Abstract
Glycosylation is an intricate process requiring the coordinated action of multiple proteins, including glycosyltransferases, glycosidases, sugar nucleotide transporters and trafficking proteins. Work by several groups points to a role for microRNA (miRNA) in controlling the levels of specific glycosyltransferases involved in cancer, neural migration and osteoblast formation. Recent work in our laboratory suggests that miRNA are a principal regulator of the glycome, translating genomic information into the glycocode through tuning of enzyme levels. Herein we overlay predicted miRNA regulation of glycosylation related genes (glycogenes) onto maps of the common N-linked and O-linked glycan biosynthetic pathways to identify key regulatory nodes of the glycome. Our analysis provides insights into glycan regulation and suggests that at the regulatory level, glycogenes are non-redundant.
Keywords: miRNA, glycosylation, glycan biosynthesis, carbohydrate pathways, glycogenes, microRNA, glycan regulation
Introduction
The cell surface is coated with glycans, complex biopolymers that form a critical interface with the extracellular world and inform diverse processes from immune recognition to cancer metastasis. Glycosylation is an intricate process requiring the integrated action of multiple proteins, including glycosylation enzymes (glycosyltransferases and glycosidases), sugar nucleotide transporters and trafficking proteins, to synthesize discrete structures appended onto serines and threonines (O-linked) or asparagines (N-linked) on proteins or onto lipids. The glycome is highly regulated during embryogenesis [1,2] and defects in glycan biosynthesis cause congenital disorders, 64 of which have been identified to date [3]. Despite the importance of glycosylation, there is currently little understanding of how glycan biosynthesis is controlled. Alteration of mRNA levels of glycosylation-related genes (glycogenes) maps onto concomitant alterations in the glycome, arguing that regulation of glycogene transcripts is important, although a direct relationship is not always observed [1].
MicroRNA (miRNA) are short endogenous noncoding RNAs ~22 nts in length that can either promote mRNA degradation or inhibit translation, downregulating associated protein levels [4]. Recently, work by several groups has pointed to a role for microRNA in controlling glycosylation [5,6,7,8,9]. To date, seven human glycogenes (C1GALT1[6], MANEA [10], FUT4 [9], FUT8 [11], GALNT1 [8], GALNT7 [7,8] and NDST1 [5]) have been validated as miRNA targets. A role for miRNA as a major regulator of glycan biosynthesis has been suggested by work in C. elegans [12] and by unpublished results in human cell lines from our laboratory. Analysis of miRISC complexes during the developmental stages of C. elegans showed robust enrichment of glycogene transcripts associated with miRNA. Work in our laboratory using the NCI-60 cancer cell set found strong correlations between the glycome and miRNA expression levels and identified three new glycogene/miRNA interactions (in submission). Taken together, the data suggests that miRNA are a principal regulator of the glycome.
Identifying the regulatory relationship between miRNA and a target gene currently relies on target prediction analysis. Although miRNA can bind anywhere within an mRNA (5′-UTR, coding region or 3′-UTR), the majority of prediction analysis focuses on the 3′-UTR, which is widely regarded as the main site of miRNA action [4]. Algorithms such as miRanda, PicTar and TargetScan rely on a combination of the thermodynamic stability of the miRNA/gene interaction and factors such as target site conservation between species to identify and prioritize miRNA interaction partners [13,14]. A single miRNA can have hundreds of predicted targets and predictions have been used to identify regulatory networks for systems as diverse as cancer [15] and schizophrenia [16]. Genes with higher numbers of predicted miRNA binding sites show increased levels of repression, faster mRNA decay rates and increased evolutionary conservation [17,18,19]. Herein we overlay predicted miRNA regulation of glycogenes onto maps of the common glycan biosynthetic pathways to identify key regulatory nodes of the glycome. Our analysis provides insights into glycan regulation and shows that at the regulatory level, glycogenes are non-redundant.
Materials and Methods
Glycogene target analysis and pathway annotation
The human miRNA target site miRSVR-miRanda prediction datasets for both conserved and non-conserved miRNAs with “good” miRSVR scores (miRSVR < −0.1) were downloaded from the latest release of microRNA.org (2010 August release) and combined to make a “total” dataset [20,21]. This dataset was then checked for duplicative entries using the built in unique function in R and checked against the original datasets. No duplications were observed. A comprehensive list of glycosylation related genes (glycogenes, 538 genes, 913 transcripts) was created based on Maupin et al [22], the Kyoto Encylopedia of Genes and Genomes (KEGG)[23] and Nairn et al [1]. This list includes trafficking proteins, glycosyltransferases, glycosidases, nucleotide sugar transporters and highly glycosylated proteins (mucins, cadherins, etc.) but excludes lectins (see Supplemental Table 1). For reference, 3′-UTR lengths were obtained from the UCSC genome browser (Build GRCh37/hg19) using the unique known gene ids for the transcripts [24]. The miRNA/glycogene target prediction data was extracted from the combined miRNA prediction dataset.
To obtain the number of total and unique miRNA/glycogene interactions we determined the frequency of each mirna/transcript interaction using the Table function in R. Summing the counts for all miRNA sites on a given transcript gave us the total hits for that glycogene (Total). For unique hits (Unique), we summed the number of unique miRNAs that interacted with a given transcript. The Total and Unique hit levels were then mapped in Cytoscape 3.0.1 [25] and overlaid onto the glycosylation pathways. For our analysis, we considered only validated mRNA sequences from the RefSeq database (designated by NM). Where no such transcript existed for the gene, the largest known transcript was used. Where more than one NM designated transcript existed for a single glycogene, the data for the transcript with the highest total number of miRNA sites were used in our pathway map. To identify highly targeted glycogenes, we first determined the average and standard deviation for the number of total hits of all genes in the dataset (Average = 131.8, Std. Dev.= 103.1). Glycogenes were then defined as “highly targeted” if their total hits were ≥ 235 (i.e. 1 standard deviation above the mean) [18,26].
Results and Discussion
General Analysis of Glycogene Regulation
Glycosylation patterns are known to be highly variable between species [27,28]. In contrast, there is high conservation among enzymes in glycosylation pathways, possibly driven by strong functional constraints [29]. This argues that the regulation of glycogenes by factors such as miRNA may be species specific, accounting for the variation in observed carbohydrate epitopes. Conservation of miRNA target sites across species is an important aspect of most target prediction algorithms. Mammalian-specific miRNA have fewer predicted conserved targets than do broadly conserved miRNA [30], arguing that species-specific regulation is not well represented in the subset of conserved target sites. However, when non-conserved sites are included in miRNA target prediction analysis, decreases in sensitivity and precision are observed, thus many analysis programs use a conservation filter [14]. In recent work, Betel et al introduced miRSVR, a new filter for miRanda predictions based on supervised learning from miRNA transfection datasets [21]. The use of miRSVR-miRanda identifies both conserved and non-conserved miRNA binding sites with improved accuracy. Thresholding miRanda predictions using a miRSVR score of −0.1 or better gives a prediction set with a reasonable probability of downregulation [21]. To study glycogene regulation, we created a dataset containing the conserved and nonconserved miRSVR-miRanda miRNA/target predictions with a miRSVR score of −0.1 or below. We then determined the total predicted number of miRNA binding sites within the 3′-UTR of an mRNA as a metric for probability of miRNA-mediated gene regulation [17,18]. Genes can be cooperatively regulated by multiple miRNAs [31,32]. The total number of predicted binding sites strongly correlates with repression levels and mRNA decay rates, irrespective of the prediction algorithm used and independent of the length of the 3′-UTR [17,18]. We observed no significant differences in the distribution of total binding sites for glycogenes compared to the entire gene set (Supplemental Figure 1).
Using this dataset, we defined a “highly regulated” glycogene subset, i.e. genes for which the total number of predicted miRNA binding sites is ≥ 235 (1 S.D. above the average for all genes, see Materials and Methods) [18,26]. A list of the top 10 most “highly regulated” glycogenes is given in Table 1. At the top of the list was CHSY3, a chondroitin sulfate synthase with an average length 3′-UTR (843 bp, Average = 1265 bp for all genes, St. Dev.= 1367). Chondroitin sulfate is a proteoglycan important in brain development [33] and recent work has shown that miRNA modulation of related chondroitin synthetase sqv-5, the C. elegans homologue of CHSY1 (another “highly regulated” gene, see Supplemental Table 1), is a critical regulator of neurodevelopment in worms. More than 70% of all miRNA are expressed in the brain and miRNA have a strong impact on brain development, suggesting a potential role for miRNA regulation of CHSY3 in this highly coordinated process [34].
Table 1.
List of top 10 most “highly regulated” glycogenes.
| Glycogene | Ref. Seq. ID | Total Hits | Function |
|---|---|---|---|
| CHSY3 | NM_175856 | 455 | chondroitin sulfate synthetase |
| CDH11 | NM_001797 | 437 | cadherin |
| GALNT1 | NM_020474 | 437 | polypeptide-GalNAc transferase |
| HAS2 | NM_005328 | 426 | hyaluronan synthase |
| SPOCK3 | NM_016950 | 399 | proteoglycan |
| GALNT7 | NM_017423 | 396 | polypeptide-GalNAc transferase |
| ITGAV | NM_002210 | 393 | integrin |
| HS6ST2 | NM_001077188 | 386 | heparin 6-O-sulfotransferase |
| GALNT3 | NM_004482 | 381 | polypeptide-GalNAc transferase |
| GLCE | NM_015554 | 380 | glucuronic acid epimerase |
To gain greater insight into regulatory hubs in glycan biosynthesis, we mapped our analysis of glycogene regulation onto the canonical N-linked, O-linked and terminal glycan biosynthetic routes. The results of this analysis are discussed in more detail in the following sections.
Predicted Regulatory Hubs in N-linked Glycosylation Pathway
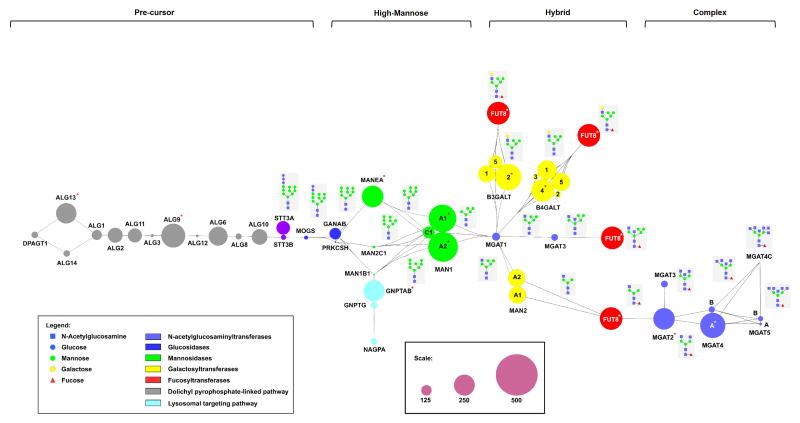
In N-linked glycosylation, a core Glc3Man9GlcNAc2-oligosaccharide is transferred onto select asparagines in a nascent polypeptide from a dolichol precursor by oligosaccharyltransferases (STT3A/B: catalytic subunits). This glycan is trimmed to Man9 and then elaborated by glycosidases and glycosyltransferases, giving rise to high mannose, hybrid and complex structures as shown in Figure 1. Glycogenes at several key points in the N-linked pathway are predicted as “highly regulated” hubs, including MAN1 genes, select MGAT genes and FUT8 (red asterisks, Figure 1). Similar results are observed when only “Unique” hits are considered (Supplemental Figure 2). The MAN1 gene family (MAN1A1, MAN1A2) encodes mannosidases that control trimming of high mannose structures to Man5, a required prerequisite for hybrid and complex epitopes. High mannose sugars are the dominant N-linked epitope on both embryonic and pluripotent human stem cells [35,36] and differentiating mouse stem cells show a loss of high mannose concomitant with increased levels of MAN1 transcripts [1], suggesting that their regulation is important in early embryogenesis. Work in our laboratory has validated MAN1A2 as a target of at least three miRNA, arguing that MAN1 transcripts are a hub for miRNA based regulation (unpublished results).
Figure 1. MiRNA regulatory map for glycogenes in the N-linked glycosylation pathway.
A schematic illustration of the N-linked glycan pathway with the relevant glycosylation enzymes is shown. Synthetic pathways for Glc2Man9GlcNAc2-dolichol precursor, high-mannose, hybrid and complex oligosaccharides are indicated. The multicomponent oligosaccharyltransferase (OST) is represented by STT3A and STT3B (purple), the genes that encode for the catalytic subunits. The size of the circles (nodes) represents the total number of miRNA binding sites (both conserved and non-conserved) predicted within the 3′UTR of the indicated glycogenes (see Methods for details, scale shown in inset). Color of circle corresponds to the enzyme class (see inset legend). Where multiple isoforms of the same glycogene exist, the isoform is indicated within the circle). Linkages between the nodes illustrate the various paths of the biosynthetic pathway and the resulting glycan structures are shown in the grey boxes. “Highly regulated” glycogenes are denoted with an asterisk (*).
The MGAT genes encode a series of enzymes responsible for transferring N-acetylglucosamine (GlcNAc) onto core mannose residues to create complex branched structures. The branching patterns of N-glycans play a crucial role in cellular differentiation and altered branching patterns are associated with immune recognition and cancer [37,38]. The predicted miRNA regulation across the MGAT family is highly variable. MGAT2 and MGAT4A, which catalyze formation of biantennary and triantennary branched glycans, respectively, are “highly regulated” hubs. In contrast, MGAT5, which creates tetraantennary glycans essential in immunity, is not a major target of miRNA regulation despite an average 3′-UTR length (1550 bp). MGAT5 activity is exquisitely regulated by available UDP-GlcNAc levels, which can be tuned by the expression levels of upstream MGAT enzymes with higher affinity for the sugar nucleotide including MGAT2 [37]. This may negate the need to regulate MGAT5 through miRNA-mediated mechanisms.
The FUT8 gene encodes the fucosyltransferase which adds α-1,6 fucose onto the core GlcNAc of N-linked glycans. Alterations in core fucosylation have been linked to cancer and emphysema [11,39,40] and recent data points to a role in controlling TGFβ signaling [41]. Our analysis predicts that FUT8 is “highly regulated” by miRNA. Recently, FUT8 was shown to be a target of two miRNAs downregulated in hepatocarcinoma, a cancer associated with high levels of core fucose [11]. Taken together, the data suggest that miRNA regulation of FUT8 is a major mechanism for controlling levels of core fucosylation.
Predicted Regulatory Hubs in O-linked Glycosylation Pathway
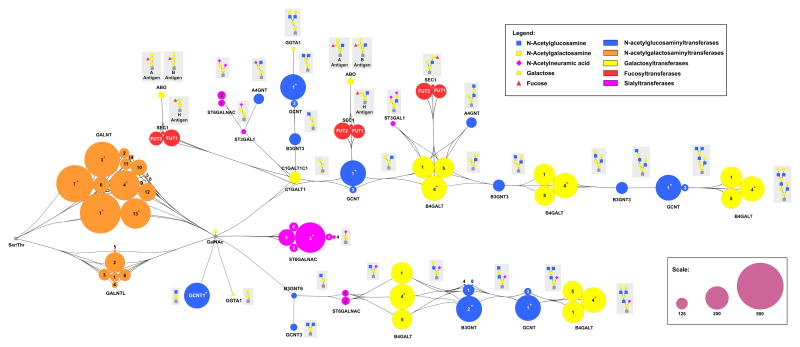
In contrast to N-glycans, canonical O-linked glycans have a single common sugar, α-linked N-acetylgalactosamine (GalNAc), which forms the core structure attached to serines or threonines. This glycan is then elaborated by a host of glycosyltransferases making more complex epitopes. As shown in Figure 2, multiple “highly regulated” genes are predicted by our analysis including several of the enzymes that initiate O-linked glycosylation (GALNTs), the core-2 synthesizing enzyme GCNT1, and the sialyltransferase ST6GALNAC3 (which synthesizes α-2,6-sialylGalNAc-α-Ser/Thr (sialylTn antigen)). The GALNT family is a large family of ~20 enzymes that transfer the core GalNAc residue onto serines and threonines in the polypeptide backbone. They have distinct but overlapping requirements for peptide sequence [42]. Little is known of the regulatory mechanisms controlling mRNA and protein levels of these enzymes, although dynamic changes in both have been observed. Based on our analysis, GALNTs display isoform dependent miRNA regulation. Several GALNT’s are among the top 10 most “highly regulated” glycogenes, which is not due to a significantly larger 3′-UTR for these genes (GALNTs 1, 3 and 7, Table 1, Figure 2). Gaziel-Sovran et al identified both GALNT1 and GALNT7 as targets of miR-30b/d, miRNA associated with metastasis in melanoma [8]. GALNT7 was also found to be a target of miR-378, with a role in osteoblast differentiation [7], showing that multiple miRNA target GALNT7 in line with our analysis. Although no miRNA targeting GALNT3 have yet been identified, levels of this gene alter in response to inorganic phosphate, calcium and vitamin D, in line with the type of dynamic regulation that miRNA provide [42]. In contrast to the highly regulated GALNTs, there are multiple family members that are predicted to have very low levels of regulation (<60 predicted hits, GALNTs 5, 8, 9 and 14, Figure 2). This may be due to the length of the gene transcripts, which are all below the average (range 52–734 bases). The disparity in predicted miRNA-based regulation of these genes belies their presumed redundancy. Knockouts of GALNTs, including GALNT3, cause subtle phenotypes in mice, attributed to functional redundancy [2,42]. The presence of subtle phenotypes however argues that the redundancy is not total, this is primarily attributed at present to substrate specificity differences however differences in miRNA regulation of transcripts, such as those predicted by our analysis, may also play a role.
Figure 2. MiRNA regulatory map for glycogenes in the O-linked glycosylation pathway.
A schematic illustration of the O-linked glycan pathway with the relevant glycosylation enzymes is shown. Pathway is annotated as in Figure 1. “Highly regulated” glycogenes are denoted with an asterisk (*).
Predicted Regulatory Hubs Affecting Terminal Glycan Epitopes on both N- and O-linked Glycans
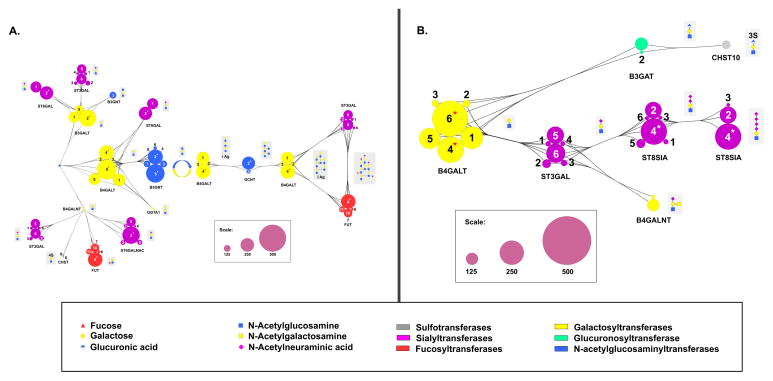
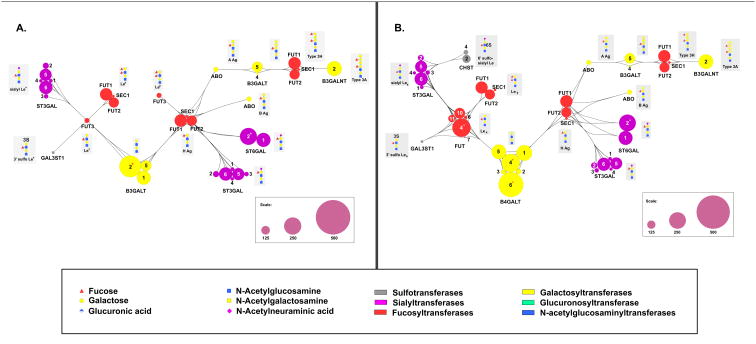
Both N- and O-linked glycans are elaborated with common terminal modifications. These include poly-N-acetyl lactosamine chains (poly-LacNAc) which can be Type I (β-1,3 linked) or Type II (β-1,4 linked, Figure 3A), sialic acids (Figure 3), blood groups and Lewis Antigens (Figure 4). Our analysis identifies isoform specific regulation of the genes involved in these complex biosynthetic pathways. Isoforms involved in both Type I and Type 2 polyLacNAc synthesis pathways that are “highly regulated” genes include the initiating galactosyltransferases B3GALT2 (Type 1), B4GALT4 and B4GALT6 (Type 2) and elaborating GlcNAc transferases (Type 2: B3GNT2, B3GNT5, Figure 3A). Only three sialyltransferases, ST6GALNAC3, ST6GAL2 (Figure 3A) and ST8SIA4 (Figure 3B) are predicted to be “highly regulated”. Of these, ST8SIA4 (PST) is particularly interesting because it is one of two sialyltranferases that synthesize polysialic acid, a critical modification of the neural cell adhesion molecule (NCAM) in the brain. ST8SIA4 is expressed in adult brain and has recently been shown to play a role in dendritic cell maturation, while the other polysialic acid enzyme ST8SIA2 (STX) is only expressed in early stages of embryogenesis. The need to constantly modulate ST8SIA4 levels in maturing dendritic cells may explain why this gene is more regulated than the ST8SIA2 isoform [43]. Few of the genes involved in Lewis structure and blood group formation are predicted to be “highly regulated”, with the exception of FUT4, an enzyme involved in Lewis x and Lewis y formation. FUT4, also known as CD15, is again one of the few known glycogene targets of miRNA [9].
Figure 3. MiRNA regulatory map for glycogenes involved in polyLacNAc and sialoside biosynthesis.
A schematic illustration of the glycogenes involved in the terminal modification processing steps leading to (A) polyLacNAc structures and (B) sialosides. Pathway is annotated as in Figure 1. “Highly regulated” glycogenes are denoted with an asterisk (*).
Figure 4. MiRNA regulatory map for glycogenes involved in blood group and Lewis antigens.
A schematic illustration of the glycogenes involved in the terminal modification processing steps leading to blood group and Lewis antigen structures on (A) Type I and (B) Type II LacNAc cores. Pathway is annotated as in Figure 1. “Highly regulated” glycogenes are denoted with an asterisk (*).
Summary
MiRNA are relatively unexplored as regulators of the glycome. Our recent work has shown that these non-coding RNA play a critical role in controlling glycosylation. Overlaying our predicted miRNA/glycogene interaction levels onto the glycan biosynthetic pathways points to major regulatory hubs, including the early steps of both the N-linked (MAN1A1, MAN1A2) and O-linked (GALNTs) pathways. In line with our analysis, five of the seven glycogenes identified as miRNA targets to date are predicted to be “highly regulated” genes (GALNT1, GALNT7, FUT4, FUT8, MANEA). We found glycogene regulation to be isoform specific, with only some isoforms designated as “highly regulated” genes. This type of isoform-specific regulation may explain why, even when functionally redundant enzymes are present, knockouts show subtle phenotypes often in the brain (behaviour) or the immune system, where miRNA play important roles. Changes in miRNA may underlie the altered glycosylation observed in dynamic processes such as cancer metastasis and embryogenesis, opening new opportunities to unravel the glycocode.
Supplementary Material
Highlights.
MicroRNA are an underappreciated regulator of glycosylation.
Key glycosylation enzymes are highly regulated by microRNA.
“Redudant” glycosylation enzymes are non-redundant in microRNA regulation.
Acknowledgments
The authors acknowledge financial support from the National Institutes of Health (7 DP2 OD004711-02).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nairn AV, Aoki K, dela Rosa M, Porterfield M, Lim JM, Kulik M, Pierce JM, Wells L, Dalton S, Tiemeyer M, Moremen KW. Regulation of glycan structures in murine embryonic stem cells: combined transcript profiling of glycan-related genes and glycan structural analysis. The Journal of biological chemistry. 2012;287:37835–37856. doi: 10.1074/jbc.M112.405233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 3.Hennet T. Diseases of glycosylation beyond classical congenital disorders of glycosylation. Biochimica et biophysica acta. 2012;1820:1306–1317. doi: 10.1016/j.bbagen.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 5.Kasza Z, Fredlund Fuchs P, Tamm C, Eriksson AS, O’Callaghan P, Heindryckx F, Spillmann D, Larsson E, Le Jan S, Eriksson I, Gerwins P, Kjellen L, Kreuger J. MicroRNA-24 suppression of N-deacetylase/N-sulfotransferase-1 (NDST1) reduces endothelial cell responsiveness to vascular endothelial growth factor A (VEGFA) The Journal of biological chemistry. 2013;288:25956–25963. doi: 10.1074/jbc.M113.484360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serino G, Sallustio F, Cox SN, Pesce F, Schena FP. Abnormal miR-148b expression promotes aberrant glycosylation of IgA1 in IgA nephropathy. Journal of the American Society of Nephrology: JASN. 2012;23:814–824. doi: 10.1681/ASN.2011060567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kahai S, Lee SC, Lee DY, Yang J, Li M, Wang CH, Jiang Z, Zhang Y, Peng C, Yang BB. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS One. 2009;4:e7535. doi: 10.1371/journal.pone.0007535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert JF, Shang S, Kerbel RS, Bhardwaj N, Shao Y, Darvishian F, Zavadil J, Erlebacher A, Mahal LK, Osman I, Hernando E. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andolfo I, Liguori L, De Antonellis P, Cusanelli E, Marinaro F, Pistollato F, Garzia L, De Vita G, Petrosino G, Accordi B, Migliorati R, Basso G, Iolascon A, Cinalli G, Zollo M. The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro-oncology. 2012;14:596–612. doi: 10.1093/neuonc/nos002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan S, Cheng X, Chen H, Castro PD, Ittmann MM, Hutson AW, Zapata SK, Sifers RN. ERManI is a target of miR-125b and promotes transformation phenotypes in hepatocellular carcinoma (HCC) PloS one. 2013;8:e72829. doi: 10.1371/journal.pone.0072829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernardi C, Soffientini U, Piacente F, Tonetti MG. Effects of MicroRNAs on Fucosyltransferase 8 (FUT8) Expression in Hepatocarcinoma Cells. PloS one. 2013;8:e76540. doi: 10.1371/journal.pone.0076540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Hammell M, Kudlow BA, Ambros V, Han M. Systematic analysis of dynamic miRNA-target interactions during C. elegans development. Development. 2009;136:3043–3055. doi: 10.1242/dev.039008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B, Li J, Cairns MJ. Briefings in bioinformatics. 2012. Identifying miRNAs, targets and functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 15.Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, Cogdell D, Nykter M, Broaddus R, Rodriguez-Aguayo C, Lopez-Berestein G, Liu J, Shmulevich I, Sood AK, Chen K, Zhang W. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer cell. 2013;23:186–199. doi: 10.1016/j.ccr.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo AY, Sun J, Jia P, Zhao Z. A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC systems biology. 2010;4:10. doi: 10.1186/1752-0509-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Z. Insight into microRNA regulation by analyzing the characteristics of their targets in humans. BMC Genomics. 2009;10:594. doi: 10.1186/1471-2164-10-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome biology. 2007;8:R166. doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng C, Bhardwaj N, Gerstein M. The relationship between the evolution of microRNA targets and the length of their UTRs. BMC genomics. 2009;10:431. doi: 10.1186/1471-2164-10-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic acids research. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome biology. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maupin KA, Sinha A, Eugster E, Miller J, Ross J, Paulino V, Keshamouni VG, Tran N, Berens M, Webb C, Haab BB. Glycogene expression alterations associated with pancreatic cancer epithelial-mesenchymal transition in complementary model systems. PLoS One. 2010;5:e13002. doi: 10.1371/journal.pone.0013002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karolchik D, Hinrichs AS, Kent WJ. In: The UCSC Genome Browser, Current protocols in bioinformatics. Unit1 4. Baxevanis Andreas D, et al., editors. Chapter 1. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS computational biology. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raju TS, Briggs JB, Borge SM, Jones AJ. Species-specific variation in glycosylation of IgG: evidence for the species-specific sialylation and branch-specific galactosylation and importance for engineering recombinant glycoprotein therapeutics. Glycobiology. 2000;10:477–486. doi: 10.1093/glycob/10.5.477. [DOI] [PubMed] [Google Scholar]
- 28.Antonopoulos A, North SJ, Haslam SM, Dell A. Glycosylation of mouse and human immune cells: insights emerging from N-glycomics analyses. Biochemical Society transactions. 2011;39:1334–1340. doi: 10.1042/BST0391334. [DOI] [PubMed] [Google Scholar]
- 29.Montanucci L, Laayouni H, Dall’Olio GM, Bertranpetit J. Molecular evolution and network-level analysis of the N-glycosylation metabolic pathway across primates. Molecular biology and evolution. 2011;28:813–823. doi: 10.1093/molbev/msq259. [DOI] [PubMed] [Google Scholar]
- 30.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borgdorff V, Lleonart ME, Bishop CL, Fessart D, Bergin AH, Overhoff MG, Beach DH. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1) Oncogene. 2010;29:2262–2271. doi: 10.1038/onc.2009.497. [DOI] [PubMed] [Google Scholar]
- 32.Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, Zhan R, He X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz NB, Domowicz M. Proteoglycans in brain development. Glycoconjugate journal. 2004;21:329–341. doi: 10.1023/B:GLYC.0000046278.34016.36. [DOI] [PubMed] [Google Scholar]
- 34.Nowak JS, Michlewski G. miRNAs in development and pathogenesis of the nervous system. Biochem Soc Trans. 2013;41:815–820. doi: 10.1042/BST20130044. [DOI] [PubMed] [Google Scholar]
- 35.An HJ, Gip P, Kim J, Wu S, Park KW, McVaugh CT, Schaffer DV, Bertozzi CR, Lebrilla CB. Extensive determination of glycan heterogeneity reveals an unusual abundance of high mannose glycans in enriched plasma membranes of human embryonic stem cells. Molecular & cellular proteomics: MCP. 2012;11:M111 010660. doi: 10.1074/mcp.M111.010660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasehira K, Tateno H, Onuma Y, Ito Y, Asashima M, Hirabayashi J. Structural and quantitative evidence for dynamic glycome shift on production of induced pluripotent stem cells. Mol Cell Proteomics. 2012;11:1913–1923. doi: 10.1074/mcp.M112.020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen HL, Li CF, Grigorian A, Tian W, Demetriou M. T cell receptor signaling co-regulates multiple Golgi genes to enhance N-glycan branching. J Biol Chem. 2009;284:32454–32461. doi: 10.1074/jbc.M109.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 39.Chen CY, Jan YH, Juan YH, Yang CJ, Huang MS, Yu CJ, Yang PC, Hsiao M, Hsu TL, Wong CH. Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:630–635. doi: 10.1073/pnas.1220425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada M, Ishii T, Ikeda S, Naka-Mieno M, Tanaka N, Arai T, Kumasaka T, Gemma A, Kida K, Muramatsu M, Sawabe M. Association of fucosyltransferase 8 (FUT8) polymorphism Thr267Lys with pulmonary emphysema. Journal of human genetics. 2011;56:857–860. doi: 10.1038/jhg.2011.118. [DOI] [PubMed] [Google Scholar]
- 41.Lin H, Wang D, Wu T, Dong C, Shen N, Sun Y, Xie H, Wang N, Shan L. Blocking core fucosylation of TGF-beta1 receptors downregulates their functions and attenuates the epithelial-mesenchymal transition of renal tubular cells. American journal of physiology Renal physiology. 2011;300:F1017–1025. doi: 10.1152/ajprenal.00426.2010. [DOI] [PubMed] [Google Scholar]
- 42.Schjoldager KT, Clausen H. Site-specific protein O-glycosylation modulates proprotein processing - deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochimica et biophysica acta. 2012;1820:2079–2094. doi: 10.1016/j.bbagen.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 43.Muhlenhoff M, Rollenhagen M, Werneburg S, Gerardy-Schahn R, Hildebrandt H. Polysialic acid: versatile modification of NCAM, SynCAM 1 and neuropilin-2. Neurochemical research. 2013;38:1134–1143. doi: 10.1007/s11064-013-0979-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.