Abstract
Mycobacterium bovis is the causative agent of bovine tuberculosis (TB), a disease that affects approximately 5% of Argentinean cattle. Among the molecular methods for genotyping, the most convenient are spoligotyping and variable number of tandem repeats (VNTR). A total of 378 samples from bovines with visible lesions consistent with TB were collected at slaughterhouses in three provinces, yielding 265 M. bovis spoligotyped isolates, which were distributed into 35 spoligotypes. In addition, 197 isolates were also typed by the VNTR method and 54 combined VNTR types were detected. There were 24 clusters and 27 orphan types. When both typing methods were combined, 98 spoligotypes and VNTR types were observed with 27 clusters and 71 orphan types. By performing a meta-analysis with previous spoligotyping results, we identified regional and temporal trends in the population structure of M. bovis. For SB0140, the most predominant spoligotype in Argentina, the prevalence percentage remained high during different periods, varying from 25.5-57.8% (1994-2011). By contrast, the second and third most prevalent spoligotypes exhibited important fluctuations. This study shows that there has been an expansion in ancestral lineages as demonstrated by spoligotyping. However, exact tandem repeat typing suggests dynamic changes in the clonal population of this microorganism.
Keywords: bovine TB, Mycobacterium bovis, spoligotype, VNTR
Bovine tuberculosis (BTB) is an infectious disease primarily caused by Mycobacterium bovis, which affects a wide range of mammals, including humans (Amanfu 2006). M. bovis is a member of the Mycobacterium tuberculosis complex (MTBC), which also includes M. tuberculosis, Mycobacterium africanum, Mycobacterium canetti, Mycobacterium microti, Mycobacterium caprae and Mycobacterium pinnipedii (Cataldi & Romano 2007) and the recently described species Mycobacterium mungi (Alexander et al. 2010) and Mycobacterium orygis (van Ingen et al. 2012).
In Argentina, the rate of condemnation of carcasses exhibiting lesions consistent with BTB dropped from 6.7% to 0.7% between 1969-2011 (de Kantor et al. 2012). The last national screening for purified protein derivative (PPD) reactors was performed in 1972 (Nader & Husberg 1988). Therefore, the current prevalence of PPD reactors is unknown, but it may be higher than 0.7% and even higher in dairy cattle. The control strategies are based on the caudal fold skin test, which is used primarily in dairy herds and slaughter surveillance. M. bovis has been isolated from tuberculin-positive bovines after a post mortem inspection of reactors in the slaughterhouse.
Although there are no reports of a phenotypic method to distinguish between different M. bovis isolates, genotyping methods have achieved some distinction. Among the molecular methods available for genotyping, the most convenient are spoligotyping (a reverse line probe assay) and genotyping by variable number of tandem repeats (VNTR).
At present, the methods for genotyping M. bovis and other MTBC species include restriction fragment length polymorphism (Otal et al. 1991, Collins et al. 1993) and VNTR (Haddad et al. 2001). However, until now, spoligotyping has been the best option for large-scale screening studies on the distribution of M. bovis strains (Kamerbeek et al. 1997). The theories of molecular fingerprinting suggest that epidemiologically related isolates will have similar fingerprints, which are expected to be different from epidemiologically unrelated strains (Maslow et al. 1993). Desirable characteristics for a typing method are related to the stability of the strain and the diversity within the species (Skuce & Neill 2001). Although spoligotyping may fulfil these characteristics in some cases, other typing methods such as VNTR must be performed to discriminate among isolates, especially in settings where one spoligotype is largely predominant or has been used to investigate a recent transmission.
Previous studies have demonstrated a wide diversity of M. bovis spoligotypes for cattle populations in Latin American countries. For example, different M. bovis spoligotypes have been found in different regions of Argentina, Brazil, Uruguay and Mexico (Zumárraga et al. 1999, Milián-Suazo et al. 2002, Rodriguez et al. 2004). The distribution of circulating M. bovis spoligotypes in five Latin American countries has been analysed recently (Zumárraga et al. 2012).
The aim of this work was to assess the genetic diversity of M. bovis in Argentina and to envisage temporal and spatial trends in the clonal population of this pathogen in the context of active transmissions in cattle.
MATERIALS AND METHODS
Animals - The bovines used in this study were examined in slaughterhouses in three provinces of Argentina, namely Buenos Aires, Córdoba and Santa Fe. The animals were classified according to age and weight as follows: calves 10% (< 12 months, < 220 kg), young steers 5% (12-18 m < 350 kg), steers 14% (> 18 m, > 350 kg), heifers 8.5% (12-30 m, 260 kg), cows 60.5% (> 30 m, > 350 kg) and bulls 0.5% (> 24 m).
Bacterial cultures - Tissue samples collected from 378 bovines with lesions classified as tuberculous were detected during regular post mortem inspections in national abattoirs.
A convenience, non-probabilistic sampling was performed at each slaughterhouse with the assistance of the local animal health service staff (National Health Service and Food Quality). For bacteriological analysis, the samples were placed in sterile receptacles and stored at -20ºC until processing. Petroff’s method was used to decontaminate the samples for mycobacterial isolates. Each sample was inoculated on Stonebrink and Löwenstein-Jensen media at 37ºC for 60 days. Each culture was performed in duplicate. During this period, the cultures were examined every seven days and when growth was visible, smears were prepared and stained with Ziehl-Neelsen to observe acid-fast bacilli. The bacteriological identification of the isolates was performed on the basis of the culture media, incubation temperature, growing time, colony morphology and Ziehl-Neelsen staining.
Isolates - A total of 261 M. bovis isolates were included in this study. The isolates originated from bovines at slaughterhouses in Buenos Aires (n = 67), Córdoba (n = 112) and Santa Fe (n = 82). These isolates were obtained between 2008-2011. These three Argentinean provinces are inhabited by 60% of the cattle in the country.
Spoligotyping assays and analysis - Spoligotyping was performed according to Kamerbeek et al. (1997). Bovine tissue samples were decontaminated by using Petroff’s method and cultured in Löwenstein-Jensen and Stonebrink media at 37ºC for 60 days. A loopful of colonies was transferred into a microcentrifuge tube containing 250 µL of distilled water and heated at 96ºC for 45 min. The samples were then centrifuged at 12,000 rpm for 10 min and 5 µL of the supernatant was used for polymerase chain reaction (PCR) to amplify the DR region. Spoligotyping was performed with a spoligotyping kit (Ocimun Biosolutions, Hyderabad, India). M. tuberculosis H37Rv (ATCC 27294) and M. bovis Bacillus Calmette-Guerin (BCG) (ATCC 27289) were included as reference strains in each spoligotyping experiment. The spoligotypes were collected in binary format in an Excel database and scanned images of the films were analysed by using BioNumerics (v.3.5, Applied Maths, Sint-Martens-Latem, Belgium). Spoligotypes were compared with the M. bovis spoligotypes contained in the Biotechnology Institute of National Institute of Agricultural Tecnology database, Argentina, and in the mbovis.org database from the Veterinary Laboratories Agency (VLA) (servers hosted at the University of Sussex in the United Kingdom). The spoligotypes were named according to VLA code (SB). We defined the following criteria for the purpose of this study: a unique spoligotype or VNTR type was observed only once within a province; an orphan spoligotype was observed only once during the study and an exclusive spoligotype was clustered or not clustered, in only one province.
VNTR typing - The analysis of the six VNTR loci originally identified by Frothingham and Meeker-O’Connell (Frothingham & Meeker-O’Connell 1998) was limited to the exact tandem repeat (ETR) of ETR-A to -D loci, as previously performed by Smith et al. (2003). The VNTR genotype of a strain, which represents the number of repeat elements at each locus, is presented as a series of four integers between 1-12 that are separated by hyphens. When the variant of an integer was identified, it was marked by an asterisk (*).
Multiplex PCRs were used to combine primer pairs ETR-A/B and ETR-C/D. The PCR mix was prepared in 96-well plates using the Hot Start Master Mix Kit (Qiagen, Germany). Five nanograms of DNA were added to a final volume of 20 µL containing 0.4 µM of each primer. For each multiplex mixture, one primer of each oligonucleotide pair was tagged with a different fluorescent dye (Table I). The thermocycler programs for both multiplex reactions were identical. The PCRs were performed with an initial denaturation of 15 min at 94ºC followed by 35 cycles of denaturation at 94ºC for 30 s, annealing at 68ºC for 1 min and extension at 72ºC for 2 min, followed by a final extension at 72ºC for 10 min.
TABLE I. Primers for exact tandem repeat (ETR)-A-D typing .
| Variable number of tandem repeats locus | Primer sequence (5-3) (labelling) |
|---|---|
| ETR-A | AAATCGGTCCCATCACCTTCTTA-(FAM) CGAAGCCTGGGGTGCCCGCGATTT |
| ETR-B | GCGAACACCAGGACAGCATCAT (JOE)-GGCATGCCGGTGATCGAGTGG |
| ETR-C | GTGAGTCGCTGCAGAACCTGCAG-(HEX) GGCGTCTTGACCTCCACGAGTG |
| ETR-D | CAGGTCACAACGAGAGGAAGAGC-(FAM) GCGGATCGGCCAGCGACTCCTC |
Statistical analysis - The discriminatory index (D) described by Hunter and Gaston and expressed by the Simpson formula was calculated to determine the discriminatory power of spoligotyping in each province (insilico.ehu.es) (Rodríguez et al. 2010). This index is given by the following equation (N: total number of strains in the sample population; s: total number of types described; xj: number of strains belonging to the jth type) (Hunter & Gaston 1988):
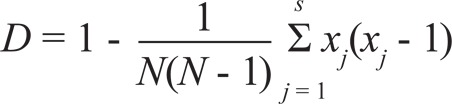
This index is based on the probability that two unrelated strains sampled from the test population will be placed into different typing groups (Hunter & Gaston 1988).
RESULTS
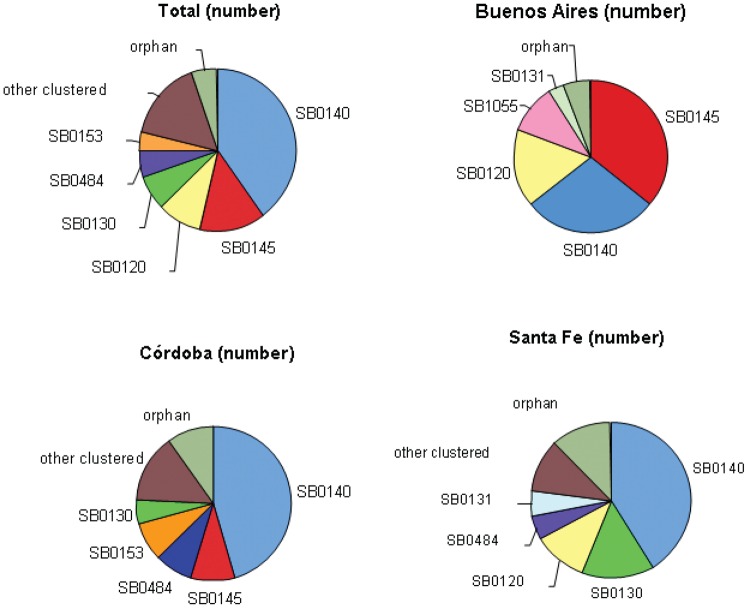
Geographical distribution of isolates and spoligotypes - Valid spoligotypes were obtained for 261 isolates across 32 patterns. Of these spoligotypes, 13 patterns were orphans and 248 (94.6%) isolates were grouped in 18 clusters of more than two isolates (Table II), with a discrimination index of 0.80. The spoligotype SB0140 represented 39.8% of the isolates. The isolates from Córdoba specifically came from slaughterhouses located in the Juarez Celman department. The numbers of spoligotypes detected in Buenos Aires, Córdoba and Santa Fe were nine, 21 and 19, respectively. A map showing the distribution of the four major spoligotypes is shown in Fig. 1A. These data can be compared to the cattle density per department (Fig. 1B) and it should be noted that in general samples were taken from departments with higher numbers of cattle. In Buenos Aires, SB0140 comprised 28.4% (n = 19) of the isolates, SB0145, 35.8% (n = 24), SB0120, 16.4% (n = 11) and SB1055, 10.4% (n = 7). In Córdoba, SB0140 comprised 45.5% of the isolates (n = 51), SB0145, 8.9% (n = 10) and SB0153 and SB0484, 8% (n = 9 each). In Santa Fe, which has a strong dairy cattle industry, SB0140 comprised 41.4% (n = 34), SB0130, 14.6% (n = 12) and SB0120, 10.9% (n = 9). Buenos Aires had the lowest number of unique spoligotypes (n = 4) followed by Córdoba (n = 11) and Santa Fe (10). Regarding orphan spoligotypes, Buenos Aires presented only one orphan spoligotype (xx138), Córdoba had seven (SB0996, 1043, 1068, 1776, xx126, xx131 and xx134) and Santa Fe had six (SB0678, 1066, 2117, 2165, 2166 and xx139). The number of major spoligotypes, the remaining clustered spoligotypes and the orphan types are shown in Fig. 2.
TABLE II. Spoligotypes distribution, number of patterns per province .
| Spoligotype (SB code) | Total of isolates | Buenos Aires | Córdoba | Santa Fe |
|---|---|---|---|---|
| SB0120 | 24 | 11 | 4 | 9 |
| SB0130 | 18 | - | 6 | 12 |
| SB0131 | 8 | 2 | 2 | 4 |
| SB0140 | 104 | 19 | 51 | 34 |
| SB0145 | 36 | 24 | 10 | 2 |
| SB0153 | 11 | - | 9 | 2 |
| SB0267 | 2 | - | 1 | 1 |
| SB0269 | 2 | - | 2 | - |
| SB0273 | 4 | - | 4 | - |
| SB0484 | 13 | - | 9 | 4 |
| SB0520 | 4 | - | 4 | - |
| SB0678 | 1 | - | - | 1 |
| SB0820 | 2 | 1 | 1 | - |
| SB0980 | 4 | - | 1 | 3 |
| SB0996 | 1 | - | 1 | - |
| SB1033 | 2 | 1 | - | 1 |
| SB1043 | 1 | - | 1 | - |
| SB1055 | 7 | 7 | - | - |
| SB1066 | 1 | - | - | 1 |
| SB1068 | 1 | - | 1 | - |
| SB1776 | 1 | - | 1 | - |
| SB1787 | 2 | 1 | - | 1 |
| SB1790 | 2 | - | - | 2 |
| SB1799 | 2 | - | 1 | 1 |
| SB2117 | 1 | - | - | 1 |
| SB2165 | 1 | - | - | 1 |
| SB2166 | 1 | - | - | 1 |
| SBxx126 | 1 | - | 1 | - |
| SBxx131 | 1 | - | 1 | - |
| SBxx134 | 1 | - | 1 | - |
| SBxx138 | 1 | 1 | - | - |
| SBxx139 | 1 | - | - | 1 |
|
| ||||
| Total | 261 | 67 | 112 | 82 |
Fig. 1A. : distribution per department of the four major spoligotypes in the three provinces (1 dot = 1 case). Department are the secondary administrative and political division of provinces in Argentina; B: number of cattle per department (source: rian.inta.gov.ar/ganaderia/stock2011).
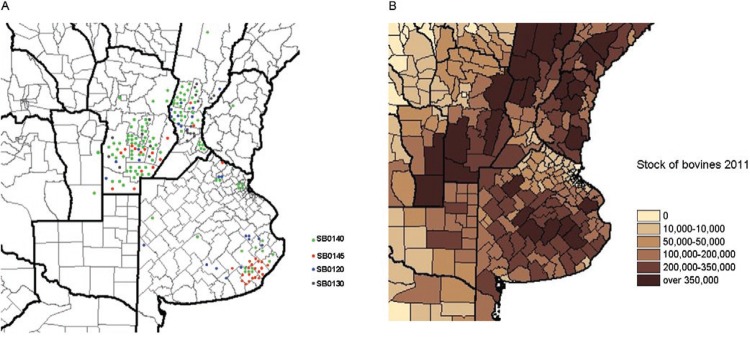
Fig. 2. : pie chart representing the major spoligotypes, the rest of clustered spoligotypes and orphan types in the three provinces and in each province.
The exclusive spoligotypes were composed of the orphans in addition to four clustered spoligotypes, with one from Buenos Aires (SB1055, n = 7), three from Córdoba (SB0269, n = 2, SB0273, n = 4 and SB0520, n = 4) and one from Santa Fe (SB1790, n = 2). The distribution of spoligotypes in each province is shown in Table II. Four spoligotypes (SB0120, SB0131, SB0140 and SB0145) were shared among the three provinces and they made up 65.9% of the isolates. Six spoligotypes were present in Córdoba and Santa Fe (SB0130, SB0153, SB0267, SB0484, SB0980 and SB1799), which represented 26.8% of the isolates from these provinces. Another spoligotype was detected in Córdoba and Buenos Aires (SB0820) (with 1.1% of the isolates from these 2 provinces) and two spoligotypes were in Buenos Aires and Santa Fe (SB1033 and SB1787) (with 2.7% of the isolates from these 2 provinces).
With regards to the distribution of spoligotypes per animal category, we observed that in the calf category (n = 27), SB0145 was predominant, with 19 isolates. The animals from this specific category were primarily sampled from Buenos Aires (81.5%). There were 13 samples in the category for young steers. All of these samples came from Córdoba and SB0140 was the predominant spoligotype (n = 5) in this category. Samples from 35 animals were included in the steer category (65.7% from Córdoba and 31.4% from Santa Fe). Out of these samples, 16 belonged to SB0140 and five to SB0145. In the heifer category, most animals came from Córdoba (83.3%). Of the 24 animals from this particular category, nine belonged to SB0140 and six to SB0145. In the cow category (n = 158) (Buenos Aires, n = 40; Córdoba, n = 49 and Santa Fe, n = 69), the following spoligotypes were described: SB0120 (19), SB0130 (13), SB0140 (68), SB0153 (5), SB0484 (11) and SB1055 (7). Only one animal, which came from Córdoba, was included in the bull category and it exhibited spoligotype SB0140. In Santa Fe, there were 11 isolates from the steer category and spoligotype SB0140 included 45.5% of the isolates. From cows, we obtained 69 isolates and the spoligotypes were distributed as follow: SB0140 (28), SB0130 (12), SB0120 (7) and SB0484 (4). In Córdoba, SB0140 predominated in all categories [young steers (38.5%), steers (47.8%), heifers (45%) and cows (49%)].
VNTR typing - A total of 197 isolates were also typed by VNTR (Buenos Aires, n = 64, Córdoba, n = 69 and Santa Fe, n = 64). There were 55 combined VNTR types that were determined by using primers directed to ETR A, B, C and D (Table III). The discrimination index was 0.92. There were 25 clusters (comprising 168 isolates) and 29 orphan types. The predominant cluster was 7-5-5-4* with 48 isolates present in the three provinces. The second most prevalent cluster was 6-5-5-4* with 20 isolates present in the three provinces, followed by 6-5-5-1 with 12 isolates from Santa Fe.
TABLE III. Variable number of tandem repeats (VNTR) types distribution, number of patterns per province .
| VNTR types | Total of isolates | Buenos Aires | Córdoba | Santa Fe |
|---|---|---|---|---|
| 2-3-5-3 | 1 | 1 | - | - |
| 2,5*-5-4,* | 1 | 1 | - | - |
| 3-10-5-4* | 1 | - | - | 1 |
| 3-3-5-1 | 5 | 5 | - | - |
| 3-5-1-4* | 1 | 1 | - | - |
| 3-5-5-1 | 1 | 1 | - | - |
| 3-5-5-4* | 1 | 3 | 1 | - |
| 4-4-5-1 | 1 | 1 | - | - |
| 4-5-5-1 | 3 | 2 | - | 1 |
| 4-5-5-4* | 3 | - | 2 | 1 |
| 5-4-5-1 | 1 | - | - | 1 |
| 5-5-1-4* | 2 | - | - | 2 |
| 5-5-5-4* | 3 | - | 2 | 1 |
| 5-8-4-4* | 1 | 1 | - | - |
| 6-1-5- 4* | 1 | - | 1 | - |
| 6-2-1-4* | 1 | - | 1 | - |
| 6-3-5-1 | 3 | - | - | 3 |
| 6-3-5-4* | 1 | - | - | 1 |
| 6-3*-5-4* | 2 | - | 1 | 1 |
| 6-4-5-4* | 1 | - | 1 | - |
| 6-5-4-4* | 1 | - | 1 | - |
| 6-5-5-1 | 12 | - | - | 12 |
| 6-5-5-3 | 2 | 1 | - | 1 |
| 6-5-5-4* | 20 | 4 | 12 | 4 |
| 6-9-4-4* | 1 | - | - | 1 |
| 6-9-5-1 | 2 | - | - | 2 |
| 6-9-5-3 | 1 | - | - | 1 |
| 6-9-5-4* | 6 | 4 | - | 2 |
| 6-9*-5-4* | 1 | 1 | - | - |
| 7-10-4-1 | 1 | - | 1 | - |
| 7-10-4-3 | 1 | - | 1 | - |
| 7-10-4-4* | 1 | - | - | 1 |
| 7-10-5-1 | 6 | 1 | - | 5 |
| 7-10-5-3 | 1 | - | - | 1 |
| 7-10-5-4* | 2 | 1 | - | 1 |
| 7-3-5-4* | 1 | - | - | 1 |
| 7-3*-5-4* | 1 | 1 | - | - |
| 7-4-4-1 | 1 | - | - | 1 |
| 7-4-5-1 | 5 | 2 | - | 3 |
| 7-4-5-4* | 6 | 4 | 1 | 1 |
| 7-5-1-4* | 2 | - | 1 | 1 |
| 7-5-2*-4 | 2 | 2 | - | - |
| 7-5-3-1 | 1 | - | - | 1 |
| 7-5-4-4* | 1 | 1 | - | - |
| 7-5-5-1 | 5 | - | 4 | 1 |
| 7-5-5-3 | 9 | 6 | 2 | 1 |
| 7-5-5-4* | 48 | 10 | 29 | 9 |
| 7-5-6-4* | 2 | - | 2 | - |
| 7-5-7*-4* | 1 | - | 1 | - |
| 7-6-4-1 | 1 | - | - | 1 |
| 7-6-5-4* | 4 | - | 4 | - |
| 7-7-5-4* | 1 | - | 1 | - |
| 7-9-5-1 | 1 | - | - | 1 |
| 7-9*-5-4* | 10 | 10 | - | - |
|
| ||||
| Total | 197 | 64 | 69 | 64 |
Subtyping of spoligotypes by VNTR - The highly predominant spoligotype SB0140 was divided in 26 VNTR types, where 7-5-5-4* included 15 of the 64 isolates (data not shown). The 34 isolates belonging to SB0145 were divided into eight VNTR types. In turn, the third most frequent spoligotype, namely SB0120 (with 21 isolates), was subdivided into 11 VNTR types. With regards to the major VNTR types, the predominant 7-5-5-4* with 48 isolates was split into 14 different spoligotypes, which were primarily distributed into the following three spoligotypes: SB0153, SB0145 and SB0140. The second most prevalent VNTR was 6-5-5-4*, with 20 isolates composing spoligotype SB0140 in 14/20 isolates and this VNTR was split into six different spoligotypes. Finally, 6-5-5-1 comprised SB0140 and SB0130 with 12 isolates each.
Combined discrimination by spoligotyping and VNTR typing - When both typing methods were combined, we detected 96 spoligotypes and VNTR types (Table IV) with 30 clusters (comprising 131 isolates) and 66 orphan types and a discrimination index of 0.98. SB0140-7-5-5-4* was the predominant cluster with 15 isolates distributed in South Córdoba, Santa Fe and Buenos Aires, followed by SB0140-6-5-5-4* with 14 isolates distributed in the three provinces. Then, SB0145-7-5-5-4* was present in Córdoba and Buenos Aires and SB0145- 7-9*-5-4* was observed only in south Buenos Aires with 10 isolates each.
TABLE IV. Distribution of combined spoligotyping-variable number of tandem repeats (VNTR) types, number of patterns per province .
| Spoligotype | VNTR | Total of isolates | Buenos Aires | Córdoba | Santa Fe |
|---|---|---|---|---|---|
| SB0120 | 2- 3- 5- 3 | 1 | 1 | - | - |
| SB0120 | 3- 5- 5- 1 | 1 | 1 | - | - |
| SB0120 | 5- 5- 1- 4* | 2 | - | - | 2 |
| SB0120 | 5- 5- 5- 4* | 1 | - | 1 | - |
| SB0120 | 7- 10- 5- 4* | 1 | 1 | - | - |
| SB0120 | 7- 3- 5- 4* | 1 | - | - | 1 |
| SB0120 | 7- 4- 5- 1 | 3 | 2 | - | 1 |
| SB0120 | 7- 4- 5- 4* | 5 | 4 | - | 1 |
| SB0120 | 7- 5- 5- 3 | 1 | - | 1 | - |
| SB0120 | 7- 5- 5- 4* | 4 | 2 | 1 | 1 |
| SB0120 | 7- 6- 5- 4* | 1 | - | 1 | - |
| SB0130 | 6- 2- 1- 4* | 1 | - | 1 | - |
| SB0130 | 6- 3- 5- 1 | 2 | - | - | 2 |
| SB0130 | 6- 3*- 5- 4* | 2 | - | 1 | 1 |
| SB0130 | 6- 5- 5- 1 | 6 | - | - | 6 |
| SB0130 | 6- 5- 5- 4* | 2 | - | 2 | - |
| SB0130 | 6- 9- 5- 4* | 1 | - | - | 1 |
| SB0130 | 7- 5- 5- 4* | 3 | - | 2 | 1 |
| SB0131 | 3- 10- 5- 4* | 1 | - | - | 1 |
| SB0131 | 4- 4- 5- 1 | 1 | 1 | - | - |
| SB0131 | 7- 4- 5- 4* | 1 | - | 1 | - |
| SB0131 | 7- 5- 3- 1 | 1 | - | - | 1 |
| SB0131 | 7- 5- 5- 1 | 1 | - | - | 1 |
| SB0131 | 7- 5- 5- 4* | 1 | 1 | - | - |
| SB0140 | 2*- 5- 5- 4* | 1 | 1 | - | - |
| SB0140 | 3- 5- 5- 4* | 2 | 1 | 1 | - |
| SB0140 | 4- 5- 5- 1 | 2 | 2 | - | - |
| SB0140 | 4- 5- 5- 4* | 2 | - | 1 | 1 |
| SB0140 | 5- 4- 5- 1 | 1 | - | - | 1 |
| SB0140 | 5- 5- 5- 4* | 1 | - | 1 | - |
| SB0140 | 5- 8- 4- 4* | 1 | 1 | - | - |
| SB0140 | 6- 1- 5- 4* | 1 | - | 1 | - |
| SB0140 | 6- 3- 5- 1 | 1 | - | - | 1 |
| SB0140 | 6- 5- 4- 4* | 1 | - | 1 | - |
| SB0140 | 6- 5- 5- 1 | 6 | - | - | 6 |
| SB0140 | 6- 5- 5- 3 | 1 | - | - | 1 |
| SB0140 | 6- 5- 5- 4* | 14 | 4 | 6 | 4 |
| SB0140 | 6- 9- 4- 4* | 1 | - | - | 1 |
| SB0140 | 6- 9- 5- 3 | 1 | - | - | 1 |
| SB0140 | 6- 9- 5- 4* | 4 | 4 | - | - |
| SB0140 | 6- 9*- 5- 4* | 1 | 1 | - | - |
| SB0140 | 7- 10- 4- 4* | 1 | - | - | 1 |
| SB0140 | 7- 10- 5- 4* | 1 | - | - | 1 |
| SB0140 | 7- 3*- 5- 4* | 1 | 1 | - | - |
| SB0140 | 7- 4- 4- 1 | 1 | - | - | 1 |
| SB0140 | 7- 5- 4- 4* | 1 | 1 | - | - |
| SB0140 | 7- 5- 5- 3 | 1 | - | - | 1 |
| SB0140 | 7- 5- 5- 4* | 15 | 1 | 10 | 4 |
| SB0140 | 7- 5- 7*- 4* | 1 | - | 1 | - |
| SB0140 | 7- 6- 4- 1 | 1 | - | - | 1 |
| SB0145 | 3- 5- 1- 4* | 1 | 1 | - | - |
| SB0145 | 6- 5- 5- 4* | 1 | - | 1 | - |
| SB0145 | 7- 4- 5- 1 | 1 | - | - | 1 |
| SB0145 | 7- 5- 2*- 4 | 2 | 2 | - | - |
| SB0145 | 7- 5- 5- 3 | 7 | 6 | 1 | - |
| SB0145 | 7- 5- 5- 4* | 10 | 5 | 5 | - |
| SB0145 | 7- 6- 5- 4* | 2 | - | 2 | - |
| SB0145 | 7- 9*- 5- 4* | 10 | 10 | - | - |
| SB0153 | 6- 3- 5- 4* | 1 | - | - | 1 |
| SB0153 | 6- 5- 5- 4* | 1 | - | 1 | - |
| SB0153 | 7- 5- 1- 4* | 1 | - | 1 | - |
| SB0153 | 7- 5- 5- 4* | 4 | - | 3 | 1 |
| SB0153 | 7- 5- 6- 4* | 2 | - | 2 | - |
| SB0153 | 7- 6- 5- 4* | 1 | - | 1 | - |
| SB0153 | 7- 7- 5- 4* | 1 | - | 1 | - |
| SB0267 | 7- 5- 5- 4* | 1 | - | - | 1 |
| SB0269 | 6- 4- 5- 4* | 1 | - | 1 | - |
| SB0273 | 7- 10- 4- 1 | 1 | - | 1 | - |
| SB0273 | 7- 10- 4- 3 | 1 | - | 1 | - |
| SB0273 | 7- 5- 5- 4* | 2 | - | 2 | - |
| SB0484 | 4- 5- 5- 1 | 1 | - | - | 1 |
| SB0484 | 7- 10- 5- 1 | 2 | - | - | 2 |
| SB0484 | 7- 5- 5- 1 | 4 | - | 4 | - |
| SB0484 | 7- 5- 5- 4* | 1 | - | 1 | - |
| SB0520 | 7- 5- 5- 4* | 3 | - | 3 | - |
| SB0820 | 7- 10- 5- 1 | 1 | 1 | - | - |
| SB0980 | 5- 5- 5- 4* | 1 | - | - | 1 |
| SB0980 | 6- 9- 5- 1 | 1 | - | - | 1 |
| SB0980 | 7- 9- 5- 1 | 1 | - | - | 1 |
| SB0996 | 6- 5- 5- 4* | 1 | - | 1 | - |
| SB1033 | 6- 5- 5- 3 | 1 | 1 | - | - |
| SB1033 | 6- 9- 5- 1 | 1 | - | - | 1 |
| SB1043 | 7- 5- 5- 4* | 1 | - | 1 | - |
| SB1055 | 3- 3- 5- 1 | 5 | 5 | - | - |
| SB1055 | 3- 5- 5- 4* | 2 | 2 | - | - |
| SB1068 | 4- 5- 5- 4* | 1 | - | 1 | - |
| SB1787 | 7- 4- 5- 1 | 1 | 1 | - | - |
| SB1787 | 7- 5- 1- 4* | 1 | - | - | 1 |
| SB1790 | 7- 10- 5- 1 | 2 | - | - | 2 |
| SB1799 | 7- 5- 5- 4* | 1 | - | 1 | - |
| SB2117 | 7- 10- 5- 1 | 1 | - | - | 1 |
| SB2165 | 6- 9- 5- 4* | 1 | - | - | 1 |
| SB2166 | 7- 10- 5- 3 | 1 | - | - | 1 |
| Spo126 | 6- 5- 5- 4* | 1 | - | 1 | - |
| Spo138 | 7- 5- 5- 4* | 1 | 1 | - | - |
| Spo139 | 7- 5- 5- 4* | 1 | - | - | 1 |
|
| |||||
| Total | - | 197 | 64 | 69 | 64 |
In addition to the orphan types, there were 18 spoligo-VNTR types that were exclusives (6 of each province), which were clustered in 60 isolates (Buenos Aires, 25, Córdoba, 15 and Santa Fe, 20).
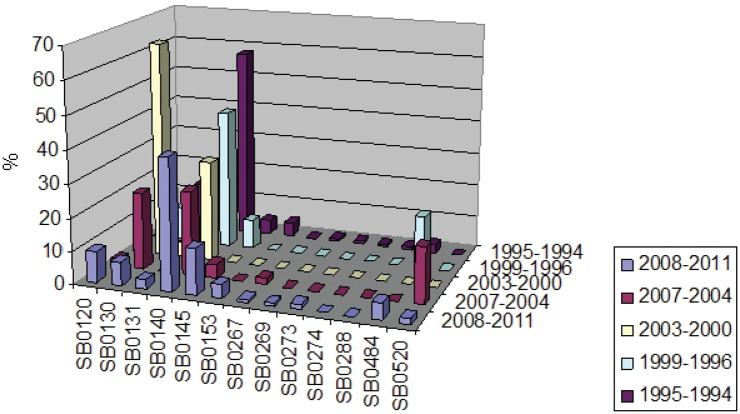
Temporal trends - A study of temporal trends was performed with a meta-analysis of spoligotypes. We used data from 1994-2011 that were recorded in our genotyping database. The analysis was performed by dividing the data into periods of four years (except for the 1st period that includes only the years 1994 and 1995). The predominant spoligotype was SB0140 and its prevalence percentage remained high throughout the different periods, varying from 25.5-57.8% (Fig. 3). SB0130 was the second most prevalent in the database registers and experienced an important fluctuation and it reached a peak of 65.2% in 2000-2003. Similarly, SB0145 also displayed fluctuations, although they were at lower prevalence levels. By contrast, other minor spoligotypes had low representation, but they were constant or sporadic. For instance, SB0131 had low representation and SB0484 was sporadically represented.
Fig. 3. : percentage of incidence of major spoligotypes in the three studied provinces along different sampling periods.
When temporal trends are taken into account by province, the most predominant spoligotype in Buenos Aires was SB0140, which is in line with the rest of the country. However, the prevalence percentage highly differs throughout the different time periods (Fig. 3), from 100-16%. SB0145, the second most predominant type in Buenos Aires, increased sharply during the last study period. Additionally, important fluctuations are observed in the periodical frequency of SB0130. In some cases, the sampling number was too low to validate the analysis (data not shown). This challenge was also the case for the analysis in Córdoba, where the number of collected isolates was too low during some periods (data not shown). In Santa Fe, SB0140 was the predominant spoligotype with a percentage above 37% during all periods, except for 2000-2003. The second most predominant spoligotype was SB0130, which varied from 0-93.75%. Finally, SB0120 rose from 0-10.2% in 2008-2011 (data not shown).
DISCUSSION
Among the 32 spoligotypes detected, 10 have not been previously included or described in Argentina and are not included in the databases from mbovis.org; therefore, they were considered to be new spoligotypes. Of these 10 spoligotypes, three formed clusters of two isolates each, that is, SB1790 of Santa Fe, SB1787 of Santa Fe and Buenos Aires and SB1799 of Santa Fe and Córdoba. The other new spoligotypes were also orphans, namely SBxx138 from Buenos Aires, SBxx131 and 1776 from Córdoba and SB2165, 2166, xx126 and xx139 from Santa Fe. Santa Fe had the majority of new spoligotypes, most likely because of the active TB eradication program in that province. This program, which was implemented across the whole country, consists of the sacrifice of infected bovines and the inspection of carcasses in slaughterhouses. In this way, when a new M. bovis strain is introduced into a herd, the infected animals are detected early and sacrificed, so a new M. bovis genotype can be detected before it can spread among the other animals. Other authors have proposed that if a human or bovine TB control program is active and long-lasting, then a reduction in predominant clones should be observed as transmission is halted (van Soolingen et al. 1991, Durr et al. 2000).
Early in the application of VNTR elements to M. bovis, the method’s discriminatory power was observed to be higher than that of spoligotyping (Roring et al. 2004). Specifically, ETR A and ETR B exhibited an important allelic variation in M. bovis (Romero et al. 2011). The ETR A-C elements were previously shown to have a discrimination index higher than 0.4 (Martinez et al. 2008, Boniotti et al. 2009, Price-Carter et al. 2011). However, the individual discrimination indexes in M. bovis for ETR A-D may be highly variable according to the geographic setting (Sahraoui et al. 2009, Sun et al. 2012). This discrimination index increases when ETRs A-D are evaluated together. Additionally, the combination of VNTR typing and spoligotyping increases the discrimination index above that of VNTR alone (Martinez et al. 2008). In contrast with the ETR-VNTR elements, minimum inhibitory concentration (MIRU)-VNTR (with the exception of MIRU26) yields a low discriminatory power for M. bovis (Rodriguez et al. 2004, Parreiras et al. 2012, Rocha et al. 2013).
One important aspect to evaluate is the discrimination conferred by VNTR typing in predominant spoligotypes. We observed that the predominant SB0140 spoligotype was split into 26 different ETR A-D types. This result is in accordance with the findings of McLernon et al. (2010) in Ireland, where SB0140 is also the predominant spoligotype and was highly subtyped by VNTR typing. Boniotti et al. (2009) observed that 408 isolates of SB0120, which is the most predominant spoligotype in Italy, were split into 64 different patterns (Boniotti et al. 2009).
The presence of combined cluster patterns is an index for the active transmission of TB. There were 66.5% of the isolates distributed into 30 different spoligotyping-VNTR combined patterns. Smith et al. (2003) suggested that the clonal expansion of a genotype is caused either by the spread of a favourable mutation or by the invasion of a geographic region by a few genotypes (Smith et al. 2003).
In this work, we detected 18 combined and clustered spoligotyping-VNTR patterns (6 in each province) that were exclusives in the sampled provinces. These combined patterns represented 30.5% of the analysed isolates. Together with the 66 orphans, these patterns yielded 84 combined and exclusive patterns. This finding suggests a pattern association with a particular geographical region. However, this putative association must be demonstrated with more isolates in the future; the association was supported by a chi-square test because of the low number of isolates (data not shown).
Our database consists of M. bovis molecular typing data from 1994 until the present. When we divided this 17-year span into regular periods, we observed that although the incidence of different spoligotypes fluctuated, the most predominant spoligotypes were unaltered. To the best of our knowledge, only one previous publication on the evolution of the M. bovis population structure has been reported (Haddad et al. 2001).
The predominance of major spoligotypes in cattle from the primary cattle farms in these Argentine provinces suggests that there is still an active transmission of the pathogen, in spite of the reduced incidence of animals with visible lesions in slaughterhouses. Spoligotypes are more stable than VNTR types. In fact, the BCGs obtained in 1921 from which the substrains originated cannot be differentiated by spoligotyping (de la Salmonière et al. 1997, Bauer et al. 1999) and VNTR subdivides BCG (Magdalena et al. 1998, Supply et al. 2000). As a consequence, the predominant spoligotypes may ancient, as suggested by the subdivision of the main spoligotypes by VNTR typing.
ACKNOWLEDGEMENTS
To Valeria Rocha, for their technical help.
Funding Statement
Financial support: National Agency of Research Promotion of Argentina (PICT 1114) KC and AC are career members of CONICET.
Footnotes
Financial support: National Agency of Research Promotion of Argentina (PICT 1114) KC and AC are career members of CONICET.
REFERENCES
- Alexander KA, Laver PN, Michel AL, Williams M, van Helden PD, Warren RM, Gey van Pittius NC. Novel Mycobacterium tuberculosis complex pathogen, M. mungi. Emerg Infect Dis. 2010;16:1296–1299. doi: 10.3201/eid1608.100314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanfu W. Tuberculosis. Vol. 86. Edinb: 2006. The situation of tuberculosis and tuberculosis control in animals of economic interest; pp. 330–335. [DOI] [PubMed] [Google Scholar]
- Bauer J, Andersen AB, Kremer K, Miorner H. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J Clin Microbiol. 1999;37:2602–2606. doi: 10.1128/jcm.37.8.2602-2606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti MB, Goria M, Loda D, Garrone A, Benedetto A, Mondo A, Tisato E, Zanoni M, Zoppi S, Dondo A, Tagliabue S, Bonora S, Zanardi G, Pacciarini ML. Molecular typing of Mycobacterium bovis strains isolated in Italy from 2000 to 2006 and evaluation of variable-number tandem repeats for geographically optimized genotyping. J Clin Microbiol. 2009;47:636–644. doi: 10.1128/JCM.01192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi A, Romano MI. Palomino JC, Rittacco V, Leao S. Tuberculosis. Amedeo; Berlin: 2007. TB caused by other members of the Mycobacterium tuberculosis complex.450 [Google Scholar]
- Collins DM, Erasmuson SK, Stephens DM, Yates GF, Lisle GW. DNA fingerprinting of Mycobacterium bovis strains by restriction fragment analysis and hybridization with insertion elements IS1081 and IS6110. J Clin Microbiol. 1993;31:1143–1147. doi: 10.1128/jcm.31.5.1143-1147.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor IN, Torres PM, Morcillo N, Imaz MS, Sequeira MD. Zoonotic tuberculosis in Argentina. Medicina. 2012;72:514–520. [PubMed] [Google Scholar]
- de la Salmonière YOG, Li HM, Torrea G, Bunschoten A, van Embden J, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:2210–2214. doi: 10.1128/jcm.35.9.2210-2214.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr PA, Clifton-Hadley RS, Hewinson RG. Molecular epidemiology of bovine tuberculosis. II. Applications of genotyping. Rev Sci Tech. 2000;19:689–701. doi: 10.20506/rst.19.3.1240. [DOI] [PubMed] [Google Scholar]
- Frothingham R, Meeker-O’Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- Haddad N, Ostyn A, Karoui C, Masselot M, Thorel MF, Hughes SL, Inwald J, Hewinson RG, Durand B. Spoligotype diversity of Mycobacterium bovis strains isolated in France from 1979 to 2000. J Clin Microbiol. 2001;39:3623–3632. doi: 10.1128/JCM.39.10.3623-3632.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdalena J, Supply P, Locht C. Specific differentiation between Mycobacterium bovis BCG and virulent strains of the Mycobacterium tuberculosis complex. J Clin Microbiol. 1998;36:2471–2476. doi: 10.1128/jcm.36.9.2471-2476.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez LR, Harris B, Black WC, Meyer RM, Brennan PJ, Vissa VD, Jones RL. Genotyping North American animal Mycobacterium bovis isolates using multilocus variable number tandem repeat analysis. J Vet Diagn Invest. 2008;20:707–715. doi: 10.1177/104063870802000601. [DOI] [PubMed] [Google Scholar]
- Maslow JN, Mulligan ME, Arbeit RD. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–162. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- McLernon J, Costello E, Flynn O, Madigan G, Ryan F. Evaluation of mycobacterial interspersed repetitive-unit-variable-number tandem-repeat analysis and spoligotyping for genotyping of Mycobacterium bovis isolates and a comparison with restriction fragment length polymorphism typing. J Clin Microbiol. 2010;48:4541–4545. doi: 10.1128/JCM.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milián-Suazo F, Banda-Ruiz V, Ramirez-Casillas C, Arriaga-Diaz C. Genotyping of Mycobacterium bovis by geographic location within Mexico. Prev Vet Med. 2002;55:255–264. doi: 10.1016/s0167-5877(02)00015-6. [DOI] [PubMed] [Google Scholar]
- Nader A, Husberg H. Rev Med Vet. Vol. 69. B. Aires: 1988. Estimación de pérdidas de producción por tuberculosis bovina en un rodeo lechero; pp. 36–43. [Google Scholar]
- Otal I, Martin C, Vincent-Levy-Frebault V, Thierry D, Gicquel B. Restriction fragment length polymorphism analysis using IS6110 as an epidemiological marker in tuberculosis. J Clin Microbiol. 1991;29:1252–1254. doi: 10.1128/jcm.29.6.1252-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parreiras PM, Andrade GI, do Nascimento TF, Oelemann MC, Gomes HM, de Alencar AP, de Assis RA, Mota PMPC, Pereira MAS, Lobato FCF, Lage AP, Suffys PN. Spoligotyping and variable number tandem repeat analysis of Mycobacterium bovis isolates from cattle in Brazil. Mem Inst Oswaldo Cruz. 2012;107:64–73. doi: 10.1590/s0074-02762012000100009. [DOI] [PubMed] [Google Scholar]
- Price-Carter M, Rooker S, Collins DM. Comparison of 45 variable number tandem repeat (VNTR) and two direct repeat (DR) assays to restriction endonuclease analysis for typing isolates of Mycobacterium bovis. Vet Microbiol. 2011;150:107–114. doi: 10.1016/j.vetmic.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Rocha VC, Figueiredo SC, Rosales CA, Grisi JHH, Filho, Keid LB, Soares RM, Ferreira JS., Neto Molecular discrimination of Mycobacterium bovis in São Paulo, Brazil. Vector Borne Zoonotic Dis. 2013;13:17–21. doi: 10.1089/vbz.2012.1035. [DOI] [PubMed] [Google Scholar]
- Rodriguez CAR, Zumárraga MJ, Oliveira EM, Cataldi AA, Romano MI, Otto HH, Bonafé VL, Ferreira JS., Neto Caracteri- zação molecular de isolados de Mycobacterium bovis do estado de São Paulo, Brasil, utilizando a técnica de spoligotyping. Arq Inst Biol. 2004;71:277–282. [Google Scholar]
- Rodríguez S, Romero B, Bezos J, Juan L, Alvarez J, Castellanos E, Moya N, Lozano F, González S, Sáez-Llorente JL, Mateos A, Domínguez L, Aranaz A, Spanish Network on Surveillance and Monitoring of Animal Tuberculosis High spoligotype diversity within a Mycobacterium bovis population: clues to understanding the demography of the pathogen in Europe. Vet Microbiol. 2010;141:89–95. doi: 10.1016/j.vetmic.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Romero B, Rodríguez S, Bezos J, Díaz R, Copano MF, Merediz I, Mínguez O, Marqués S, Palacios JJ, Viedma DG, Sáez JL, Mateos A, Aranaz A, Domínguez L, de Juan L. Humans as source of Mycobacterium tuberculosis infection in cattle, Spain. Emerg Infect Dis. 2011;17:2393–2395. doi: 10.3201/eid1712.101476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roring S, Scott AN, Hewinson RG, Neill SD, Skuce RA. Evaluation of variable number tandem repeat (VNTR) loci in molecular typing of Mycobacterium bovis isolates from Ireland. Vet Microbiol. 2004;101:65–73. doi: 10.1016/j.vetmic.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Sahraoui N, Muller B, Guetarni D, Boulahbal F, Yala D, Ouzrout R, Berg S, Smith NH, Zinsstag J. Molecular characterization of Mycobacterium bovis strains isolated from cattle slaughtered at two abattoirs in Algeria. 4BMC Vet Res. 2009;5 doi: 10.1186/1746-6148-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuce RA, Neill SD. Tuberculosis. Vol. 81. Edinb: 2001. Molecular epidemiology of Mycobacterium bovis: exploiting molecular data; pp. 169–175. [DOI] [PubMed] [Google Scholar]
- Smith NH, Dale J, Inwald J, Palmer S, Gordon SV, Hewinson RG, Smith JM. The population structure of Mycobacterium bovis in Great Britain: clonal expansion. Proc Natl Acad Sci USA. 2003;100:15271–15275. doi: 10.1073/pnas.2036554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Cao R, Tian M, Zhang X, Li Y, Xu Y, Fan W, Huang B, Li C. Evaluation of spoligotyping and MIRU-VNTR for Mycobacterium bovis in Xinjiang, China. Res Vet Sci. 2012;92:236–239. doi: 10.1016/j.rvsc.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- van Ingen J, Rahim Z, Mulder A, Boeree MJ, Simeone R, Brosch R, van Soolingen D. Characterization of Mycobacterium orygis as M. tuberculosis complex subspecies. Emerg Infect Dis. 2012;18:653–655. doi: 10.3201/eid1804.110888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D, Hermans PW, Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumárraga MJ, Arriaga C, Barandiaran S, Cobos-Marin L, Waard J, Estrada-Garcia I, Figueiredo T, Figueroa A, Giménez F, Gomes HM, Gonzalez-y-Merchand JA, Macías A, Suazo FM, Rodríguez CAR, Santillán MA, Suffys PN, Trangoni MD, Zárraga AM, Cataldi A. Understanding the relationship between Mycobacterium bovis spoligotypes from cattle in Latin American countries. Res Vet Sci. 2012;94:9–21. doi: 10.1016/j.rvsc.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Zumárraga MJ, Martín C, Samper S, Alito A, Latini O, Bigi F, Roxo E, Cicuta ME, Errico F, Ramos MC, Cataldi A, van Soolingen D, Romano MI. Usefulness of spoligotyping in the molecular epidemiology of Mycobacterium bovis - related infections in South American countries. J Clin Microbiol. 1999;37:296–303. doi: 10.1128/jcm.37.2.296-303.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]