Abstract
c-MYC (hereafter MYC) overexpression has been recognized in aggressive B-cell lymphomas and linked to adverse prognosis. MYC activation results in widespread repression of miRNA expression and associated lymphoma aggressive progression. Our recent study identified a MYC-miRNAs-EZH2 feed-forward loop linking over-expression of MYC, EZH2, and miRNA repression. Here, using a novel small-molecule BET bromodomain inhibitor, JQ1 and the EZH2 inhibitor, DZNep, we demonstrated that combined treatment of JQ1 and DZNep cooperatively disrupted MYC activation, resulting in a greater restoration of miR-26a expression and synergistically suppressed lymphoma growth and clonogenicity in aggressive lymphoma cells. Furthermore, CHIP assay demonstrated that MYC recruited EZH2 to miR-26a promoter and cooperatively repressed miR-26a expression in aggressive lymphoma cell lines as well as primary lymphoma cells. Loss or gain-of-function approaches revealed that miR-26a functioned as a tumor suppressor miRNA and mediated the combinatorial effects of JQ1 and DZNep. These findings represent a novel promising approach for silencing MYC-miRNA-EZH2 amplification loop for combinatorial therapy of aggressive B-cell lymphomas.
Keywords: B-cell lymphoma, MYC, EZH2, miRNA-26a, survival and clonogenicity
Introduction
Aggressive lymphomas include Burkitt lymphoma, MYC-associated diffuse large B-cell lymphoma (DLBCL), B-cell lymphoma, unclassifiable with features intermediate between DLBCL and Burkitt lymphoma (double-hit lymphoma), rare de novo acute lymphoblastic lymphoma/leukemia, blastic variant of mantle cell lymphoma (MCL), transformed follicular lymphoma, and plasmablastic lymphoma.1 These lymphomas are usually fast growing. Although MYC has been described as a defining feature and the driving oncogene for Burkitt lymphoma, MYC overexpression has also been recognized in other non-Hodgkin lymphomas. For example, rearrangement of MYC was detected in 9–14% of DLBCLs and has been linked to adverse prognosis with chemoresistance and shortened survival. In MCL, increased expression of MYC has been found to be associated with poor prognosis and with an aggressive, blastic variant of MCL. MYC rearrangement is a recurring genetic abnormality in these aggressive B-cell lymphomas. A recent report reveals that MYC overexpression was detected in 80% of transformed large cell lymphomas, implying the role of MYC in aggressive transformation.2 Unlike other lymphoma oncogenes, forced expression of Myc in murine lymphoid cells is sufficient to generate B-cell leukemia and lymphoma.3 In addition, MYC activation is necessary to develop lymphoma in BCL2 or CCND1 transgenic mice, further identifying MYC as a pivotal oncogene in initiating and sustaining aggressive transformation of B-cell lymphomas.4 However, mechanisms underlying the clinical aggressiveness of lymphoma are unclear.
The MYC transcriptional network has been shown to also include miRNAs. These small (20–24 nucleotides), non-protein-coding, single-stranded RNAs function as negative regulators of mRNA levels and affect virtually every aspect of tumorigenesis.5 Interestingly, although MYC can alter a large set of different miRNAs, rather than activation, the majority of identified miRNAs are repressed by MYC. Indeed, down-regulation of a subset of miRNAs is a commonly observed feature of cancers, suggesting that these molecules may act as tumor suppressors. Among these, many are putative tumor suppressors, such as miR-15a/16-1, miR-34a, and let-7 family members.6 Our recent experiments revealed loss or low expression of MYC-regulated miRNAs and reverse correlation of tumor suppressor miRNAs such as miR-15/16, miR-26a, and miR-29 with MYC overexpression in aggressive B-cell lymphomas and showed that ectopic expression of miR-29 suppresses MYC-driven lymphoma cell proliferation.7, 8 Collectively, these data support the notion that MYC activation results in widespread direct repression of miRNA expression and MYC-induced miRNA repression contributes to lymphoma aggressive progression. EZH2, the catalytic subunit of polycomb repressive complex 2 (PRC2), catalyzes trimethylation of lysine 27 on histone H3 (H3K27me3) and mediates transcriptional silencing. 9, 10 H3K27 trimethylation suppresses transcription of specific genes that are proximal to the site of histone modification. EZH2 level has been directly implicated in various cancers as well as lymphomagenesis. For example, Velichutina et al. found that EZH2 mRNA levels were directly correlated with cellular proliferation in primary DLBCLs, whereas levels of many EZH2 target genes were negatively correlated with proliferation in these same tumors.11 A somatic gain of function mutation of EZH2 was identified in large B cell lymphoma.12 Likewise, expression levels of EZH2 and the PRC2 component SUZ12 have been linked to MCL lymphomagenesis.13, 14 Overall, these data support Myc and EZH2 as a novel therapeutic target for these lymphomas.
Given the emerging evidence that points to oncogenic and tumor-suppressive roles of miRNAs in lymphoma, we recently studied explored the underlying mechanism of miRNA dysregulation, and the role of HDAC and EZH2 in miRNA expression in MCL, Burkitt lymphoma, and DLBCL cells. We demonstrated that MYC silences miR-29 and miR15-a/16 expression through recruitment of HDAC3 and EZH2.7, 8 We identified a MYC-miR-26a-EZH2 feed-forward pathway that consistently leads to MYC and EZH2 overexpression and miRNA repression and maintains tumorigenic potential of lymphoma cells.8 Furthermore, MYC and EZH2 act in concert to silence tumor suppressor miRNAs in aggressive lymphoma cells. Recently a novel BET bromodomain inhibitor, JQ1, has been developed and shown to inhibit c-Myc in multiple hematopoietic malignancies,15, 16 but its effects on aggressive lymphomas and miRNAs remain uncharacterized. Here, we applied JQ1 to explore whether combined inhibition of MYC with JQ1 and EZH2 with DZNep can: 1) cooperatively disrupt MYC-miRNA-EZH2 regulatory circuitry and interaction and 2) restore miR-26a expression, as well as the role of miR-26a in MYC-driven lymphoma cell survival and growth. Ultimately, we tested the strategy of silencing MYC with JQ1 for combinatorial therapy to subsequently suppress lymphoma growth and clonogenicity in aggressive lymphoma cells.
Materials and Methods
Cell Preparation, Antibodies, and Reagents
Lymphoma cell lines Jeko-1, Mino, HBL-2, SUDHL4 and Ramos were described as previously, 17 P493-6 cells (kindly provided by G Bornkamm),18 used to turn off MYC expression, were grown in the presence of 0.1 3g/mL tetracycline (Sigma, MO, USA) for 24–72 hours; MYC turn on cells were maintained in RPMI1640 with 10% FBS. Burkitt lymphoma and MCL cells were obtained from fresh biopsy-derived lymphoma tissues (lymph nodes) after informed consent from patients and approval by the Institutional Review Board of the University of South Florida. The details of cell preparation were described as previously. 19
Antibodies included anti-MYC (sc-764 X; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-β-actin (A1978; Sigma, MO, USA), and anti-EZH2 (07–689; Millipore, MA, USA). JQ1 was kindly provided by James E. Bradner (Boston, MA), and DZNep was kindly provided by Victor E. Marquez (Frederick, MD).
Cell Transfection
Optimized nucleofection protocols generated by Amaxa (Amaxa, Gaithersburg, MD) were followed for the transfection of HBL-2 (Nucleofector KIT V, program O-017) and SUDHL4 (Nucleofector Kit V, Program X-001) with miRNA precursors or anti-miRNA inhibitors (Applied Biosystems, CA, USA), respectively. miRNA precursor molecules and miRNA inhibitors in miRNA function experiments were purchased from Applied Biosystems (CA, USA).
CCK8 (Cell Viability) Assay and Colony Formation Assay
Cell viability was measured using cell counting kit-8 (CCK-8, Dojindo molecular technologes, MA, USA), and quantified by Gen5 data analysis software. For assessment of synergistic or additive effects of combined drug treatment, combination index (CI) was used. CI for each drug combination was calculated by median dose-effect analyses using the commercially available software CalcuSyn (Biosoft, Cambridge, UK). CI values <1.0 represent synergism of the 2 drugs in the combination.
For the colonogenic assay, 2×103 HBL-2 or SUDHL4 cells (0.05 mL) were added to 0.5 mL MethoCult® (STEMCELL, WA, USA) per well in 24-well plates. After 10–14 days of culture, the number of colonies was counted, with images of colonies recorded daily.
Chromatin Immunoprecipitation (ChIP) and Quantitative Real-Time Polymerase Chain Reaction
ChIP was carried out as described previously;7 Primer sequences used in ChIP assay are as follows:
26a1D-Chip-F: GGAGAGACTGGGAGCGAGTGT,
26a1D-Chip-R: CAAACTCACAACCTCCCGGT;
26a2S-Chip-F: CTCCATCTGTGAGCGGCC,
26a2S-Chip-R: AAAATAGCAAAGCTCCCGACTG;
26a2U-Chip-F: CAACCTTCGAATCCCGAAAG,
26a2U-Chip-R: GAGTCCTAGGTCCGCCCAC;
26a1NC-Chip-F: AGCTACCCAGCACCAGTGTCCAA,
26a1NC-Chip-R: GGGAATTGGGGGTGGACATCACA.
Quantitative reverse-transcribed PCR was performed according to the manufacturer’s instructions (Applied Biosystems, CA, USA). miR-26a (000405), RNU-44 primers (4373384), and GAPDH primers (Hs99999905_m1) were purchased from Applied Biosystems. Pri-miR-26a1 and pri-miR-26a2 primers were synthesized by IDT (OR, USA). Primer sequences are as below:
Pri-miR-26a1-F: GGAGAGGCTGCCCAATGGCAT,
Pri-miR-26a1-R: GCAGTGGGCAGGCCAGTCAT;
Pri-miR-26a2-F: GTTCCCCCATGCGTCTCAGGAACT,
Pri-miR-26a2-R: GGCTGCTCTGCTCTTCCTCAGGT.
miRNA and mRNA levels were separately normalized with RNU44 or GAPDH, and the relative expression level of specific miRNA and mRNA was presented by 2−Δ ΔCt.
miRNA Array
Jeko-1 cells were treated with JQ1 (1 μM) for 48 hours; total RNA was then extracted and used for array analysis. miRNA microarray analysis was performed by LC Sciences, as previously described.7 The GEO database accession number for the microarray data is GSE40019.
Statistical Analysis
SPSS 11.0 software was used for statistical analyses. Significant differences between values obtained with different experimental conditions were determined using Student t test. Significance was set at P < 0.05. Statistical analysis for cell proliferation was carried out by ANOVA.
Results
Silencing MYC with JQ1 disrupts the MYC-miRNA-EZH2 regulatory circuitry and inhibits lymphoma cell growth in aggressive lymphoma cell lines
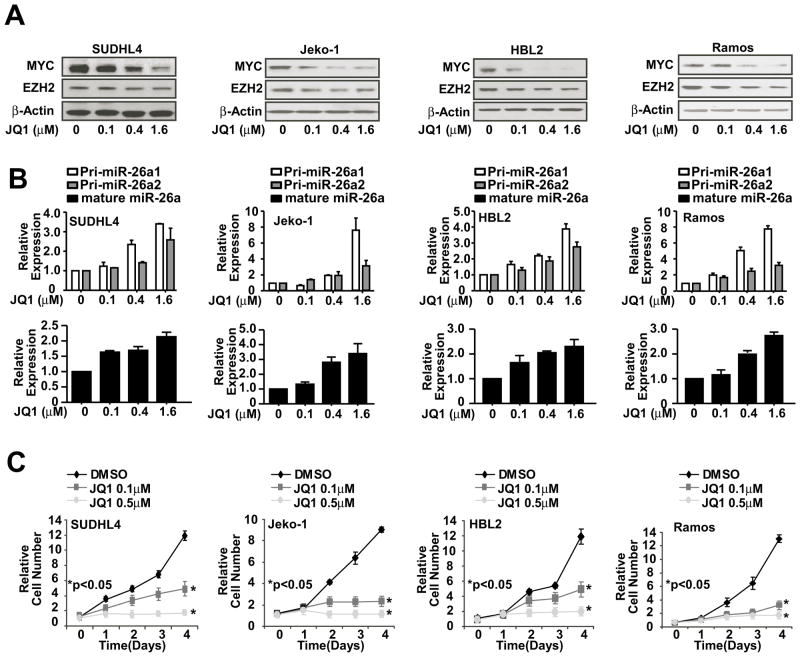
Since the discovery of the cancer-promoting gene MYC in the late 1970s, researchers have worked toward developing drugs that inhibit its function. Due to the diverse mechanisms drivingMYC activation and the difficulty of disrupting protein-DNA interactions, efforts to target MYC activity have been unsuccessful.20 Recently, a small molecule termed JQ1, a substituted 6H-thieno[3,2-f][1,2,4]triazolo[4,3-a]azepine, was identified to specifically prevent bromodomain from binding to acetylated histone and silence MYC transcription, thus preventing transactivation of MYC target genes.15, 16 JQ1 has been shown to have growth inhibitory effects on multiple myeloma, acute myelogenous leukemia, and Burkitt lymphoma cells.21 We recently reported that MYC contributes to EZH2 upregulation via repressing EZH2-targeting miR-26a and that EZH2 in turn induces MYC expression via MYC-targeting miR-494, thereby generating a positive MYC-miR-26a-EZH2 feedback loop to ensure persistent high protein levels of MYC and EZH2, and suppression of miR-26a.8 Here, we first investigate whether JQ1 indeed silences MYC expression, disrupts the MYC-miR-26a-EZH2 circuit, and inhibits lymphoma cell growth in aggressive lymphomas. Using four aggressive lymphoma cell lines HBL-2, Jeko-1 (blastic variant of MCL), SUDHL4 (transformed large cell lymphoma) and Ramos (Burkitt lymphoma), we explored the effect of JQ1 on MYC as well as EZH2 expression by Western blot, miR-26a using qRT-PCR and lymphoma cell growth with CCK8 assay in these cell lines. As shown in Figure 1 and Figure S1, JQ1 induced a dose-dependent down-regulation of MYC and EZH2, up-regulation of miR-26a and inhibition of lymphoma cell growth. In addition, we examined the effect of JQ1 on other known MYC target genes and miRNAs such as p21(Cip1/WAF1), miR-494 and miR-27a. We found that JQ1 also induced expression of p21, miR-494 and miR-27a, supporting that JQ1 regulates miR-26a through inhibition of MYC transactivation (Figure S1A–B). These data validated the inhibitory role of JQ1 in MYC expression and MYC dependence of EZH2, miR-26a expression and lymphoma cell growth.8, 15
Figure 1. Silencing MYC with JQ1 disrupts the MYC-miRNA-EZH2 regulatory circuitry and suppresses tumor cell growth and clonogenecity.
(A) Western blot shows JQ1-induced a dose-dependent downregulation of MYC and EZH2 protein expression in SUDHL4, Jeko-1, HBL2 and Ramos cells after 48-hour treatment. (B) qRT-PCR shows JQ1-induced dose-dependent expression of primary and mature miR-26a in SUDHL4, Jeko-1, HBL2 and Ramos cells after 48-hour treatment. Pri-miR-26a level was normalized to GAPDH, and mature miR-26a expression was normalized to RNU44. (C) Cell proliferation assay (CCK8) shows JQ1 induced a dose-dependent suppression of cell growth in SUDHL4, Jeko-1, HBL2 and Ramos cells. Results are either representative of 3 independent experiments (A) or are means ± SD from at least 3 biological replicates (B and C). (See also Figure S1)
JQ1 synergizes with inhibition of EZH2 to suppress MYC expression, lymphoma cell survival and clonogenic growth
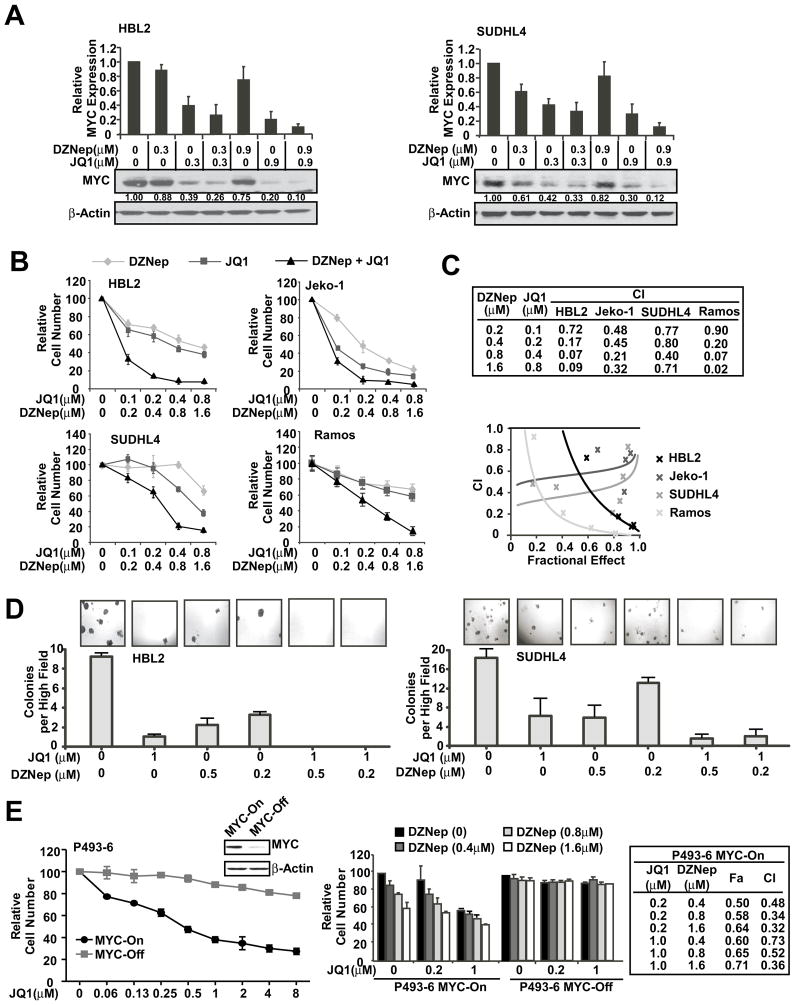
As previously demonstrated, MYC expression is also regulated by EZH2, and both MYC and EZH2 pathways are significant survival and proliferative signaling pathways and are constitutively activated in aggressive lymphomas.8 Thus, simultaneous inhibition of these pathways by combined MYC and EZH2 inhibition may be required for optimal anti-lymphoma activity. Furthermore, MYC and EZH2 were shown to act in concert to silence tumor suppressor miRNAs in aggressive lymphoma.8 These prompted us to study the potential of combined treatment with JQ1 and the EZH2 inhibitor DZNep to 1) further disrupt the MYC-miRNA-EZH2 cross-talk, resulting in more dramatic downregulation of MYC and suppression of lymphoma cell growth and clonogenicity, and 2) co-operatively reactivate tumor suppression miRNA(s) to synergistically regulate lymphoma cell survival. Compared with each agent alone, co-treatment with JQ1 and DZNep induced significantly lower expression of MYC (Figure 2A and S2A). Moreover, this combined treatment resulted in enhanced inhibition of cell survival and clonogenic growth (Figure 2, B–D). We further evaluated these results using median effect analysis (CalcuSyn, Biosoft, Great Shelford, UK) to determine whether the greater growth inhibitory activity of combined treatment reflected an additive or synergistic effect. We found all combinations of MYC and EZH2 inhibitors to be synergistic in comparison to single-agent treatment (Figure 2C). In addition we examined the selectivity of JQ1 and DZNep effects on the viability of transformed and non-transformed lymphocytes. We used P493-6 cells, a transformed human B-cell line featuring a tetracycline-repressible MYC gene that allows MYC expression to be turned on or off without altering the survival of these cells.22 As shown in Figure 2E, exposure of MYC-On P493-6 cells to DZNep and/or JQ1 dose-dependently induced a significant and synergistic cytotoxicity; in contrast, minimal cytotoxicity was noted in MYC-Off P493-6 cells, supporting the selectivity over the transformed MYC-associated lymphoma cells. Together, these results indicate that cytotoxicity triggered by DZNep and JQ1 is mediated, at least in part, via MYC-dependent pathway(s).
Figure 2. JQ1 and DZNep co-treatment synergistically suppresses MYC expression and inhibits lymphoma cell growth and clonogenicity.
(A) Western blot shows JQ1 and DZNep co-treatment more dramatically suppresses MYC protein expression than each agent alone in HBL2 and SUDHL4 cells. The relative level of MYC expression was measured by quantitative densitometry using the Software Quantity One and is indicated below each lane and mean+SD of 3 independent experiments are presented (top panel). (B) Cell proliferation assay (CCK8) shows JQ1 and DZNep co-treatment synergistically inhibits lymphoma cell growth. Cells were treated with DZNep and/or JQ1 as indicated for 48 hours, and CCK8 assay was performed. (C) Combination index (CIs) for drug combinations were obtained with CalcuSyn software using percent inhibition (fraction affected, Fa) resulting from combined action of the 2 drugs versus effects of either drug alone. CI values <1.0 indicate synergism of the 2 agents. (D) Colony formation assay shows combined treatment of JQ1 and DZNep resulted in enhanced inhibition of cell clonogenicity in HBL2 and SUDHL4 cells. Micrographs show colonies in methycellulose gels at low power. (E) CCK8 assay shows that MYC-On P493-6 cells are more sensitive than MYC-Off P496-3 cells to JQ1 and DZNep treatment (left). P493-6 cells were treated with JQ1 and/or DZNep for 48 hours, and cytotoxicity was assessed by CCK8 assay. CI values were calculated as in C. Results are representative of 3 independent experiments or are means ± SD from at least 3 biological replicates.(See also Figure S2)
To confirm that the above DZNep effect is indeed through EZH2 inhibition, we next performed siRNA experiments to more specifically inhibit EZH2, and investigated knockdown of EZH2 on JQ1 activity against MYC and on its anti-lymphoma effects. As shown in Figures S2B–C, knockdown of EZH2 with siRNA significantly enhanced JQ1 effect on Myc protein expression and lymphoma survival supporting DZNep functions via inhibition of EZH2.
MYC and EZH2 cooperatively regulate miR-26a Expression
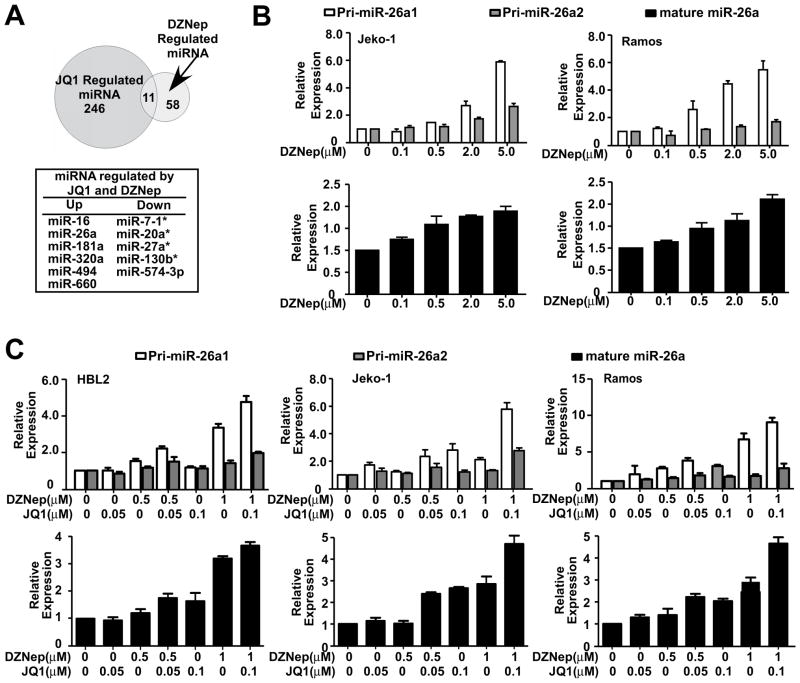
Next, we examined whether silencing MYC cooperates with EZH2 inhibition to induce (reactivate) miRNA(s) expression and subsequently contributes to suppression of lymphoma cell survival. To investigate which miRNAs are regulated by both JQ1 and DZNep, miRNA expression was explored by using microarray analysis. The expression profiles of Jeko-1 cells after 48-hour JQ1 (1 μM) treatment was determined and compared with expression profiles of Jeko-1 cells after DZNep treatment.8 As shown in Figure 3A, we identified a set of miRNAs that were co-regulated by JQ1 and DZNep: six that were up-regulated and five that were down-regulated. Among these miRNAs, we focused on miR-26a since this miRNA has been reported as a tumor suppressor and are down-regulated and inversely correlated with MYC and EZH2 expression in aggressive lymphomas.8 Induction of miR-26a by JQ1 from this array experiment is in agreement with qRT-PCR experiment shown in Figure 1 and further validated in DZNep-treated lymphoma cells showing DZNep-induced miR-26a expression in various aggressive lymphoma cell lines (Figure 3B and S3A). When compared with each agent alone, JQ1 and DZNep co-treatment induced significantly higher expression of pri-miR-26a1/2 and mature miR-26a in HBL2, Jeko-1, SUDHL4 and Ramos cells (Figure 3C and S3B).
Figure 3. MiR-26a is co-regulated by MYC and EZH2.
(A) Venn diagram illustrates the overlap between JQ1 and DZNep-regulated miRNAs, with table listing genes in the overlap. (B) DZNep treatment for 48 hours dose-dependently increased primary and mature miR-26a expression levels in Jeko-1 and Ramos cells. (C) JQ1 and DZNep cooperatively increased primary and mature miR-26a expression levels in HBL2, Jeko-1and Ramos cells after 48-hour treatment. (See also Figure S3)
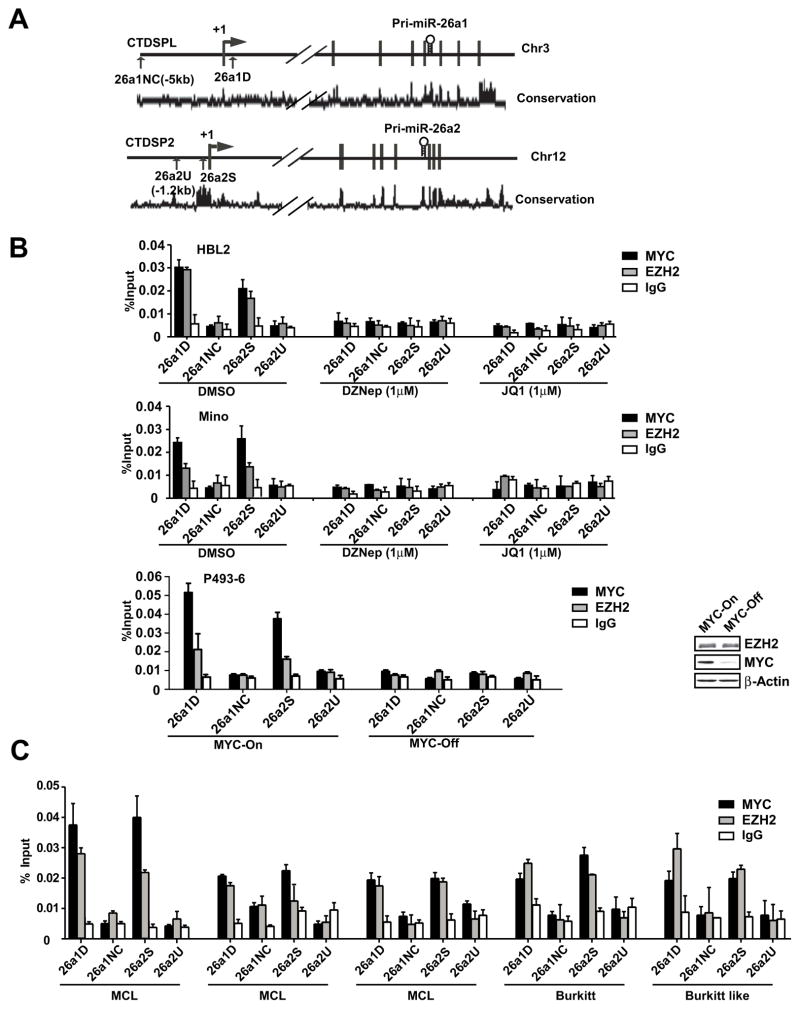
To determine whether the reactivation effect of JQ1 and DZNep is indeed attributed to direct binding of MYC and EZH2 to miR-26a gene promoter, we analyzed the upstream region (−5kb) of the miR-26a harboring gene CTDSPL (for pri-miR-26a1) and CTDSP2 (for pri-miR-26a2) for transcriptional factor binding sites and identified two E-box MYC binding sites, 26a1D and 26a2S (Figure 4A). ChIP assay was performed to explore whether EZH2 could be recruited to the miR-26a promoters by MYC and whether EZH2 binding is MYC dependent. Figure 4 revealed that antibodies against both MYC and EZH2 efficiently immunoprecipitated the miR-26a promoter regions. EZH2 binding is MYC dependent since EZH2 binding is abolished in MYC-Off P493-6 cells. This result further supports the recruitment role of MYC, and MYC cooperates with EZH2 to regulate miR-26a expression. Additional ChIP experiments were performed to determine the MYC and EZH2 bindings after JQ1 and DZNep treatment. Minimal or no enrichment of MYC and EZH2 to miR-26a promoter after JQ1 and DZNep treatment was observed, supporting that MYC recruited EZH2 to repress miR-26a expression (Figure 4B). To validate the MYC and EZH2 binding to miR-26a promoter in primary lymphoma samples, five high MYC lymphoma samples of three blastic MCLs, one Burkitt and one Burkitt like (double-hit) lymphomas were used for ChIP assay. The high MYC expression levels in these samples were further confirmed by florescence in situ hybridization (FISH) and immunohistochemical stains (not shown). Figure 4C and S4A reveals consistent enrichment of MYC and to a lesser extent EZH2 in miR-26a promoter regions in MYC-associated lymphomas and diminished MYC and EZH2 binding after JQ1 and EZH2 treatment, further providing that these interactions are operative in primary lymphoma cells and MYC-dependent. Taken together, these results confirm that MYC is required and are in agreement with a previous report that miR-26a expression is MYC-dependent, thus providing further support of the underlying mechanistic of MYC-induced miR-26a repression. 8
Figure 4. MYC recruits EZH2 to miR-26a promoters to repress miR-26a expression in lymphoma cell lines and primary lymphoma cells.
(A) Schematic diagram shows location of MYC-binding sites of pri-miR-26a1 (CTDSPL) and pri-miR-26a2 (CTDSP2) regulatory region. 26a1D and 26a2S represent MYC-binding site with E-box sequence. 26a1NC, negative control (located in the −5 kb of pri-miR-26a1 promoter and without E-box in this region). Both pri-miR-26a1/2 are highly conserved in their putative promoter region. (B) ChIP assay shows MYC and EZH2 enrichment on pri-miR-26a1 and pri-miR-26a2 promoters in HBL2, Mino cells and P493-6 cells. Low panel; ChIP assay showing MYC and EZH2 enrichment on pri-miR-26a1 and pri-miR-26a2 promoters and dependence of EZH2 binding on MYC in P493-6 MYC-on and MYC-off cells. Insert, Western blots showing protein level of MYC and EZH2 in MYC-on and MYC-off (24 hr tet treatment) P493-6 cells. (C) ChIP assay shows MYC and EZH2 enrichment on pri-miR-26a1 and pri-miR-26a2 promoters in primary patient samples. %Input was calculated with 2(Ct [1% of input]−Ct [ChIP]). For ChIP assays, IgG was used as negative control. Results are means ± SD from at least 3 biological replicates. (See also Figure S4)
MiR-26a functions as a tumor-suppressor miRNA and is required for the effect of MYC and EZH2 on lymphoma cell growth
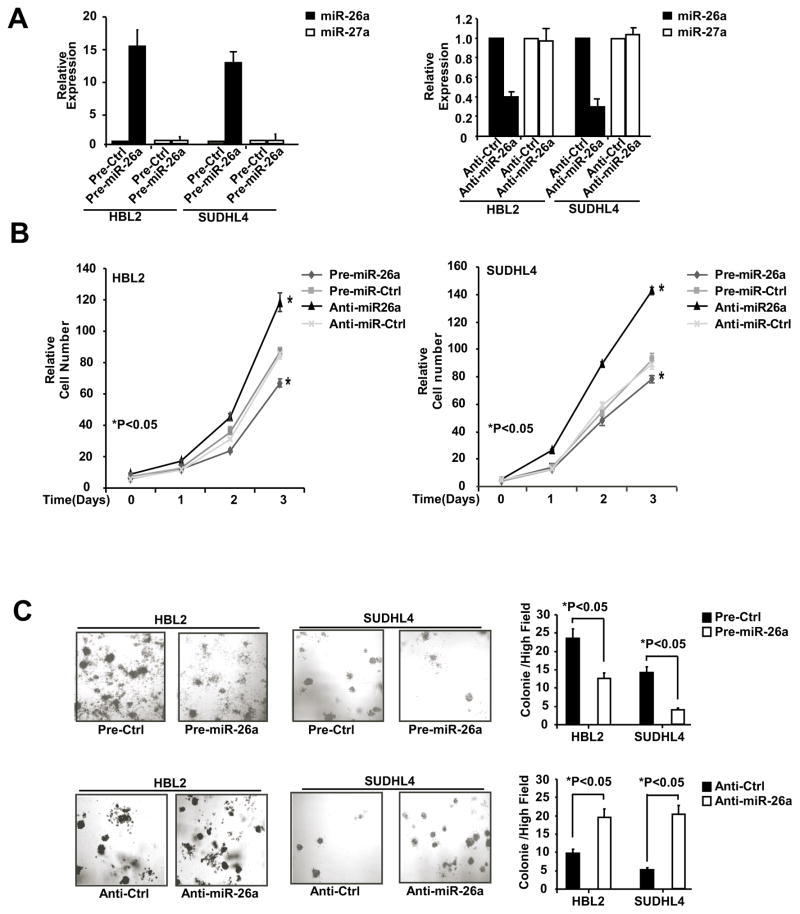
Using loss- and gain-of function assays, we evaluated whether miR-26a contributes to lymphoma cell survival and anchor-independent growth and determined whether miR-26a is required for JQ1 and DZNep-induced suppression of lymphoma cell growth. First, as shown in Figure 5A–C, over-expression of miR-26a by enforced ectopic expression of pre-miR-26a (A) specifically increased miR-26a (but not miR-27a) expression, significantly inhibited lymphoma cell survival (B) as well as anchor-independent growth in HBL-2 and SUDHL4 cells (C); in contrast, knock-down of miR-26a through anti-miR-26a specifically decreased miR-26a expression and further increased lymphoma cell growth as well as clonogenic growth. Collectively, these data support miR-26a as a tumor-suppressor miRNA in these aggressive lymphoma cells.
Figure 5. MiR-26a functions as a tumor suppressor miRNA to negatively regulate growth and colonogenicity of lymphoma cell.
(A) MiR-26a and miR-27a expression level after pre-miR-26a or anti-miR-26a transfection. miR-26a and miR-27a level of cell transfected with pre-Ctrl or anti-Ctrl is set to 1. (B) Cell proliferation assay (CCK8) shows that ectopic miR-26a expression by pre-miR-26a transfection inhibits cell growth of HBL2 and SUDHL4 cells, while knock-down of miR-26a through anti-miR-26a transfection increases cell growth. (C) Colony formation assay shows that ectopic miR-26a expression inhibits the cell colony formation ability (clonogenicity) in HBL2 and SUDHL4 cells, whereas inhibition of miR-26a through anti-miR-26a transfection increases cell clonogenicity. The numbers of tumor colonies were enumerated microscopically after 2-week incubation.
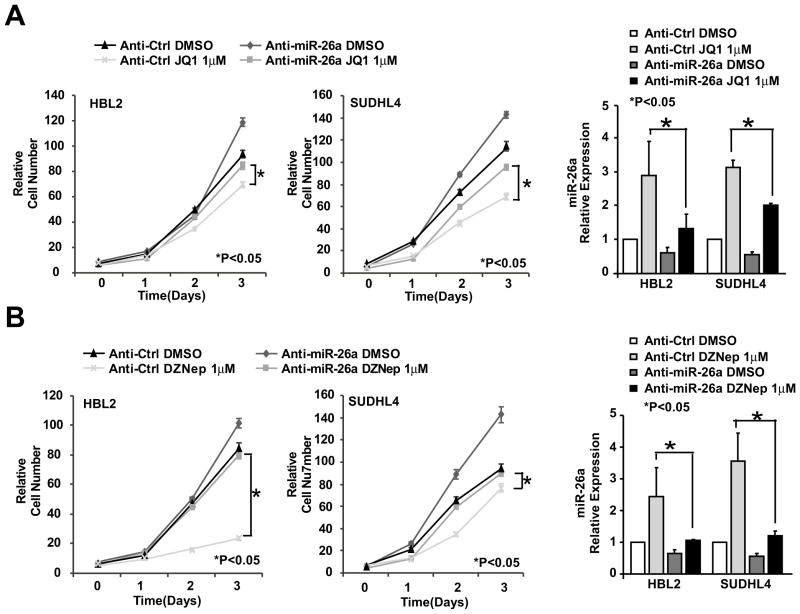
Next, we explored whether miR-26a is required for the effect of MYC and EZH2 on lymphoma cell growth. We pre-transfected these lymphoma cells with Anti-miR-26a to block JQ1- and EZH2-induced miR-26a induction and performed CCK8 assay. We explored whether miR-26a knock-down subsequently suppress JQ1- and EZH2-induced miR-26a expression and rescue JQ1- and EZH2-triggered lymphoma growth inhibition. As shown in Figure 6A–B, we found that knock-down of miR-26a with Anti-miR-26a significantly decreased miR-26a expression levels, suppressed JQ1- and EZH2-induced miR-26a expression, and partially rescued JQ1- and DZNep-induced suppression of lymphoma cell survival. Collectively, these results support that miR-26a functions as a tumor suppressor miRNA and contributes to inhibitory effect of JQ1 and EZH2 on aggressive lymphoma cell survival and growth.
Figure 6. MiR-26a is required for the effect of MYC and EZH2 on lymphoma cell growth.
Knock-down of miR-26a with anti-miR-26a blocks JQ1-(A) and EZH2-(B) induced miR-26a expression and attenuates JQ1- and EZH2-triggered lymphoma growth inhibition. Right panels show miR-26a expression levels of the corresponding cell groups. miR-26a level of cell transfected with anti-Ctrl treated with DMSO is set to 1. miR-26a expression was normalized to RNU44. Results are means ± SD from at least 3 biological replicates.
Discussion
MYC is a pleiotropic transcription factor that both activates and represses a broad range of target genes and is indispensable for cell growth. Rather than activation, the majority of identified miRNAs are repressed by MYC. Among these repressed miRNAs, many are putative tumor suppressors, such as miR-15a/16-1, miR-34a, and let-7 family members.6 Our recent experiments have revealed loss or low expression of MYC-regulated miRNAs in aggressive B-cell lymphomas and reverse correlation of tumor suppressor miRNAs such as miR-15/16, miR-26a, miR-29 with MYC and EZH2 overexpression as well as cell proliferation, CDK6, and IGF-1R expression.7, 8 MiRNA repression is a result of MYC/HDAC3 or/and EZH2 interaction and contributes to aggressive clinical outcome of MYC-associated lymphomas. We further observed the underlying mechanism of persistent MYC activation in these aggressive lymphomas through a MYC-miRNA-EZH2 positive feedback loop. Collectively, these studies indicated that MYC-induced EZH2-mediated miRNA repression plays a key role in lymphoma cell growth and progression. This study was undertaken to validate the inhibitory effect of bromodomain inhibitor, JQ1 on MYC expression, and to determine whether co-treatment with JQ1 and EZH2 inhibitor DZNep synergistically disrupt the MYC-miRNA-EZH2 regulatory circuitry, resulting in enhanced miRNA expression, and greater suppression of lymphoma cell growth.
In contrast to DZNep that indirectly targets MYC through histone trimethylation and silencing of miRNA(s) such as miR-494, 8 JQ1 has been developed and shown to inhibit MYC by binding competitively to the acetyl-lysine motif of the BET bromodomains15, 16. These suggest that JQ1 and DZNep regulate MYC via different signal transduction pathways and thus may underlie the cooperative effect of DZNep and JQ1 on MYC level. Moreover, our findings indicate that miR-26a repression is a result of MYC and EZH2 interaction and contributes to MYC-driven lymphoma cell growth. We revealed co-localization of MYC and EZH2 in the promoters of miR-26a,. This co-regulation is likely attributed to MYC recruitment of EZH2 to the promoter regions of some suppressor genes evidenced by Co-IP and CHIP experiments.8 This further supports our previous study that MYC and EZH2 form a repressive complex tethered to miRNA promoter elements to epigenetically repress miRNA transcription in MYC-expressing lymphoma cells. 8 The biological function of miR-26a is in line with previous studied demonstrating that miR-26a functions as a tumor suppressor miRNA.23–26 We demonstrated that JQ1 induced miR-26a expression, MYC suppression and potent activity against a range of human aggressive B-cell lymphoma lines. These findings further support the idea that inhibition of MYC activity by targeting bromodomains may be lethal to aggressive B-cell lymphomas. Furthermore, using P493-6 MYC-on and MYC-off cells, we revealed that JQ1 is selectively more potent against MYC-associated lymphoma cells.
JQ1 functions as a specific inhibitor of BET proteins including bromodomain-containing protein 4 (BRD4) and BRD2 through interfering the acetyl-lysine recognition domains (bromodomains). However, BRD4 not only regulates MYC but may also affect other oncogenic proteins. CHIP revealed that BRD4 is strongly enriched at immunoglobulin heavy-chain (IgH) enhancer.21 There is evidence that BRD4 is associated with immunoglobulin heavy chain gene enhancer and BRD4 inhibition can reduce the expression oncogenes that translocated to IgH locus.16 Given B-cell lymphomas are frequently associated with chromosomal translocation involving many genes such as BCL2, BCL6 that are switched on by immunoglobulin enhancers, we therefore can not conclude that the observed effect of JQ1 in this study are exclusively attributed to the effect on MYC. Similarly, a recent study demonstrated that DZNep may not be a specific EZH2 inhibitor; it may also affect other histone methyl transferases and contribute to the DZNep effect especially at high concentration.27 However, our experiments using siRNA demonstrated that specific knockdown of EZH2 induced miR-26a expression, suppression lymphoma cell growth and enhanced JQ anti-lymphoma effect. These results support the oncogenic role of EZH2 in lymphoma and corroborate the recent reports revealing EZH2 as a novel target for lymphoma therapy. 28–31
Recently, somatic heterozygous mutations of Y641 and A677 residues within the catalytic SET domain of EZH2 were found in up to 22% of germinal centre B-cell DLBCL and follicular lymphoma harbouring mutations. 31–35 These mutations enable EZH2 to more efficiently add a third methyl group to H3K27 and are responsible for H3K27 hyper-trimethylation and drive the lymphomagenic proliferation and survival.36 These findings have rapidly led to the development of small molecule inhibitors that reverse the defective processes caused by these mutations.29–31 Two independent groups using high-throughput screening for inhibitors of the PRC2 complex followed by medicinal chemistry optimization reported low nanomolar potency small molecule EZH2 inhibitors.30, 31 These small molecules displayed remarkable selectivity for EZH2 with similar efficacy against wild-type and mutant forms of EZH2. These EZH2 inhibitors were most effective against DLBCLs, especially those with EZH2 point mutations by inducing apoptotic cell death and proliferation arrest. More recently, using similar screening technology, Qi et al developed another Ezh2 inhibitor, EI1, which also act in a S-adenosyl methionine (SAM)-competitive manner and are highly selective over Ezh1. EI1-treated cells exhibit genome-wide loss of H3K27 methylation and activation of PRC2 target genes. Furthermore, inhibition of Ezh2 by EI1 in DLBCL cells carrying the Y641 mutations results in decreased proliferation, cell cycle arrest, and apoptosis. Overall, these results provide strong validation of Ezh2 as a potential therapeutic target for the treatment of EZH2 mutant and some EZH2 overexpression lymphomas. Thus, EZH2 lymphoma cells are critically dependent on EZH2 enzymatic activity for proliferation and survival (oncogen addiction).
As EZH2 mutations often coincide with other mutations and oncogen dysregulations in lymphomas, EZH2 may also cooperate with other oncogenes such as Myc to promote lymphoma aggressive progression through regulation of tumor suppressors including miRNAs. 8 When combining the expression of EZH2 Y641F with the over-expression of MYC by crossing the transgenic line with Emu-myc transgenic mice, a dramatic acceleration of lymphoma development with the combination of MYC and EZH2 Y641F as compared to MYC alone was observed. This indicates that EZH2 Y641F can collaborate with Emu-myc to promote lymphoma induction.37 Our finding that combined treatment of JQ1 and DZNep induced a synergistic anti-proliferation effect also supports that MYC and EZH2 work in concert to promote lymphoma progression. Moreover, our data that knock-down of miR-26a rescued JQ1- and DZNep-induced suppression of lymphoma cell survival implicates MYC-induced miR-26a repression at least partially underlies the molecular mechanism of the JQ1 therapeutic effects. In this study, we demonstrated that combined inhibition of MYC and EZH2 cooperatively disrupted MYC activation, resulting in greater restoration of miR-26a expression and synergistically suppressed lymphoma growth and clonogenicity in these aggressive lymphoma cells. Taken together, our findings indicate that miR-26a is a tumor suppressor miRNA and can be epigenetically targeted through manipulation of MYC and histone methylation. To achieve maximal anti-lymphoma effects, inhibition of both MYC and EZH2 pathways may be necessary. Silencing MYC with JQ1 for combinatorial therapy represents a promising, novel approach for aggressive B-cell lymphomas.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Cancer Institutes (R01 CA137123, to JT), Maher Fund (to JT), and Lymphoma Research Foundation (to JT) and supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (to VM). JQ1 was kindly provided by James E. Bradner (Boston, MA), and DZNep was kindly provided by Victor E. Marquez (Frederick, MD).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary information is available online at Leukemia website.
References
- 1.Slack GW, Gascoyne RD. MYC and aggressive B-cell lymphomas. Adv Anat Pathol. 2011;18(3):219–228. doi: 10.1097/PAP.0b013e3182169948. [DOI] [PubMed] [Google Scholar]
- 2.Craig VJ, Cogliatti SB, Imig J, Renner C, Neuenschwander S, Rehrauer H, et al. Myc- mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011;117(23):6227–6236. doi: 10.1182/blood-2010-10-312231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c- myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 4.Lovec H, Grzeschiczek A, Kowalski MB, Moroy T. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. EMBO J. 1994;13(15):3487–3495. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Chen X, Lin J, Lwin T, Wright G, Moscinski LC, et al. Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene. 2012;31(24):3002–3008. doi: 10.1038/onc.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Zhao X, Fiskus W, Lin J, Lwin T, Rao R, et al. Coordinated silencing of MYC- mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell. 2012;22(4):506–523. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Radulovic V, de Haan G, Klauke K. Polycomb-group proteins in hematopoietic stem cell regulation and hematopoietic neoplasms. Leukemia. 2013;27(3):523–533. doi: 10.1038/leu.2012.368. [DOI] [PubMed] [Google Scholar]
- 10.Popovic R, Licht JD. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov. 2012;2(5):405–413. doi: 10.1158/2159-8290.CD-12-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116(24):5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visser HP, Gunster MJ, Kluin-Nelemans HC, Manders EM, Raaphorst FM, Meijer CJ, et al. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br J Haematol. 2001;112(4):950–958. doi: 10.1046/j.1365-2141.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- 14.Martin-Perez D, Sanchez E, Maestre L, Suela J, Vargiu P, Di Lisio L, et al. Deregulated expression of the polycomb-group protein SUZ12 target genes characterizes mantle cell lymphoma. Am J Pathol. 2010;177(2):930–942. doi: 10.2353/ajpath.2010.090769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J, Lwin T, Zhao JJ, Tam W, Choi YS, Moscinski LC, et al. Follicular dendritic cell- induced microRNA-mediated upregulation of PRDM1 and downregulation of BCL-6 in non-Hodgkin’s B-cell lymphomas. Leukemia. 2011;25(1):145–152. doi: 10.1038/leu.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, et al. Control of cell growth by c-Myc in the absence of cell division. Current biology : CB. 1999;9(21):1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 19.Lwin T, Lin J, Choi YS, Zhang X, Moscinski LC, Wright KL, et al. Follicular dendritic cell-dependent drug resistance of non-Hodgkin lymphoma involves cell adhesion-mediated Bim down-regulation through induction of microRNA-181a. Blood. 2010;116(24):5228–5236. doi: 10.1182/blood-2010-03-275925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pajic A, Spitkovsky D, Christoph B, Kempkes B, Schuhmacher M, Staege MS, et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int J Cancer. 2000;87(6):787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 23.Di Lisio L, Gomez-Lopez G, Sanchez-Beato M, Gomez-Abad C, Rodriguez ME, Villuendas R, et al. Mantle cell lymphoma: transcriptional regulation by microRNAs. Leukemia. 2010;24(7):1335–1342. doi: 10.1038/leu.2010.91. [DOI] [PubMed] [Google Scholar]
- 24.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112(10):4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 25.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71(1):225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 26.Alajez NM, Shi W, Hui AB, Bruce J, Lenarduzzi M, Ito E, et al. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis. 2010;1:e85. doi: 10.1038/cddis.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8(6):1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodor C, O’Riain C, Wrench D, Matthews J, Iyengar S, Tayyib H, et al. EZH2 Y641 mutations in follicular lymphoma. Leukemia. 2011;25(4):726–729. doi: 10.1038/leu.2010.311. [DOI] [PubMed] [Google Scholar]
- 29.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109(52):21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8(11):890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 31.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 32.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan RJ, Nitta M, Borger D, Zukerberg LR, Ferry JA, Harris NL, et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS One. 2011;6(12):e28585. doi: 10.1371/journal.pone.0028585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107(49):20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berg T, Yap D, Thoene S, Wee T, Schoeler N, Umlandt P, et al. Mutated EZH2 Collaborates with Myc in Inducing Lymphoma in a Mouse Model. Blood. 2011;118(21):104–104. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.