3,4-b]indole hydrochloride (Sigma-Aldrich, St. Louis, MO), a bivalent MALDI matrix, solvated in a solution of chloroform:methanol:water (5:10:4, v:v:v) was spray-coated onto slides. For those experiments targeting proteins or peptides, slides were spray coated with 30 mg mL−1 sinapinic acid (SA) in 70% acetonitrile, 0.1% trifluoroacetic acid, or 12 mg mL−1 alpha-cyano-4-hydroxycinnamic acid (CHCA) in 50% acetonitrile, 0.3% trifluroacetic acid. All matrices were applied using a Bruker Daltonics ImagePrep spray chamber (Billerica, MA). Unless otherwise specified, all reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Mass spectrometry and image analysis
MS data were collected in positive mode (detection range: m/z 400–900 for lipids, m/z 2,000–20,000 for proteins, and m/z 1,000–3,000 for peptides) and negative mode (detection range: m/z 400–900 for lipids), raster width 50 μm, 500 shots per raster, on a Bruker Daltonics Ultraflex Extreme Matrix-Assisted Laser Desorption Ionization Time-of-Flight/Time-of-Flight Mass Spectrometer (MALDI-TOF/TOF MS) using flexControl software (version 3.4.105). Subsequently, data were analyzed using the software packages flexImaging (version 3.4.54) and flexAnalysis (version 3.4.57). All MALDI-MSI specific materials, equipment, instruments, and software were obtained from Bruker Daltonics (Billerica, MA).
Post-MSI histology
Tissue sections were analyzed by MALDI-MSI and then stripped of matrix in 70% ethanol. Post-processed tissues were stained with one of two traditional histological stains, H&E (hematoxylin and eosin from Sigma-Aldrich, St. Louis, MO) or Masson's Trichrome (Polysciences, Warrington, PA) according to manufacturer's protocols. Images were captured on a ScanScope CS2 slide scanner at 20X resolution and exported as high-resolution .tiff files using ImageScope software (Aperio, Vista, CA). Images were processed (rotation and cropping) in the GNU Image Manipulation Program (GIMP, version 2.8.3, freeware).
Data analysis and prediction software
Molecular predictions for lipid-like ions were made using the Lipid Mass Structure Database (LMSD), available from Lipid Metabolites and Pathways Strategy (LIPID MAPS Consortium, La Jolla, CA). Peptide identifications were performed by processing MS/MS peptide mass fingerprints through the MASCOT Database (Matrix Sciences, Boston, MA).
Results
MALDI-MSI workflow
Tissues were extracted and processed for MALDI-MSI according to the workflow diagrammed in Fig. 2. Based on sample optimization reports, the tissues were cryosectioned unfixed and unembedded at 12-μm thickness (Yang and Caprioli 2011). Tissue samples to be analyzed for lipids were not washed in alcohols. For the analysis of peptides as well as other molecular classes, tissue samples were washed and dehydrated due to the potential for lipid signals to convolute small peptide mass signatures (Casadonte and Caprioli 2011; Shanta et al. 2011). Multiple MALDI matrices were used in combination with targeted solvent systems to optimize detection of disparate molecular classes: Sinapinic acid in a solution of acetonitrile and trifluoroacetic acid for proteins; cyano-4-hydroxycinnamic acid in a solution of acetonitrile and trifluoroacetic acid for peptides; and 9H-pyrido[3,4-b]indole hydrochloride in a chloroform, methanol, and water solution for lipids.
MALDI-MSI maps unique ions to histological features in mouse lung
Following radiation exposure, the lungs can undergo slow and permanent damage. The most notable long-term effects in the lung include perivascular lymphocytic cuffing and fibrosis via extracellular matrix deposition, both of which contribute to decreased physiological function (Jackson et al. 2012). Given this delayed, organ-specific response and the potential for meaningful damage mitigation by MCM, the lung was an obvious starting point for testing the suitability of MALDI-MSI to detect and map unique ion signatures.
Lungs from control (age matched, non-irradiated) and sublethally irradiated (6.5 Gy) adult mice were necropsied two days post-irradiation and processed for MALDI-MSI. Initially, two sets of tissue sections were prepared to target lipid ions in either negative ion mode or positive ion mode, using a solvent mixture of chloroform and methanol to apply a bivalent MALDI matrix. Scanning in both positive and negative modes is advantageous and results in detection of a wider variety of phospholipids due to the diverse chemical nature and polarity of the phospholipid head-groups. A series of negative and positive lipid-like ions were detected and spatially correlated by intensity back to the tissue (Fig. 3a and b). Identities of lipid-like ions were predicted in silico using database tools available from Lipid Metabolites and Pathways Strategy (Lipid MAPS, www.lipidmaps.org). The dominant lipid types were predicted to be glycerophospholipids (PL) of various classes defined by the polar head group; PLs can be further characterized by the length and degree of unsaturation of the component fatty acids. Of interest are the unique distribution types observed in negative MALDI mode in the lung (Fig. 3a), including ions localized primarily in the lining and interior of the major airways (bronchi) (m/z 1451.9, cardiolipin); ions mapping to smaller airways (bronchioles) [red, m/z 885.7, phosphatidylinositol; PI 38:4 (phospholipid class carbons:unsaturations)]; and ions distributed throughout the tissue parenchyma but excluded from the large airways (green, m/z 746.4, phosphatidylethanolamine, PE 36:0).
Fig. 3.
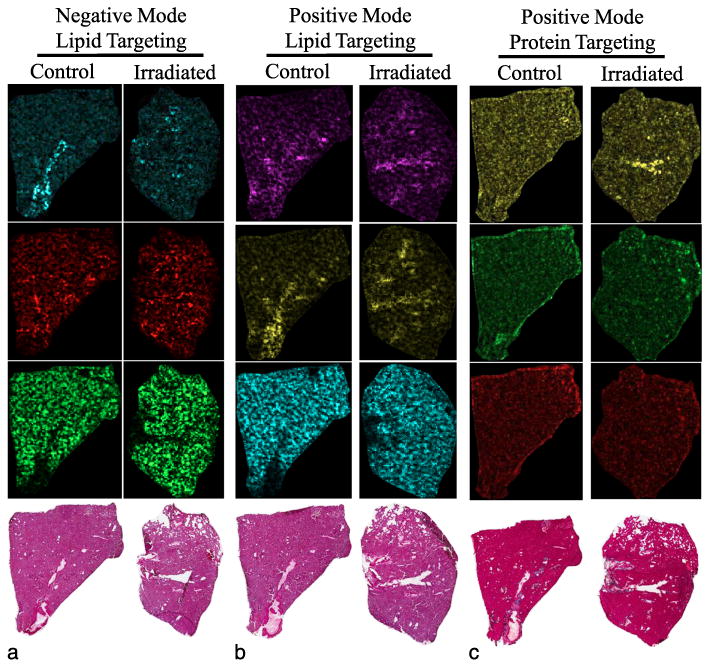
Differentially mapped molecules of interest in mouse lungs. Lung tissue from mouse lungs processed by MALDI-MSI targeting (a) lipids in negative ion mode, (b) lipids in positive ion mode, and (c) small proteins in positive ion mode, and three unique m/z signals are shown for each. Lipids were targeted by solvating a lipid-appropriate matrix in a solution of chloroform and methanol; proteins were targeted by first washing away lipids followed by protein-appropriate matrix application in a solution of acetonitrile. Pixel saturation representative of relative abundance of the ion. (a) False color, [M-H}-mass/charge ratio, predicted identity: cyan, m/z 1451.9, cardiolipin; red, m/z 885.7, phosphatidylinositol; green, m/z 746.4, phosphatidylethanolamine. (b) False color, [M+H}+ mass/charge ratio, predicted ion identity: magenta, m/z 782.5, phosphatidylcholine; yellow, m/z 806.5, phosphatidylcholine; cyan, m/z 706.5, phosphatidylcholine. (c) False color, positive mode mass/charge ratio: yellow, m/z 17,785; green, m/z 15,533; red, m/z 14,546.
Additional lipid-like ions were detected in positive ion mode with similar tissue correlation patterns (Fig. 3b). Fatty acid length variants of PLs can be predicted as m/z Δ28 (C2H4) changes, and unsaturations can be predicted as m/z Δ2 (loss of two hydrogens) decreases from the molecular mass of the saturated fatty acids; both features can contribute to fine-tuning of membrane fluidity and permeability (Spector and Yorek 1985). Three predicted acyl variants of the PL class phosphatidylcholine (PC) are represented in Fig. 3b. PC is an abundant PL in eukaryotic membranes and was predicted to be a large proportion of the relative PL constituency (Henneberry et al. 2002). Two PC ion signatures, m/z 782.5 (magenta, PC 36:4) and m/z 806.5 (yellow, PC 38:6), were both detected in the bronchi and bronchioles but in low abundance in the rest of the tissue, whereas a third ion signature, m/z 706.5 (cyan, PC 30:0), mapped to regions of the lung directly associated with oxygen exchange. This unique pattern of tissue distribution may provide further insight into the individual role of each of these predicted PC molecules. Dipalmitoyl-PC (DPPC, PC 32:0) ions, a lipid made by type II pneumocytes and a known component of lung surfactant, were also detected in abundance in the lung (Schlame 1988). As expected, DPPC signatures were localized largely in the oxygen exchange regions, supporting the use of MALDI-MSI to link ion identity with physiological function (data not shown). Together, these results highlight the use of MALDI-MSI for understanding the mechanism of disease by supplementing molecular identity with histological information.
In addition to targeting lipid-like molecules, total, intact proteins were also queried. Mouse lung tissue was analyzed from a serial section from the lipid slide series. Using a solution of acetonitrile (ACN) and trifluoroacetic acid (TFA) to apply a matrix appropriate for whole proteins (sinapinic acid), several unique ion signals in the lung, m/z 17,785,15,533, and 14,546 were detected (yellow, green, red, respectively), shown in Fig. 3c. While it was possible to detect ion signatures throughout the lung tissue, the technical problems with identifying and characterizing intact proteins were clear, especially the narrow mass detection range (roughly less than 25,000 daltons), troublesome for routine MALDI-MSI experiments. However, these issues illustrate the power of redundancy built into this biomarker discovery scheme (Fig. 1); if a protein identity was predicted by mass spectrometry profiling (bottom-up proteomics), then the protein should be easier to identify and address in MALDI-MSI either by predicting m/z or by alternative MALDI-MSI techniques, such as on-tissue proteolytic digestion.
Early molecular response in irradiated mouse ileum
Another tissue that can undergo dramatic and lasting damage and reconstruction following radiation exposure is the GI tract. GI, specifically ileum, the distal-most region of the small intestine, was the other natural choice to evaluate the use of MALDI-MSI as a discovery tool for biomarkers.
Ileum from control and sublethally irradiated (6.5 Gy) mice showed no obvious tissue damage at 2 d post-irradiation by gross histology, as expected (Booth et al. 2012a). Fig. 4a and b represent ileum tissue processed for lipid targeting by MALDI-MSI in negative and positive mode, respectively. Of note, these GI samples were not flushed of lumenal contents prior to cryopreservation. The spatial distribution of three unique lipid-like ions identified in negative mode is demonstrated in Fig. 4a. An ion, m/z 885.7 (cyan, PI 38:4), localized to the gut lining but was not observed in the lumenal contents. Similarly, the ion m/z 632.7 (red, PE 28:1) is distributed throughout the gut lining and lumenal contents in control tissue, but this ion is observed in notably lower abundance within the gut lining and lumenal contents of the irradiated tissue. Interestingly, differential abundance of an ion, m/z 1451.9 (yellow, cardiolipin), is observed between control and irradiated ileum. The predicted cardiolipin ion is evenly distributed through lumenal contents and localizes in modest abundance and intensity in the ileum lining of control tissue, whereas in irradiated tissue, the lumenal abundance is decreased and the tissue-localized abundance and intensity are increased. Further investigation of this ion distribution pattern is pending; however, these types of differential distribution patterns highlight the importance of the imaging aspect of MALDI-MSI. These results illustrate the possibility that the concentration of this ion after tissue extraction might be equal between the control and irradiated sample, and this differential result may be lost.
Fig. 4.
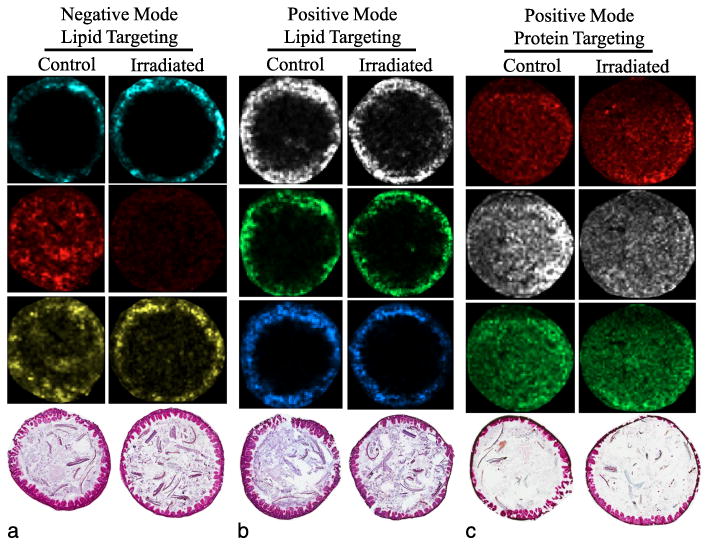
Localization of unique molecular targets in mouse ileum. Ileum tissue from mouse processed by MALDI-MSI targeting (a) lipids in negative ion mode, (b) lipids in positive ion mode, and (c) low molecular weight proteins in positive ion mode; three unique m/z signals are shown for each. Lipids were targeted by solvating a lipid-appropriate matrix in a solution of chloroform and methanol; proteins were targeted by first washing away lipids followed by protein-appropriate matrix application in a solution of acetonitrile. Pixel saturation representative of relative abundance of the ion. (a) False color, [M-H}-mass/charge ratio, predicted identity: cyan, m/z 885.7, phosphatidylinositol; red, m/z 632.7, phosphatidylethanolamine; yellow, m/z 1451.9, cardiolipin. (b) False color, [M+H]+ mass/charge ratio, predicted ion identity: white, m/z 762.6, phosphatidylcholine; green, m/z 734.6, phosphatidylcholine; azure, m/z 786.6, phosphatidylcholine. (c) False color, positive mode mass/charge ratio: red, m/z 3710; white, m/z 14,330; green, m/z 8,615.
Fig. 4b demonstrates the tissue-localized abundance of three predicted PC ions detected in positive mode: m/z 762.6 (white, PC 34:0), m/z 734.6 (green, PC 32:0), and m/z 786.6 (azure, PC 36:2), all likely representing acyl length variants of PC. These three ions have been reported previously to be present in abundance in the gut (Tyurina et al. 2008). As expected, these ions localize predominantly to the ileum tissue and not to the lumenal contents, further highlighting the capacity of imaging to discriminate not only ion abundance but also spatial correlation.
The complexity of a MALDI-MSI experiment increases when the mass and molecular character of targets is highly variable. Fig. 4c demonstrates some of the complexities present when targeting intact proteins by MALDI-MSI. Three ion signatures [m/z 3,710 (red), m/z 14,330 (white), and m/z 8,615 (green)] are detected across the tissue and in the lumenal contents. Ion m/z 3,710 appears to localize with higher intensity to the ileum tissue in irradiated animals compared to controls. A second larger ion, m/z 14,330, is detected across both the control and irradiated tissue but may be present in higher abundance in the controls. Finally, a third ion signature, m/z 8,715, is observed in the tissue of both control and irradiated samples. Positive identification of these ions is possible but can be difficult without additional information. Smaller range ions (m/z 2,000–10,000) could be representing larger proteins carrying multiple charges or could be endogenous peptides; in either case, with more information, positive identification is possible. Larger range ions (m/z 10,000–20,000) could represent small, intact proteins, but identification remains difficult unless other methods are applied (on-tissue digestion, correlation to total protein extracts). Spatial mapping of intact proteins may be better suited for targeted imaging experiments where the ion identity is predefined. Correlation of spatial mapping and protein identity can be facilitated best by on-tissue protein digestion, which has been used to identify unknown targets (Casadonte and Caprioli 2011).
Protein identification by peptide digestion
Given the inherent challenges with identifying unknown intact proteins, alternative methods are employed to identify proteins positively while maintaining the rich, spatial information afforded by MALDI-MSI. Several protocols have been reported for on-tissue proteolytic digestion coupled to MALDI-MSI; simply, in situ trypsin digest and tandem mass spectrometry (MS/MS) for peptide identification. Tissue sections are prepared and then washed (to reduce lipid interference and semi-fix the tissue), and a mass spectrometry-grade protease solution is applied to the tissue by nebulization (Fig. 5). Following an incubation period, matrix appropriate for peptide ionization is applied to the tissue, and the MALDI-MSI process follows as in Fig. 2. After a peak-of-interest is selected, peptide identification is achieved by first fragmenting the peptide by tandem mass spectrometry, then predicting peptide identity by querying a peptide MS/MS database such as Mascot. These results can then be validated through other proteomic techniques or antibody-based detection techniques including western blotting and immunohistochemistry.
Fig. 5.
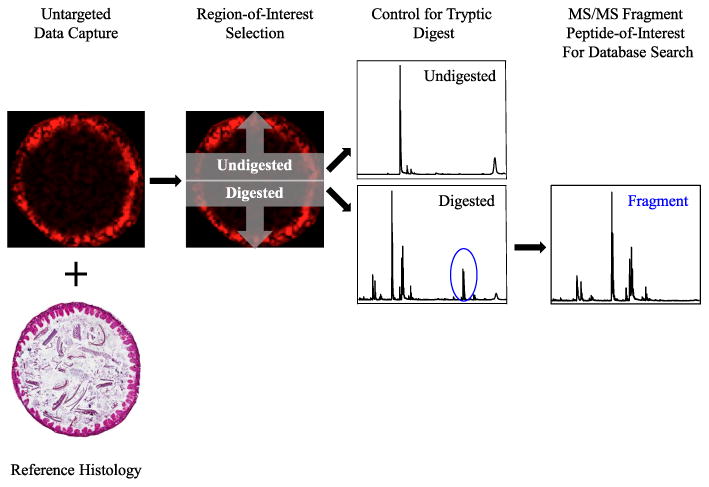
On-tissue proteolytic digest facilitates protein identification. (a) Distribution of m/z signal (representative) in control mouse ileum and reference histology (H&E). (b) Illustrated control experiment with half of tissue excluded from protease application (top) and half incubated with protease (bottom). (c) Peptide mass signatures present in abundance in protease-incubated tissue (note m/z representative ion, circled in blue), positive identification of protein targets through database search of peptide MS/MS spectra.
Discussion
Proposed is a systematic, parallelized pathway to biomarker discovery. The approach of complimentary rounds of mass spectrometry profiling and MALDI-MSI present MCART with the power to succeed in biomarker identification and validation. Targeting specific questions in radiation biology, they have described a series of methods to discover molecular correlates of radiation dose, onset of ARS and DEARE, predictors of syndrome severity, and indicators of successful mitigation. The discovery approaches include MALDI-MSI and mass spectrometry profiling; both techniques allow for characterization of proteins, lipids, metabolites, and other molecular target types.
Proof-of-concept studies were performed to characterize the MALDI-MSI aspect of this complimentary biomarker discovery approach. Here, detection of unique lipid profiles in mouse tissues is shown, along with a sample of intact protein survey data and a working pathway to identify protein targets through on-tissue tryptic digest. Notably, the histological information is retained in rich detail, considering these tissues were unfixed and stained following MALDI-MSI and matrix stripping. The unique ability to map these diverse molecules supports the logic of using MALDI-MSI as a component of a cyclic biomarker discovery pathway to solve problems in radiation biology.
In mouse lung and ileum tissue, ions were detected corresponding to diverse phospholipid classes, including CL, PE, PI, and PC. Within these subclasses of phospholipid, acyl-length variants were also identified, most strikingly with the abundant PCs 30:0, 32:0, 36:4, and 38:6. Additionally, mono- and poly-unsaturations were identified on a fatty acid chain within these phospholipid subclasses by investigating neighboring m/z values differing by two mass unit decreases (in the case of singly charged ions) from the predicted saturated fatty acid (PC X:0). Using these predictive techniques, the authors have observed oxidized phospholipids (Shirey et al. 2013) that have been shown to be present differentially in irradiated tissue compared to control (Tyurina et al. 2008). Alterations in phospholipid abundance and presence of oxidative lesions thereof can serve as downstream sentinels of global cellular response and stress from reactive oxygen species (Sparvero et al. 2010). Therefore, the ability to detect and characterize lipid responses is crucial to this biomarker discovery pathway. By applying this approach to dose range studies and timing tissue collection to the nadir of each specific sequelae, the authors aim to correlate tissue damage with specific molecular responses that can be validated as biomarkers and serve as powerful research tools for the radiation research community.
Conclusion
The authors present a rational, complimentary approach to biomarker discovery. By pulling from the strengths of the individual techniques, MALDI-MSI and traditional analytical mass spectrometry, one can search for novel correlates of both ARS and DEARE in diverse species and tissues. Subsequently, identification of biomarkers will empower researchers to study the correlation to radiation dosage and develop molecular readouts for medical countermeasure efficacy. When combined with the realistic, well-defined model systems described previously by the MCART consortium, the true discovery power of these complimentary untargeted (MALDI-MSI) and targeted (MS) systems will be clear. With this approach, it will be possible to link key sequelae and organ-specific syndromes across model systems at the physiological and histological levels as well as the molecular level.
Acknowledgments
The authors would like to thank the members of the MCART consortium for their admirable dedication, collegial interactions, and insightful guidance in developing this biomarker discovery pathway. Additionally, we would like to extend our gratitude to the members of the Ernst and Kane laboratories for productive discussions and critical reading of this manuscript.
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272201000046C.
Footnotes
The authors declare no conflicts of interest.
References
- Angel PM, Caprioli RM. Matrix-assisted laser desorption ionization imaging mass spectrometry: in situ molecular mapping. Biochem. 2013;52:3818–3828. doi: 10.1021/bi301519p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth C, Tudor G, Tonge N, Shea-Donohue T, MacVittie TJ. Evidence of delayed gastrointestinal syndrome in high-dose irradiated mice. Health Phys. 2012a;103:400–410. doi: 10.1097/hp.0b013e31826530e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth C, Tudor G, Tudor J, Katz BP, MacVittie TJ. Acute gastrointestinal syndrome in high-dose irradiated mice. Health Phys. 2012b;103:383–399. doi: 10.1097/hp.0b013e318266ee13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand J, Sistla S, Mériaux C, Dorrestein PC, Gaasterland T, Ghassemian M, Wisztorski M, Fournier I, Salzet M, Macango E, Bafna V. Automated querying and identification of novel peptides using MALDI mass spectrometric imaging. J Proteome Res. 2011;10:1915–1928. doi: 10.1021/pr101159e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadonte R, Caprioli RM. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nature Protocols. 2011;6:1695–1709. doi: 10.1038/nprot.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casales C, Rustow B, Rabe H, Kunze D, Schlame M. Molecular species of phosphatidylcholine and phosphatidylglycerol in rat lung surfactant and different pools of pneumocytes type II. Biochemical J. 1988;253:209–215. doi: 10.1042/bj2530209. CCBRHRDK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazares LH, Troyer DA, Wang B, Drake RR, Semmes JO. MALDI tissue imaging: from biomarker discovery to clinical applications. Anal Bioanal Chem. 2011;401:17–27. doi: 10.1007/s00216-011-5003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua HL, Plett PA, Sampson CH, Joshi M, Tabbey R, Katz BP, MacVittie TJ, Orschell CM. Long-term hematopoietic stem cell damage in a murine model of the hematopoietic syndrome of the acute radiation syndrome. Health Phys. 2012;103:356–366. doi: 10.1097/HP.0b013e3182666d6f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner BM, Hahne H, Mazur PK, Trajkovic-Arsic M, Maier S, Esposito I, Kalideris E, Michalski CW, Kleef J, Rauser S, Schmid RM, Kuster B, Walch A, Siveke JT. MALDI imaging mass spectrometry for in situ proteomic analysis of preneoplastic lesions in pancreatic cancer. PLoS One. 2012;7:e39424. doi: 10.1371/journal.pone.0039424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberry AL, Wright MM, McMaster CR. The major sites of cellular phospholipid synthesis and molecular determinants of fatty acid and lipid head group specificity. Mol Biol Cell. 2012;13:3148–3161. doi: 10.1091/mbc.01-11-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson IL, Xu P, Hadley C, Katz BP, McGurk R, Down JD, Vujaskovic Z. A preclinical rodent model of radiation-induced lung injury for medical countermeasure screening in accordance with the FDA animal rule. Health Phys. 2012;103:463–473. doi: 10.1097/HP.0b013e31826386ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVittie TJ. The MCART Consortium animal models series. Health Phys. 2012;103:340–342. doi: 10.1097/HP.0b013e318261175a. [DOI] [PubMed] [Google Scholar]
- MacVittie TJ, Bennett A, Booth C, Garofalo M, Tudor G, Ward A, Shea-Donohue T, Gelfond D, McFarland E, Jackson W, Lu W, Farese A. The prolonged gastrointestinal syndrome in rhesus macaques: the relationship between gastrointestinal, hematopoietic, and delayed multi-organ sequelae following acute, potentially lethal, partial-body irradiation. Health Phys. 2012a;103:427–453. doi: 10.1097/HP.0b013e318266eb4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVittie TJ, Farese AM, Bennett A, Gelfond D, Shea-Donohue T, Tudor G, Booth C, McFarland E, Jackson W. The acute gastrointestinal subsyndrome of the acute radiation syndrome: a rhesus macaque model. Health Phys. 2012b;103:411–426. doi: 10.1097/HP.0b013e31826525f0. [DOI] [PubMed] [Google Scholar]
- Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev. 2013;113:2309–2342. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plett PA, Sampson CH, Chua HL, Joshi M, Booth C, Gough A, Johnson CS, Katz BP, Farese AM, Parker J, MacVittie TJ, Orschell CM. Establishing a murine model of the hematopoietic syndrome of the acute radiation syndrome. Health Phys. 2012;103:343–355. doi: 10.1097/HP.0b013e3182667309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schone C, Hofler H, Walch A. MALDI imaging mass spectrometry in cancer research: combining proteomic profiling and histological evaluation. Clin Biochem. 2013;46:539–545. doi: 10.1016/j.clinbiochem.2013.01.018. [DOI] [PubMed] [Google Scholar]
- Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry—looking beyond classical histology. Nature Reviews Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- Shanta SR, Zhou LH, Park YS, Kim YH, Kim Y, Kim KP. Binary matrix for MALDI imaging mass spectrometry of phospholipids in both ion modes. Anal Chem. 2012;83:1252–1259. doi: 10.1021/ac1029659. [DOI] [PubMed] [Google Scholar]
- Shirey KA, Lai W, Scott AJ, Lipsky M, Mistry P, Pletneva LM, Karp CL, McAlees J, Gionnani TL, Weiss J, Chen WH, Ernst RK, Rossignol DP, Gusovsky F, Blanco JCG, Vogel SN. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature. 2013;497:498–502. doi: 10.1038/nature12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvero LJ, Amoscato AA, Kochanek PM, Pitt BR, Kagan VE, Bayir H. Mass-spectrometry based oxidative lipidomics and lipid imaging: applications in traumatic brain injury. J Neurochem. 2010;115:1322–1336. doi: 10.1111/j.1471-4159.2010.07055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector AA, Yorek MA. Membrane lipid composition and cellular function. J Lipid Res. 1985;26:1015–1035. [PubMed] [Google Scholar]
- Touboul D, Laprevote O, Brunelle A. Micrometric molecular histology of lipids by mass spectrometry imaging. Current Opinion Chemical Biol. 2011;15:1–8. doi: 10.1016/j.cbpa.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Tyurin VA, Epperly MW, Greenberger JS, Kagan VE. Oxidative lipidomics of gamma-irradiation-induced intestinal injury. Free Radic Biol Med. 2008;44:299–314. doi: 10.1016/j.freeradbiomed.2007.08.021. [DOI] [PubMed] [Google Scholar]
- vanHove ERA, Smith DF, Heeren RMA. A concise review of mass spectrometry imaging. J Chromatography A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- Yang J, Caprioli RM. Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal Chem. 2011;83:5728–5734. doi: 10.1021/ac200998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nature Chemical Biol. 2009;5:1–3. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]