Abstract
Clonality analysis of viral vector-transduced cell populations represents a convincing approach to dissect the physiology of tissue and organ regeneration, to monitor the fate of individual gene-corrected cells in vivo, and to assess vector biosafety. With the decoding of mammalian genomes and the introduction of next-generation sequencing technologies, the demand for automated bioinformatic analysis tools that can rapidly process and annotate vector integration sites is rising. Here, we provide a publicly accessible, graphical user interface-guided automated bioinformatic high-throughput integration site analysis pipeline. Its performance and key features are illustrated on pyrosequenced linear amplification-mediated PCR products derived from one patient previously enrolled in the first lentiviral vector clinical gene therapy study. Analysis includes trimming of vector genome junctions, alignment of genomic sequence fragments to the host genome for the identification of integration sites, and the annotation of nearby genomic elements. Most importantly, clinically relevant features comprise the determination of identical integration sites with respect to different time points or cell lineages, as well as the retrieval of the most prominent cell clones and common integration sites. The resulting output is summarized in tables within a convenient spreadsheet and can be further processed by researchers without profound bioinformatic knowledge.
Arens and colleagues provide an automated, high-throughput, publicly accessible, graphical user interface-guided bioinformatic tool for analysis of viral vector integration sites. Analyses include trimming of vector genome junctions, alignment of genomic sequence fragments to the host genome for the identification of clinically relevant integration sites, and the annotation of nearby genomic elements.
Introduction
The ability of integrating viral vectors to become a persistent part of the host genome makes them valuable tools in clinical gene therapy, as these gene transfer agents can provide long-term correction of diseases. Profound advances in ex vivo retroviral gene therapy resulted in the correction of a variety of inherited disorders, including adenosine deaminase (ADA) deficiency (Aiuti et al., 2002, 2009), X-linked severe combined immunodeficiency (X-SCID) (Gaspar et al., 2004; Hacein-Bey-Abina et al., 2002, 2010), chronic granulomatous disease (CGD) (Ott et al., 2006; Bianchi et al., 2009), Wiskott-Aldrich syndrome (WAS) (Boztug et al., 2010), and β-thalassemia (Cavazzana-Calvo et al., 2010). In the first lentiviral vector-based clinical trial for adrenoleukodystrophy (ALD), the therapeutic aim to stop the progression of the disease could be reached (Cartier et al., 2009).
Despite the benefits of gene therapy and the increased interest of pharmaceutical partners for the development of new treatments (Ylä-Herttuala, 2011), the integration of therapeutic vectors into patients' genomes still bears risks. In some of the patients enrolled in gammaretroviral gene therapy trials, regulatory elements of the integrated vector led to overexpression of proto-oncogenes, causing clonal dominance and even malignant transformation of the affected gene-corrected cells (Hacein-Bey-Abina et al., 2003, 2008; Ott et al., 2006; Howe et al., 2008; Cavazzana-Calvo et al., 2010; Stein et al., 2010; Grez et al., 2011; Paruzynski et al., 2011).
As vector locations in the host genome can be used as molecular markers to monitor the fate of affected cells, analyses of vector integration sites (ISs) allow long-term follow-up of the gene-corrected cell pool and the dissection of individual clonal contributions. In addition, such clonality analyses may uncover potential progression and dominance of individual cell clones in patients—one of the major safety issues in gene therapy studies. The availability of the complete human genome sequence and the implementation of next-generation sequencing (NGS) technologies have dramatically influenced the possibilities for in-depth analyses of the clonal repertoire in preclinical and clinical studies. Yielding more than 1 million raw sequence reads in a single pyrosequencing run (genome sequencer; 454 Life Sciences/Roche, Branford, CT), the analyses of IS data require automated bioinformatic programs.
The identification of ISs followed by NGS can be divided into several steps (Paruzynski et al., 2010): (1) host genomic DNA flanking the integrated vector is accessed, commonly by PCR-based methods; (2) for sequencing, molecular barcodes and pyrosequencing-specific oligonucleotides are incorporated into the PCR amplicons to allow simultaneous sequencing of numerous samples; (3) the resulting raw sequence reads are bioinformatically processed; followed by (4) downstream analysis by scientists without profound bioinformatic knowledge. We, and others, focused on determining the integration profile of both low and frequent integrating vector systems used in, or suitable for, gene therapy in various mammalian host genomes (Gabriel et al., 2009, 2011; Bartholomae et al., 2011). These efforts uncovered preferences of distinct gene transfer vectors to target gene coding regions, CpG islands, or transcriptional start sites (Schroder et al., 2002; Wu et al., 2003; Mitchell et al., 2004).
Here, we describe our high-throughput insertion site analysis pipeline (HISAP, available at http://hisap.nct-heidelberg.de/HISAP), its underlying components, and key features, and the broad range of its applicability. HISAP is designed for the assessment of oncoretroviral or lentiviral vector integrations in mouse, rat, or human genomes. Its key features are designated for clinical in vivo monitoring. We demonstrate IS analysis by HISAP on linear amplification-mediated (LAM)-PCR-amplified and pyrosequenced ALD patient samples from our lentiviral vector gene therapy study (Cartier et al., 2009). Moreover, HISAP is also suitable for the analysis of vector genome junctions derived by nonrestrictive LAM-PCR (Gabriel et al., 2009; Paruzynski et al., 2010), inverse PCR (Silver et al., 1989), and ligation-mediated PCR (Mueller and Wold, 1989).
Materials and Methods
HISAP
HISAP was developed with the Perl (Perl version 5.10.1) and Java (Java 6) programming languages. The graphical user interface (GUI) was created with the Google Web Toolkit (GWT, version 2.1). To generate the result spreadsheet the Perl modules Spreadsheet-WriteExcel (version 2.37) and Spreadsheet-ParseExcel (version 0.58) are used. Mapping of the trimmed sequence reads to identify ISs is accomplished with the BLAST-like alignment tool (BLAT; Kent, 2002). Genomic data underlying the HISAP analysis are downloaded from the University of California Santa Cruz (UCSC) Genome Bioinformatics resource. Annotations are obtained via the UCSC Table Browser (Karolchik et al., 2004) and stored locally to obtain stable annotations, whereas the chromosomal sequences are downloaded from the UCSC FTP server (Table 1). HISAP runs on any server environment with a minimal memory capacity of 6 gigabytes (GB). To create the figures illustrating the HISAP results, the tables were converted into charts with the Excel program (Microsoft, Redmond, WA). The Venn diagram (see Fig. 2b) was created with the R package gplots (venn {gplots}).
Table 1.
Genomic Sequences and Elements Used within HISAP
| Assembly | Sequenced genome (bp) | RefSeq genes | CpG islands | Repeats |
|---|---|---|---|---|
| Homo sapiens (hg19) | 2,861,327,131 | 33,309 | 27,719 | 5,232,242 |
| Mus musculus (mm9) | 2,558,509,480 | 26,738 | 15,991 | 4,881,442 |
| Rattus norvegicus (rn4) | 2,477,053,777 | 16,521 | 15,303 | 4,550,842 |
HISAP, high-throughput insertion site analysis pipeline.
Note: Sequenced genome, the number of nucleotides unambiguously encoded in the corresponding genome assembly.
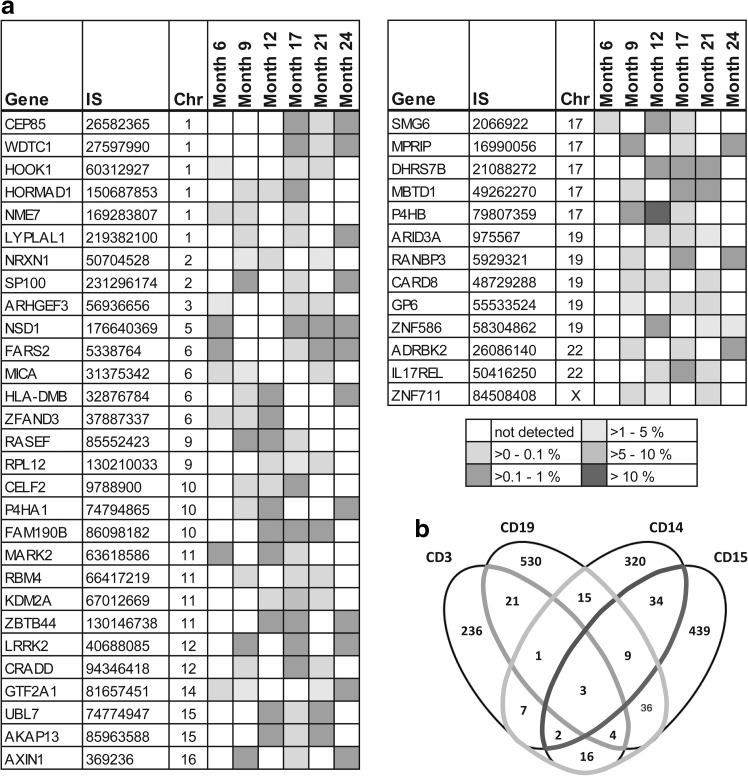
FIG. 2.
Identical integration sites (ISs) with respect to time and across lineage (a) IS clones are tracked with respect to time. The reoccurrence of ISs is monitored for various lengths of time. All ISs present at three or four time points are listed. (b) In the adrenoleukodystrophy (ALD) study, blood cells were sorted for the surface markers CD3 and CD19 (lymphocytes) as well as CD14 and CD15 (myeloids) before IS determination. Therefore, ISs across various blood cell fractions could also be compared. The light grey line encloses the number of ISs found in both lineages; the dark grey line encloses those that are shared between monocytes (CD14) and granulocytes (CD15); the middle grey line encloses identical ISs within T lymphocytes (CD3) and B lymphocytes (CD19).
LAM-PCR, pyrosequencing, and HISAP analysis of ALD patient 1
To isolate the vector host genome junctions from cellular DNA we performed LAM-PCR as previously described (Schmidt et al., 2007). Analysis was performed on cell fractions from peripheral blood (CD14, CD15, CD3, and CD19) and bone marrow (CD34). Samples up to 24 months after gene therapeutic treatment were analyzed by 3′ LAM-PCR on 1–1000 ng of DNA, using the restriction enzyme Tsp509I, NlaIII, or HpyCHIV as previously described (Cartier et al., 2009). After a linear PCR step, biotinylated PCR products were enriched via magnetic beads, followed by second-strand synthesis and restriction digest using ligation of a linker cassette and two additional exponential amplifications. LAM-PCR amplicons were purified. For multiplexed sequencing LAM-PCR amplicons were tagged, using fusion-primers with incorporated barcodes (Paruzynski et al., 2010). Fusion-primer design and sequencing were done according to the manufacturer's protocols on the 454/Roche genome sequencer (Margulies et al., 2005) (GS20, FLX, and titanium chemistry). In total, 125,359 raw LAM-PCR amplicons were obtained after sequencing and the amplified ISs were located in the human genome by HISAP within 15 hr afterward. The ISs of ALD patient 1 were annotated with molecular biological data obtained on May 16, 2011.
Results
Structure of HISAP
A prerequisite for HISAP was the capability to identify fusion sequences that are composed of a known sequence fragment (e.g., long terminal repeats [LTRs] of a retroviral vector) and an unknown sequence fragment such as a part of the host genome. In addition, the host genomic fragment may be additionally flanked at the 3′ end by another known sequence fragment (i.e., a linker cassette; Supplementary Fig. S1 [supplementary data are available online at http://www.liebertpub.com/hgtb]). In the following, we refer to LAM-PCR analysis and pyrosequencing of lentiviral vector ISs in clinical gene therapy.
To enable the simultaneous sequencing of hundreds of samples in a single sequencing run, short DNA barcodes are incorporated into the LAM-PCR amplicons together with the Roche/454 pyrosequencing-specific amplification and sequencing nucleotides (fusion-primers) (Paruzynski et al., 2010). When sequencing is completed, raw sequences are sorted according to their barcodes (e.g., by the command line tools sfffile and sfffinfo provided by Roche/454) and project affiliation. Each independent project is then further processed bioinformatically by HISAP.
HISAP determines the precise genomic integration locus and reports relevant information about the surrounding genomic DNA (see below) as well as general vector characteristics and across-sample comparisons (Fig. 1). By providing a web-based GUI on top of the underlying HISAP core, submissions are easy and straightforward (Supplementary Fig. S2).
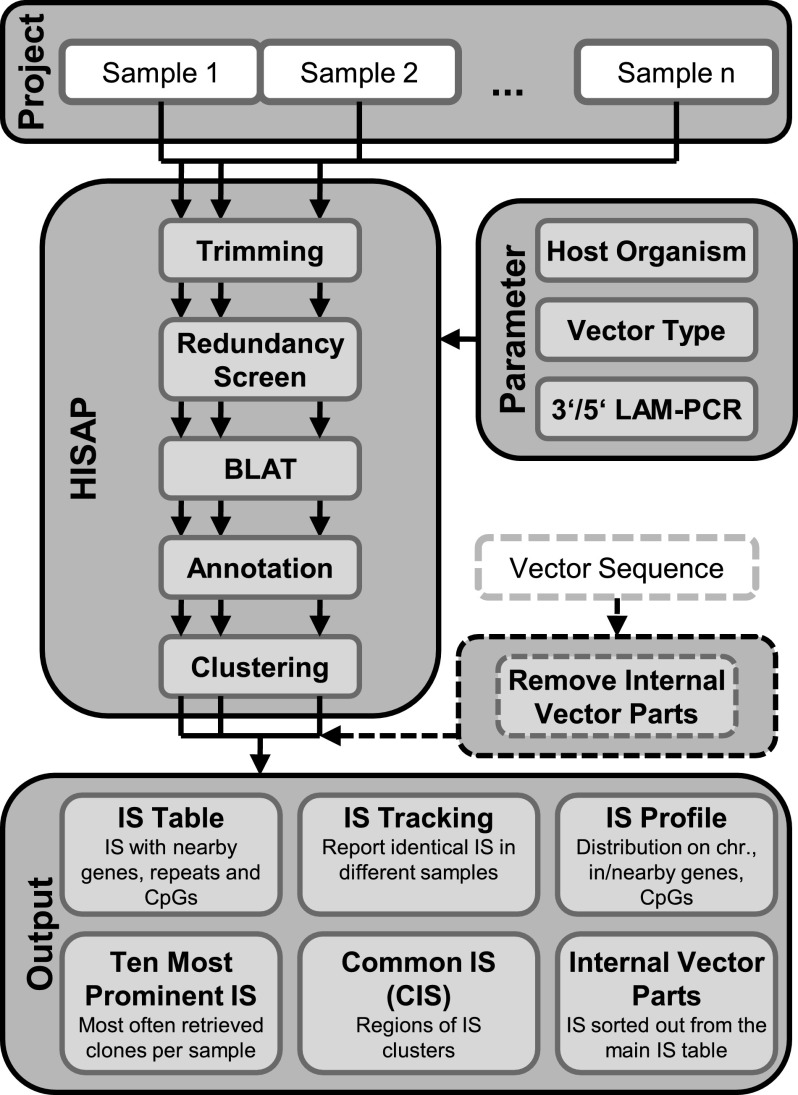
FIG. 1.
High-throughput integration site analysis pipeline (HISAP) organization and structure. HISAP is capable of analyzing whole integration site (IS) projects within one submission. The project-specific parameters can be provided via a graphical user interface and the analysis will be performed accordingly. Initially, the ISs from all individual samples are determined and across-sample comparison is performed in a second process. BLAT, BLAST-like alignment tool; LAM-PCR, linear amplification-mediated PCR.
Sample submission and processing
The HISAP GUI guides the user through the submission process. For an HISAP submission, only user and project information as well as the multi-FASTA formatted sequence files for the various, barcode-sorted samples of the project need to be provided. Certain project parameters are the vector type used for cell transduction, the remaining terminal vector sequence, and the application of 3′ or 5′ LTR primers used for LAM-PCR (Table 2). The GUI can easily be extended with primers that differ from those cited in Table 2.
Table 2.
Target Sequences Used for Trimming within HISAP
| Parameter | 3′/5′ LAM-PCR | Vector-specific fusion-primer sequence | Terminal vector sequence |
|---|---|---|---|
| Lenti | 3′ | TGTGTGACTCTGGTAACTAG | AGATCCCTCAGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCA |
| Lenti | 5′ | AAGCAGATCTTGTCTTCG | TTGGGAGTGAATTAGCCCTTCCA |
| nrLenti | 3′ | GATCCCTCAGACCCTTTTAGTC | AGTGTGGAAAATCTCTAGCA |
| MLV | 3′ | GTCTCCTCTGAGTGATTGAC | TACCCGTCAGCGGGGGTCTTTCA |
| MLV | 5′ | CCTTGCAAAATGGCGTTACT | TAAGCTAGCTTGCCAAACCTACAGGTGGGGTCTTTCA |
| SFFV | 3′ | GTCTCCTCTGAGTGATTGAC | TGCCCACCTCGGGGGTCTTTCA |
| SFFV | 5′ | CCTTGCAAAATGGCGTTACT | GCAGCTAGCTTGCCAAACCTACAGGTGGGGTCTTTCA |
HISAP, high-throughput insertion site analysis pipeline; Lenti, lentivirus; nrLenti, Lenti and nrLenti, lentivirus; MLV, murine leukemia virus; SFFV, spleen focus-forming virus.
For each sample one multi-FASTA file, which contains all obtained sequences, needs to be provided. In the first step, all sequences are trimmed. During trimming, vector- and linker-specific nucleotides are removed from raw sequences in order to retain solely the host genomic fragment of the sequenced LAM-PCR amplicons. The resulting trimmed sequences allow for the determination of the exact location of ISs within the host genome.
Unspecific sequences, for example, primer multimers or nonmappable sequences shorter than 20 bp after trimming, are removed from downstream analysis. Identical, and therefore redundant, sequences are combined and their frequency is recorded. The trimmed sequences are then mapped to the corresponding host genome via BLAT (Kent, 2002). Only sequences with a sequence identity greater than 95% to the reference genome are further processed by adding nearby RefSeq gene (Pruitt et al., 2009), Repeat Masker (F.A. Arian, R. Hubly, unpublished data), and CpG island annotations. Notably, sequencing errors at the precise vector integration locus (e.g., insertion or deletion of single nucleotides) are addressed by combining all ISs that can be found in a 3-bp window and sequences that have an overall sequence identity greater than 90%. These ISs are summarized in a single table entry as previously described (Paruzynski et al., 2010).
The analysis of 3′ and 5′ LTR LAM-PCR amplicons differs slightly. Because sequencing proceeds in the 5′-to-3′ direction, the vector genome junction is always located at the 5′ end of the raw sequence reads. Therefore, the reverse complementary sequence of 5′-LTR LAM-PCR amplicons is analyzed via HISAP.
To eliminate false positive ISs, it is recommended that all retrieved IS sequences are compared with the respective vector sequence. This task can be performed by HISAP implicitly by uploading the vector sequence together with the sample sequences. Sequences that map to the vector genome are removed from further downstream analysis and are provided in a separate spreadsheet.
On completion of data processing by HISAP, the user receives a download link for the results via e-mail. The resulting Excel spreadsheet summarizes the sample name (provided by the name of the respective FASTA file), the trimmed sequences, the number of deleted bases at the vector end (e.g., the LTR), and the resulting genomic coordinates of the IS (chromosome, position, and orientation) together with the quality of the sequence alignment (sequence identity and alignment length with respect to the total sequence length) and the number of sequence reads for each individual IS. Furthermore, details on the annotations and distances to nearby genomic elements such as the nearest RefSeq genes, CpG islands, and repetitive elements are provided.
First, the ISs derived from one sample are analyzed individually and are subsequently combined with the results obtained from other samples of interest within the same submission (see below). These meta-analyses are provided in separate sheets within the result spreadsheet. The latter has a profound impact for the in vivo monitoring of gene-corrected hematopoietic cells. On the one hand, the detection of identical ISs over a time course and across different cell lineages provides information about the self-renewal potency and multilineage capacity of the initially transduced cells. On the other hand, the abundance of individual ISs and the clustering of ISs within a certain genomic area may help to determine potential side effects, including clonal outgrowth. As such, HISAP employs an automated process to define (1) the presence of identical ISs over time and across lineages, (2) the 10 most prominent ISs for each analyzed sample, based on sequence counts, and (3) the clustering of ISs termed common integration sites (CISs).
Identical ISs over time and across lineages
The complete IS data sets of the various samples within a project are screened for identical ISs. The results are displayed as a matrix. On the vertical axis, each individual IS identified within the entire project is listed. On the horizontal axis, the relative frequency of each individual IS for every sample is reported. As an example, we performed this analysis on sorted hematopoietic cells derived at various time points after treatment (6, 9, 12, 17, 21, and 24 months) of a patient involved in the ALD gene therapy study (Cartier et al., 2009). Analyzing the cell fractions up to 24 months after treatment revealed 1737 uniquely mappable ISs, of which 197 could be detected at multiple time points. The occurrence of identical ISs at different time points (Supplementary Table S1) indicates the long-term activity of transduced cell lines, particularly in the case of short-lived CD15 granulocytes. Figure 2 displays the comparison of ISs derived at various time points posttransplantation (Fig. 2a) and the ISs found in various blood cell fractions (Fig. 2b). After combining all fractioned samples from the different time points, we retrieved 93 identical ISs derived from lymphoid as well as myeloid cell fractions, suggesting the successful ex vivo transduction of early progenitor or stem cells.
The 10 most prominent clones
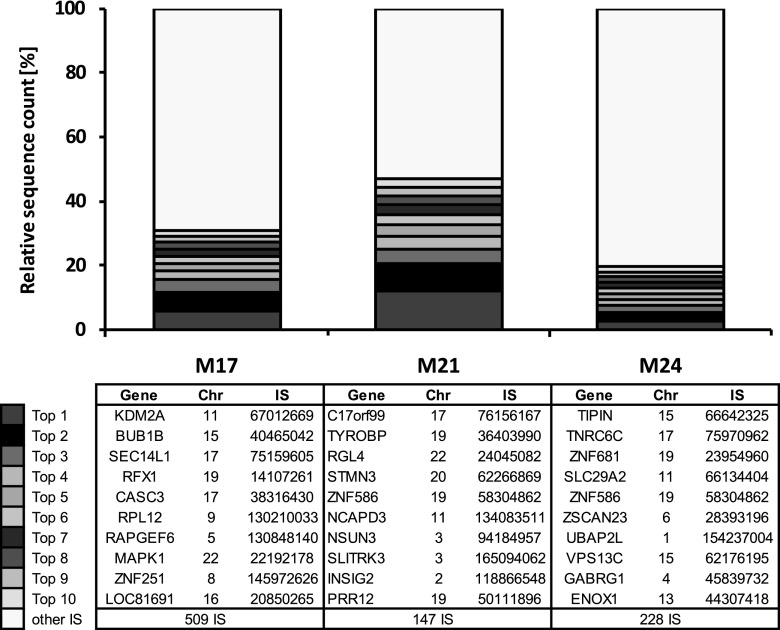
For each sample, a list of the 10 most prominent clones according to their sequence count is provided. This table is ordered with respect to the abundance of an individual IS within one analyzed sample. When performing this analysis on polyclonal ALD patient samples (as an estimate, we propose to use polyclonal samples consisting of at least 50 unique IS sequences), we observed a highly variable list of the potentially 10 most prominent clones. None of the 10 most prominent clones could be detected at more than one time point, showing the polyclonal hematopoietic repopulation in this patient (Fig. 3).
FIG. 3.
10 most prominent clones of the last 3 time points. The 10 most prominent (frequent) clones of polyclonal samples are listed according to the number of supporting sequences for the corresponding IS. In contrast to some gammaretroviral vector studies, no dominant clones could be detected in the ALD study. A table for the 10 most prominent clones at each time point is displayed below the chart. The most abundant clone is listed on top. Chr, chromosome; IS, integration site.
Common integration sites
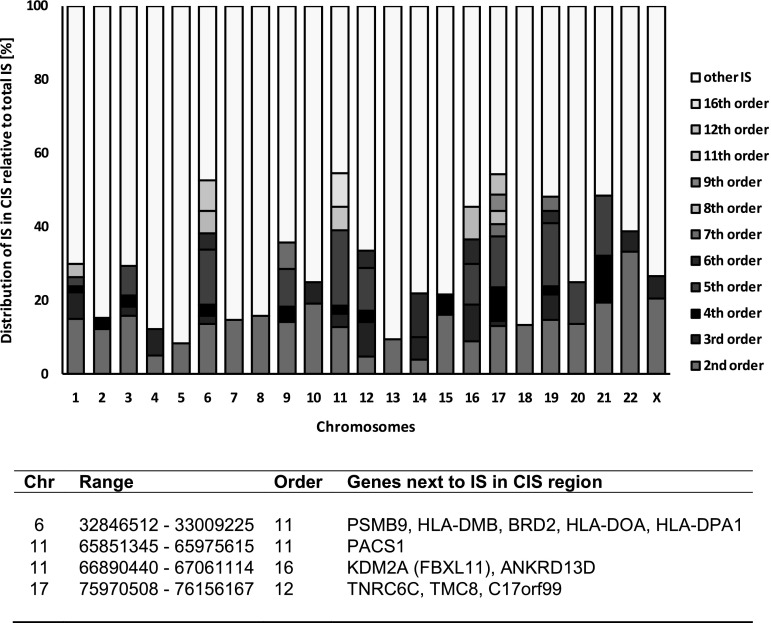
Another characteristic of the different vector types is their tendency to cluster in specific genomic areas. Such IS clusters are termed CISs and have been successfully used to identify oncogenes in tumor prone mouse models or to uncover clonal skewing in clinical gene therapy. CISs of the second, third, or fourth order are defined as 2, 3, or 4 ISs within a window of 30, 50, and 100 kb, respectively. CISs of the fifth and higher orders are defined as 5 or more ISs in a window of 200 kb (Suzuki et al., 2002; Abel et al., 2007, 2011). A table summarizing all CISs found within a study, as well as their respective order, is provided by HISAP. For patient 1 of the ALD trial, CISs of highest orders (>10th order) were found on chromosomes 6, 11, and 17 and coincide with genes such as KDM2A (alias FBXL11), PACS1, and the HLA locus (Fig. 4) (Cartier et al., 2009). As shown, these results were in line with the preferred lentivirus-specific IS distribution in the megabase range (Biffi et al., 2011).
FIG. 4.
CIS distribution on different chromosomes. The number of IS found within CIS as well as the respective CIS orders vary between different chromosomes (Biffi et al., 2011). For patient 1 from the ALD study, CIS of highest orders could be found on chromosomes 6, 11 and 17. All CIS of order 11 and higher are listed in the provided table. The same chromosomes also contain the highest portion of IS that comprise CIS. No CIS could be detected on the Y chromosome. Chr, chromosome.
Discussion
In clinical gene therapy, in vivo monitoring of clonal dynamics provides insight into the physiology of hematopoietic repopulation and is decisive in identifying potential clonal skewing and outgrowth of individual gene-corrected cell clones in patients. Following the occurrence of severe side effects in clinical studies, the demand for characterizing the clonal inventory in treated patients increased enormously. Highly sensitive IS analyses coupled with NGS devices produce high amounts of raw sequence data such that a sophisticated analysis of the retrieved integrome is not feasible without substantial bioinformatic support. Our publicly available HISAP tool is developed for LAM-PCR in combination with pyrosequencing and provides a complete straight forward application for the analysis of vector IS in preclinical and clinical gene therapy studies. In addition, HISAP is capable of analyzing any raw sequence reads that are composed of an “unknown–known” nucleotide sequence as are generated by other IS amplification and sequencing protocols.
Sample submission is simplified by the web-based HISAP GUI. After processing of the raw sequences, HISAP lists the precise vector locations in the host genome together with information on nearby genetic elements. Chromosomal distribution and other integration preferences of the viral vector are given in a tabular overview. A hallmark of HISAP is the incorporation of potentially biological and clinical relevant key features. These comprise an across-sample analysis tool that includes the tracking and progression of individual clones with respect to time as well as their presence in different cell fractions or tissues. Although tracking with respect to time easily reveals persistent clones, the comparison of blood cells, sorted according to their specific cell surface markers, can provide an insight into the hierarchical organization of the human hematopoietic system (Schmidt et al., 2005; Cavazzana-Calvo et al., 2011). This feature can also be used for the molecular follow-up of a gene-corrected cell pool in transplantation experiments (Lemischka and Jordan, 2001; Glimm et al., 2005). Cross sample contaminations (Noonan et al., 2006; Cartier et al., 2009) which can be introduced by the sequencing technologies used, can also be identified with this downstream analysis tool and eliminated from the analysis. Another tool incorporates an overview of the 10 most abundant IS for each sample. By comparing the 10 most prominent clones from different samples over a time course, this tool allows for the detection of shifts from polyclonal to oligoclonal or even monoclonal IS patterns and the fast detection of persistent and increasing dominant clones within a patient. To complete the set of downstream analysis tools, the incorporated CIS feature highlights regions in the host genome which reveal any clustering of IS. CIS have been reported in numerous retroviral gene therapy studies and may be applicable to serve as an indicator for in vivo clonal skewing (e.g., MDS1-EVI1 in the CGD trial) (Ott et al., 2006; Stein et al., 2010). Nevertheless, HISAP does not aim to give statistical evaluations of these CIS regions. For this purpose a number of HISAP independent programs exist, that focus on answering the statistical significance of CIS events (de Ridder et al., 2006; Abel et al., 2007, 2011).
The duration of an HISAP analysis is highly dependent on the nature and composition of different IS projects and their corresponding samples. Crucial factors of HISAP analysis duration, besides the number of raw sequences obtained after sequencing, are the polyclonality of samples as well as the number of IS located in repeat regions.
In addition to HISAP, other integration site analysis pipelines capable analyzing viral integration sites obtained via Roche/454 high-throughput sequencing exist. QuickMap (Appelt et al., 2009) and SeqMap 2.0 (Hawkins et al., 2011) are also publicly available. Although the general strategy of IS detection is similar for all pipelines, they differ in the individual steps performed during analyses, parameter optimization, use of underlying bioinformatic tools, and biological annotation resources. Beyond the fast annotation of IS, HISAP uniquely adds value to the gene therapy field by providing a project-based analysis approach including the incorporation of features that allow straight forward downstream analyses. HISAP has been applied to preclinical biosafety studies such as the investigation of differences in lentiviral vector profiles in actively dividing and postmitotic cells (Bartholomae et al., 2011), the integration characteristics of integrase deficient lentiviral vectors (Matrai et al., 2011), tracking of clones in serial transplantation experiments (Dieter et al., 2011), and the detection of zinc finger nuclease off-target activity (Gabriel et al., 2011). Clinical trials that used HISAP for IS identification and corresponding annotation have recently been published and include retroviral treatment of CGD (Kang et al., 2011) and WAS (Boztug et al., 2010) as well as lentiviral vector treatment of ALD (Cartier et al., 2009). Moreover, HISAP uniquely provides significant and novel downstream analysis aspects that go beyond the mere identification of IS and can be applied to a broad range of nonviral gene therapy and even non-gene therapy applications such as mutant screening (Bessereau, 2006), transgenetics (Newman and Lardelli, 2010), cancer research (Copeland and Jenkins, 2010), and virology (e.g., HTLV-1 [Gillet et al., 2011] and HIV).
Supplementary Material
Acknowledgments
The authors acknowledge their colleagues of the Translational Oncology Research Group at the German Cancer Research Center and National Center for Tumor Diseases (Heidelberg, Germany) involved in integration site analysis for valuable feedback and beta testing of HISAP. The authors also thank their cooperation partners from GATC Biotech (Konstanz, Germany) for fruitful discussions. This work was supported by the Deutsche Forschungsgemeinschaft (grant SPP1230), the Bundesministerium für Bildung und Forschung (iGene), and European Union VIth and VIIth Framework Program CONSERT and PERSIST.
Author Disclosure Statement
No competing financial interests exist.
References
- Abel U. Deichmann A. Bartholomae C., et al. Real-time definition of non-randomness in the distribution of genomic events. PLoS One. 2007;2:e570. doi: 10.1371/journal.pone.0000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel U. Deichmann A. Nowrouzi A., et al. Analyzing the number of common integration sites of viral vectors—new methods and computer programs. PLoS One. 2011;6:e24247. doi: 10.1371/journal.pone.0024247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiuti A. Slavin S. Aker M., et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- Aiuti A. Cattaneo F. Galimberti S., et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N. Engl. J. Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- Appelt J.U. Giordano F.A. Ecker M., et al. QuickMap: A public tool for large-scale gene therapy vector insertion site mapping and analysis. Gene Ther. 2009;16:885–893. doi: 10.1038/gt.2009.37. [DOI] [PubMed] [Google Scholar]
- Bartholomae C.C. Arens A. Balaggan K.S., et al. Lentiviral vector integration profiles differ in rodent postmitotic tissues. Mol. Ther. 2011;19:703–710. doi: 10.1038/mt.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessereau J.-L. Transposons in C. elegans. WormBook. The C. elegans. 2006. http://www.wormbook.org/chapters/www_transposons/transposons.html. http://www.wormbook.org/chapters/www_transposons/transposons.html Research Community. [DOI] [PMC free article] [PubMed]
- Bianchi M. Hakkim A. Brinkmann V., et al. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood. 2009;114:2619–2622. doi: 10.1182/blood-2009-05-221606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A. Bartolomae C.C. Cesana D., et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117:5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- Boztug K. Schmidt M. Schwarzer A, et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N. Engl. J. Med. 2010;363:1918–1927. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N. Hacein-Bey-Abina S. Bartholomae C.C., et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M. Payen E. Negre O., et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo M. Fischer A. Bushman F.D., et al. Is normal hematopoiesis maintained solely by long-term multipotent stem cells? Blood. 2011;117:4420–4424. doi: 10.1182/blood-2010-09-255679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland N.G. Jenkins N.A. Harnessing transposons for cancer gene discovery. Nat. Rev Cancer. 2010;10:696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- de Ridder J. Uren A. Kool J., et al. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput. Biol. 2006;2:e166. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieter S.M. Ball C.R. Hoffmann C.M., et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 2011;9:357–365. doi: 10.1016/j.stem.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Gabriel R. Eckenberg R. Paruzynski A., et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat. Med. 2009;15:1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- Gabriel R. Lombardo A. Arens A., et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- Gaspar H.B. Parsley K.L. Howe S., et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- Gillet N.A. Malani N. Melamed A., et al. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood. 2011;117:3113–3122. doi: 10.1182/blood-2010-10-312926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimm H. Schmidt M. Fischer M., et al. Efficient marking of human cells with rapid but transient repopulating activity in autografted recipients. Blood. 2005;106:893–898. doi: 10.1182/blood-2004-07-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grez M. Reichenbach J. Schwable J., et al. Gene therapy of chronic granulomatous disease: The engraftment dilemma. Mol. Ther. 2011;19:28–35. doi: 10.1038/mt.2010.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Le Deist F. Carlier F., et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Von Kalle C. Schmidt M., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Garrigue A. Wang G.P., et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Hauer J. Lim A., et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2010;363:355–364. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins T.B. Dantzer J. Peters B., et al. Identifying viral integration sites using SeqMap 2.0. Bioinformatics. 2011;27:720–722. doi: 10.1093/bioinformatics/btq722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe S.J. Mansour M.R. Schwarzwaelder K., et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.J. Bartholomae C.C. Paruzynski A., et al. Retroviral gene therapy for X-linked chronic granulomatous disease: Results from phase I/II trial. Mol. Ther. 2011;19:2092–2101. doi: 10.1038/mt.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D. Hinrichs A.S. Furey T.S., et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemischka I.R. Jordan C.T. The return of clonal marking sheds new light on human hematopoietic stem cells. Nat. Immunol. 2001;2:11–12. doi: 10.1038/83115. [DOI] [PubMed] [Google Scholar]
- Margulies M. Egholm M. Altman W.E., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrai J. Cantore A. Bartholomae C.C., et al. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. 2011;53:1696–1707. doi: 10.1002/hep.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R.S. Beitzel B.F. Schroder A.R., et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P.R. Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Newman M. Lardelli M. A hyperactive Sleeping Beauty transposase enhances transgenesis in zebrafish embryos. BMC Res. Notes. 2010;3:282. doi: 10.1186/1756-0500-3-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan J.P. Coop G. Kudaravalli S., et al. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M.G. Schmidt M. Schwarzwaelder K., et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Paruzynski A. Arens A. Gabriel R., et al. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat. Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- Paruzynski A. Boztug K. Ball C., et al. Molecular follow-up of the German WAS clinical gene therapy trial [abstract 343] Mol. Ther. 2011;19:S133. [Google Scholar]
- Pruitt K.D. Tatusova T. Klimke W., et al. NCBI Reference Sequences: Current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. Hacein-Bey-Abina S. Wissler M., et al. Clonal evidence for the transduction of CD34+ cells with lymphomyeloid differentiation potential and self-renewal capacity in the SCID-X1 gene therapy trial. Blood. 2005;105:2699–2706. doi: 10.1182/blood-2004-07-2648. [DOI] [PubMed] [Google Scholar]
- Schmidt M. Schwarzwaelder K. Bartholomae C., et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- Schroder A.R. Shinn P. Chen H., et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Silver J. Keerikatte V. Novel use of polymerase chain reaction to amplify cellular DNA adjacent to an integrated provirus. J. Virol. 1989;63:1924–1928. doi: 10.1128/jvi.63.5.1924-1928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S. Ott M.G. Schultze-Strasser S., et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- Suzuki T. Shen H. Akagi K., et al. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- Wu X. Li Y. Crise B., et al. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Ylä-Herttuala S. Gene therapy moves forward in 2010. Mol. Ther. 2011;19:219–220. doi: 10.1038/mt.2010.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.