Abstract
Background
Polyhydroxyalkanoates (PHAs) are a group of biodegradable plastics that are produced by a wide variety of microorganisms, mainly as a storage intermediate for energy and carbon. Polyhydroxybutyrate (PHB) is a short-chain-length PHA with interesting chemical and physical properties. Large scale production of PHB is not wide-spread mainly due to the downstream processing of bacterial cultures to extract the PHB. Secretion of PHB from Escherichia coli could reduce downstream processing costs. PHB are non-proteinaceous polymers, hence cannot be directly targeted for secretion. Phasin, PhaP1, is a low molecular weight protein that binds to PHB, reducing PHB granule size. In this study PHB is indirectly secreted with PhaP1 from E. coli via type I secretion using HlyA signal peptides.
Results
This study demonstrated the successful secretion of phasin and phasin bound PHB outside of the cell and into the culture medium. The secretion of PHB was initiated between 24 and 48 h after induction. After 48 h of culturing, 36% of the total PHB produced in the secreting strain was collected in the secreted fraction and 64% remained in the internal fraction. To further support the findings of this study, the PHB secretion phenomenon was observed using SEM.
Conclusions
From this study, the ability to use type I secretion to: 1) secrete phasin and 2) successfully secrete PHB has been shown.
Keywords: Polyhydroxyalkanoates, PHA, PHB, Secretion, Phasin, Translocation, HlyA, Synthetic biology
Background
Fossil derived plastics are non-biodegradable and toxic to the environment. Based on an United States Environmental Protection Agency study in 2011, there was an increase in non-biodegradable plastic accumulation in municipal solid waste systems from 0.5% to 12.4% during 1960 to 2010 [1]. Alternative means of producing plastics in large quantities that are both economically and environmentally friendly have recently gained considerable attention [2].
Replacing traditional plastic with biodegradable plastic such as polyhydroxyalkanoates (PHAs) can potentially reduce total waste by up to 20% [3]. PHAs are produced by a variety of microorganisms as an intercellular storage medium for energy and carbon and can accumulate up to 90% of the cell dry weight [4]. PHAs are biodegradable polymers. The biodegradability can range from days [5] to months [6] with degradation either taking place extracellularly or intracellularly. Extensive review on degradation of PHAs can be found in [7,8]. There are 155 different confirmed types of PHA monomer subunits, each with varying monomer repeat number and side groups [9]. Additionally, PHAs have melting temperatures between 50-180°C and crystallinity of 30–70% [10]. Thus, PHAs have a variety of possible applications, that could replace traditional plastics derived from petroleum [11]. Some of the possible applications are highlighted in previous studies [12,13] and include: packaging, medical uses [14], agricultural uses, and in carbon nanotubes [15].
Polyhydroxybutyrates (PHB) are a short-chain-length (scl) PHA polyester with between 3–5 carbon monomers [9]. The production of PHB in recombinant systems such as Escherichia coli has been made possible by the isolation of the phaCAB operon from Ralstonia eutropha (Cupriavidus necator) and cloning into pBluescriptSK- to generate plasmid pBHR68. pBHR68 has been widely used for recombinant production of PHB in E. coli. The phaCAB operon is a three-step enzymatic process by which acetyl-CoA is converted to PHB: phaC (PHA synthase), phaA (β-ketothiolase), and phaB (acetoacetyl-CoA reductase) [4,16]. After production of PHB, the PHB polymer forms spherical granules with a hydrophobic core and attached proteins at the surface, including PHA synthase and phasin, PhaP1 [17-19].
The cost of producing PHAs is approximately US$ 4-6/ kg and one of the major bottlenecks in scaling up recombinant PHA production systems is the isolation of the PHAs [12,20]. Current techniques that are used to isolate PHB from E. coli are invasive, including mechanical, chemical, and biological treatments. These techniques involve lysing the cellular membrane prior to isolation of the PHBs. There are a variety of methods that have been suggested for recovery of PHBs from microorganisms and a detailed review of the different PHB isolation methods is outlined in [20].
The development of a secretion mechanism eliminates the need for cell disruption by mechanical or chemical means, and may lead to continuous (or semi-continuous) PHB production systems. In gram-negative bacteria like E. coli, compounds are exported via six major secretory pathways [21-24]. Recombinant proteins can be targeted to type I and II secretory pathways through genetic fusion with signal peptide targeting sequences.
PHBs are non-proteinaceous polyesters and therefore cannot be directly targeted for translocation by signal peptide fusion. Phasins are low-molecular weight proteins that play a role in PHB granule formation by binding to the PHB granule surface [19,25]. Phasins are structural proteins found in organisms that naturally produce PHAs and are similar in function to oleosins which are found in plants [19,26]. Currently, oleosins are used to purify various compounds such as pharmaceuticals from plants [27].
Translocation of PHBs is possible through optimization of granule size, which reportedly varies from 50 to 1,000 kDa based on growth parameters and host strain [4]. One of the functions of phasin is to increase the surface-to-volume ratio of granules so that higher accumulation levels can be achieved [25,28,29]. Therefore, the size of the PHB granule can be decreased significantly through phasin overexpression. In one such study, overexpression of phasin resulted in a decrease of PHB granules from 70–310 nm diameter to 20–60 nm [28].
Type I secretion is a simple one step secretion system that can translocate proteins from the cytoplasm to the extracellular medium without protein interaction with the periplasm [30]. Proteins of nearly 900 kDa (large adhesion protein, LapA) have reportedly been secreted to the extracellular milieu by the type I secretory mechanism of gram-negative bacteria [31,32]. Specifically using the Hemolysin (HlyA) secretion mechanism proteins such as β-galactosidase (117 kDa) [33], β-gal-OmpF (56 kDa) [34], and green fluorescent protein (GFP) (27 kDa) [35] have been secreted by E. coli. The physical characteristics of the secretion channel are approximately 3.5 nm in diameter with a length of 14 nm as reported by Fernández et al., which makes the secretion phenomena of large proteins very interesting [36].
Our group has previously used a synthetic biological engineering approach to demonstrate the feasibility of HlyA, GeneIII, PelB, and TorA secretion systems in E. coli with the use of GFP [35]. From the aforementioned study, the type I secretion system using the HlyA signal peptide yielded the best results for secretion of GFP outside of the cell and into the medium [35]. The objective of this study was to demonstrate that phasin can be used to secrete PHB from E. coli using type I secretion machinery.
Results and discussion
Initial studies were carried out to demonstrate expression and then successful translocation of phasin, PhaP1, into the extracellular medium. Once this was demonstrated, PHB secretion experiments were conducted that included: growth studies, PHB production in secreted and non-secreted fractions, and visualization with scanning electron microscopy (SEM).
Analysis of phasin translocation
For E. coli cells expressing pCMEL1 and pLG575, a phasin band is observed at 22–26 kDa in the cytoplasmic fraction, the periplasmic fraction, the membrane fraction, and the concentrated extracellular media (Figure 1). This polyacrylamide gel and corresponding immunoblot demonstrated: 1) the ability for E. coli to produce non-codon optimized Phasin (from R. eutropha), a protein not naturally expressed in E. coli and 2) translocation of phasin into different fractions of the cell. Compared to other studies that focused non-translocated phasin, the phasin band sizes in all of the different fractions are similar [19,29]. Since the translocation of phasin was successful, secretion of PHB using phasin was then attempted.
Figure 1.
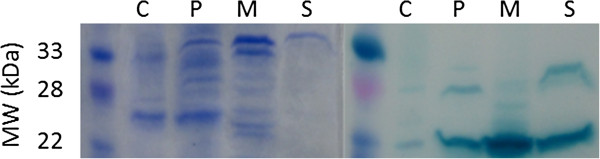
SDS polyacrylamide gel and corresponding immunoblot of subcellular fractions for PhaP1:HlyA. C – cytoplasmic fraction, P – periplasmic fraction, M – membrane fraction, S – concentrated supernatant (media) fraction. The position of phasin bands varies from roughly 22–26 kDa.
Growth studies
The PHB secreting strain consisted of pCMEL3 + pLG575, whereas the non-secreting strain consisted of pBHR68 + pLG575. From the CFU/mL vs. time graph (Figure 2) stationary phase was reached at approximately 8–12 h for both non-secreting and secreting strains. The non-secreting and secreting strains had the highest overall CFU/mL at approximately 9×1012 and 5×1012 CFU/mL, respectively, after 12 h. There was no significant difference between CFU/mL for the non-secreting and secreting strains after 24 and 48 h (p > 0.05). This statistical analysis on the CFU/mL demonstrated that there were no significant differences in E. coli growth between the two samples at times when PHB analysis was conducted (24 and 48 h). Furthermore, this suggests that the secretion of PHB does not affect cell viability.
Figure 2.
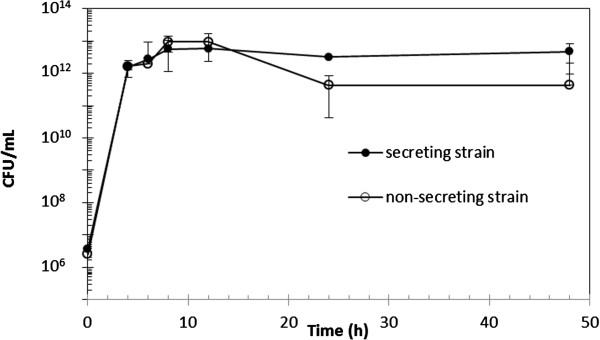
CFU/mL vs. Time (hours) for secreting and non-secreting strains of PHB producing E. coli , averaged from triplicate experiments (one standard deviation shown).
PHB production analysis
PHB production analysis was carried out 24 and 48 because after 24 h the E. coli harboring the plasmid systems were in stationary phase. Previous studies demonstrated that PHB did not accumulate to significant levels during the exponential growth phase. Acetyl-CoA is required for cell synthesis during the exponential phase but is diverted to produce PHB in the stationary phase, thus, there is a delay between carbon source utilization and PHB production [37].
PHB measured inside the cell is defined as the internal fraction and PHB collected by the CaCl2 precipitation method is the secreted fraction. 24 h after induction, the internal fraction of the cells demonstrated PHB production for both the secreting and non-secreting strains. While the non-secreting strain accumulated approximately 41.93 ± 13.5% of PHB in the dry cell weight at 24 h, the secreting strain accumulated approximately 28.85 ± 0.41% (Table 1). 48 h after induction the non-secreting and secreting strains had accumulated 47.24 ± 6.0% and 38.80 ± 15.5% PHB, respectively. There was no significant statistical difference seen in internally accumulated PHB in either the non-secreting or secreting strains at 24 and 48 h (p > 0.05) after induction. The internal levels of PHB accumulation in the non-secreting and secreting strains are comparable to those seen in other studies such as [38], where the authors showed accumulation of 42% PHB in E. coli DH5α harboring pBHR68 after 24–48 h [38].
Table 1.
Production of PHB in secreting (pCMEL3 + pLG575) and non-secreting (pBHR68 + pLG575) strains of E. coli at 24 and 48 h
| |
|
% mass of PHB in dry mass |
Production g/L PHB |
|
|||
|---|---|---|---|---|---|---|---|
| Strain | Time (h) | Secreted fraction | Non- secreted fraction | Secreted fraction | Non-secreted fraction | Total production | g PHB/g Glucose |
| Non-secreting |
24 |
0.31 ± 0.35 |
41.93 ± 13.5 |
0.40 ± 0.27 |
5.65 ± 1.1 |
6.05 ± 1.1 |
0.40 ± 0.07 |
| |
48 |
0.72 ± 0.89 |
47.24 ± 6.0 |
0.50 ± 0.41 |
5.43 ± 1.7 |
5.93 ± 1.8 |
0.40 ± 0.12 |
| Secreting |
24 |
0.69 ± 0.18 |
28.85 ± 0.41 |
0.40 ± 0.06 |
3.42 ± 0.33 |
3.82 ± 0.3 |
0.25 ± 0.02 |
| 48 | 28.29 ± 7.2* | 38.80 ± 15.5 | 2.57 ± 0.75* | 4.58 ± 2.47 | 7.15 ± 2.6 | 0.48 ± 0.17 | |
*Indicates statistical significance within column (p < 0.05).
PHB secreted fractions were analyzed at 24 and 48 h after induction for the secreting and non-secreting strains. PHB harvested in the secreted fraction of the non-secreting strains was 0.31 ± 0.35% and 0.72 ± 0.89%, respectively. The PHB present in the secreted phase demonstrated that some lysed cells containing PHB were found in this fraction after CaCl2 precipitation. This is to be expected from a differential centrifugation technique, such as that used in this study. 24 and 48 h after induction, the secreting strain produced 0.69 ± 0.18% and 28.29 ± 7.2% respectively in the secreted fraction. The level of PHB seen in the secreted fraction of the secreting strain was statistically significant after 48 h post induction, compared to the non-secreting strain (p < 0.05). This increase in PHB present in the secreted phase after 48 h indicates that the secretion system is functioning and producing higher amounts of PHB, with a small amount of non-secreted PHB ending up in the secreted fraction. These results demonstrate that PHB secretion is initiated 24 h after induction.
Secretion of PHB can help in downstream processes by aiding in PHB separation from biomass. Of the total PHB produced by the secreting strain after 48 h, 36% was collected in the secreted fraction and the remaining 64% was in the internal fraction. The secreting strain had a total PHB production of 7.15 g/L compared to 5.93 g/L for the non-secreting strain after 48 h post induction.
It has been demonstrated from previous studies that PHB can accumulate in larger quantities in E. coli when using a bioreactor compared to a shaker flask. A fed-batch bioreactor study by Choi et al. 1999, reported accumulation of up to 77% PHB of dry cell weight [39] and another study reported accumulation of up to 80% [37]. Future studies will be performed to determine how well the secreting strain performs under similar bioreactor growth conditions.
SEM analysis
SEM is not widely used for PHB analysis since a surface topographical analysis is typically of little use. SEM has however been used to visualize PHA granules produced from recombinant E. coli[40] and PHA degradation from a variety of organisms [41]. In the case of secretion, SEM can provide images of what is occurring at the surface of E. coli during the secretion process. Figure 3A shows a PHB non-secreting E. coli strain harboring the pBHR68 plasmid. Figure 3B shows E. coli that is accumulating PHB and overexpressing phasin. Figure 3C shows the full secretion system in E. coli (pCMEL3 + pLG757). Figure 3C suggests that PHB is being secreted outside of the bacteria and into the medium. These observations further demonstrate the functioning of the PHB secretion system.
Figure 3.
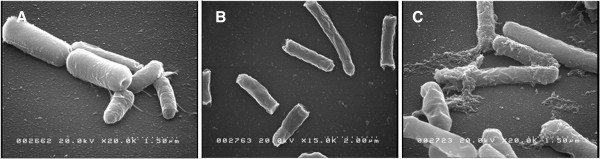
SEM images taken from overnight cultures of E. coli XL1Blue harboring different plasmid systems: A) pBHR68 (non-secreting), B) pCMEL3 (non-secreting with phasin overexpression), and C) pCMEL3 + pLG575 (complete PHB production and secretion system).
The SEM photo in Figure 3C suggests that secretion of PHB occurs at the polar regions of the cell. Interestingly it has been found in previous studies [42,43] that PHB granule formation occurs at the cell poles in E. coli when both phasin and PHB are being produced. When PHB is being produced the interpretation is that PHA synthase is active, and it has previously been suggested that PHA synthase has polar targeting information [43].
It is interesting that E. coli is able to secrete PHB through pore sizes of 3.5 nm [36] in diameter. From the results of this this study not all the PHB produced is being secreted, suggesting there could be differences between the secreted and non-secreted PHB.
Conclusions
This study demonstrated the successful expression of phasin, PhaP1, and its translocation using type I secretion in E. coli. Once translocation of PhaP1 was successful, a recombinant synthetic biological system to secrete PHBs was designed and tested. Initiation of PHB secretion occurs between 24 and 48 h after induction of PHB synthesis. Of the total PHB produced by the secreting strain, 36% was collected in the secreted fraction and 64% remained in the internal fraction after 48 h.
Secretion of PHB should help in downstream processing whereby PHB is separated from the cell mass, which can aid in PHB recovery and purification. Future studies will include a detailed comparison of secreted versus non-secreted PHB characteristics.
Methods
Strains and plasmids
Genetic parts were constructed in accordance with the BioBrick and BioFusion technical standards [44,45]. The BioFusion standard was specifically used to create phasin and signal peptide parts because this genetic fusion system is compatible with the original BioBrick standard [45,46].
Descriptions of strains and plasmids used to study PHB and phasin production and translocation are provided as Table 2. Completed BioBrick parts were transformed in BL21-Gold (DE3) competent E. coli (Agilent Technologies, Santa Clara, CA) for protein expression studies. PHB production and secretion studies were carried out in E. coli XL1 Blue (Agilent Technologies, Santa Clara, CA). Plasmid pLG575 includes the coding regions for proteins HlyB and HlyD [47,48]. Plasmids pSB1AK3, pSB1A3, and pSB3K3 are BioBrick standard vectors for assembly and expression of BioBrick genetic devices [49].
Table 2.
Strains, plasmids, and oligonucleotides used in this study
| Strains | Relevant characteristics | Reference |
|---|---|---|
|
Strains |
|
|
| BL21-Gold (DE3) |
E. coli B F– ompT hsdS(rB– mB–) dcm+ TetR gal λ(DE3) endA Hte |
Agilent technologies |
| XL1 Blue |
recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F’ proAB lacIqZΔM15 Tn10 (Tetr)] |
Agilent technologies |
| Cupriavidus necator H16 |
Wild type, PHA producing |
ATCC 17699 |
|
Plasmids |
|
|
| pLG575 |
pACYC184 derivative, HlyBD, p15A origin, CmR |
[47] |
| pBHR68 |
pBluescript SK-, phbCAB genes from R. eutropha |
[16] |
| pSB1AK3 |
High copy BioBrick vector, pMB1 origin, AmpR and KanR |
[49] |
| pSB1A3 |
High copy BioBrick vector, pMB1 origin, AmpR |
[49] |
| pSB3K3 |
Medium copy BioBrick standard vector, p15A origin, KanR |
[49] |
| pCMEL1 |
phaP1, C-terminal BioFusion with HlyA signal peptide, Lac promoter (BBa_R0010), RBS(BBa_B0034), in pSB1A3 |
This study |
| pCMEL2 |
phaP1, C-terminal BioFusion with HlyA signal peptide, Lac promoter (BBa_R0010), RBS(BBa_B0034), in pSB3K3 |
This study |
| pCMEL3 |
phaP1, C-terminal BioFusion with HlyA signal peptide, Lac promoter (BBa_R0010), RBS(BBa_B0034), in pBHR68 |
This study |
|
Oligonucleotides |
|
|
| PhaP1FOR |
5′-gaattcgcggccgcttctagaatgatcctcaccccggaaca-3′ |
This study |
| PhaP1REV |
5′- ctgcagcggccgctactagttcaggcagccgtcgtcttct-3′ |
This study |
| g114t |
5′-cgtcgagctgaaccttcaggtcgtcaagact-3′ |
This study |
| g114t_antisense | 5′-agtcttgacgacctgaaggttcagctcgacg-3′ | This study |
BioBrick and plasmid construction
All restriction enzymes and related reagents were purchased from Thermo Fisher Scientific Inc. (Glen Burnie, MD). The signal peptide HlyA was made through synthetic design and construction as mention in Linton et al. 2012 (DNA 2.0, Menlo Park, CA) [35]. A BioFusion-compatible phasin BioBrick was constructed by isolating phaP1 from the genomic DNA of R. eutropha using PCR with primers PhaP1FOR and PhaP1REV that included the BioFusion prefix and suffix as overhanging ends (Table 2). The 620 bp PCR product was isolated by gel electrophoresis, digested with EcoRI and SpeI, and ligated into pSB3K3. A PstI site was removed from phaP1 (while conserving amino acid sequence) using a QuikChange II Site-Directed Mutagenesis Kit and the QuikChange® Primer Design Program (Agilent Technologies, Santa Clara, CA). The designed primers (g114t and g114t_antisense) for site-directed mutagenesis are shown in Table 2. The PstI mutation in phaP1 was successfully carried out and confirmed by sequence analysis.
Step-wise assembly of composite BioBrick devices was primarily carried out in pSB1AK3. Completed devices were subsequently ligated into pSB1A3 and pSB3K3. The lac promoter (BBa_R0010) and ribosome binding site (BBa_B0034) were used as described in Linton et al. 2012 [35]. The pCMEL1 plasmid was used for studies on phasin translocation because its origin of replication (pMB1) was compatible with the origin of replication of pLG575 (p15A). BL21-Gold (DE3) was co-transformed with pCMEL1 and pLG575. The pCMEL3 plasmid was used for studies on type I secretion of PHA. The composite part containing the promoter, RBS, coding region, and terminator were cloned into pBHR68 from pCMEL2 by digestion with EcoRI and XhoI. XL1-Blue was co-transformed with pCMEL3 and pLG575 for PHA secretion studies. A schematic of the secretion system is presented in Figure 4.
Figure 4.
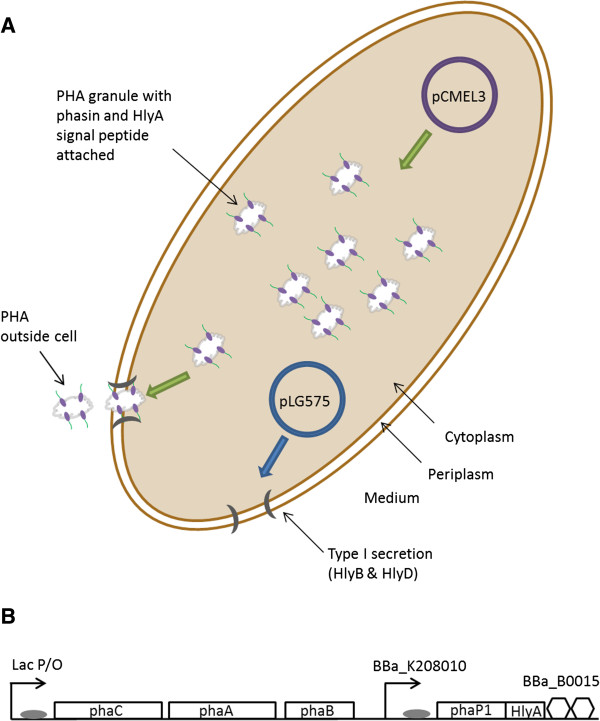
PHB secretion. A. Schematic for PHB secretion containing dual plasmid system pCMEL3 and pLG575. Phasin with attached signal peptide binds to PHB granule surface and the PHB-phasin-signal peptide complex is targeted for type I secretion. B. pCMEL3 plasmid consisting of phaC, phaC, and phaB genes from pBHR68. pCMEL3 also contains the genes needed for phasin-HylA production.
Cellular fractionation and western blotting
Overnight cultures of E. coli BL21-Gold (DE3) harboring pCMEL1 and pLG575 were used to inoculate (1:100 v/v dilution) 100 ml of LB media containing 25 μg/ml chloramphenicol (Acros Organics, Fair Lawn, NJ) and 50 μg/ml ampicillin (IBI Scientific, Peosta, IA) [50]. The lysozyme/EDTA/osmotic shock and chloroform-based cellular fractionation procedures and methods for analyzing supernatant fractions are previously discussed in Linton et al. 2012 [35]. Briefly, 50 mL of cell culture were centrifuged and the pellet was resuspended in a buffer and subjected to osmotic shock. Centrifugation was used to separate the periplasm from spheroplast. Cytoplasm and membranes were fractionated via ultracentrifugation.
Approximately 40 μg of protein from each of the respective fractioned samples were separated using precast 12% SDS-polyacrylamide tris-glycine gels (Jule Inc., Milford, CT). Electrophoresis operating conditions were used as specified by the manufacturer. Phasin immunoblotting was carried out using a PhaP1 specific antibody [51,52]. An Anti-Rabbit IgG HRP conjugated secondary antibody was purchased from Promega (Madison, WI). Respective primary and secondary antibody concentrations of 1:50,000 v/v and 1:2,500 v/v were used.
PHB secretion studies
All secretion studies were carried out in triplicate. Secretion studies were conducted with the secreting system (pCMEL3 + pLG575 in XL1 Blue) and non-secreting system (pBHR68 + pLG575 in XL1 Blue). The non-secreting system does not contain phasin-HlyA.
Media formulation and growth conditions
R. eutropha was cultured following the methods outlined in Linton et al. 2012 [53]. M9 salts (Becton, Dickinson and Co, Sparks, MD) supplemented with 1.5% (w/v) glucose (ACS grade, Acros Organics, Fair Lawn, NJ) and 0.2% (w/v) yeast extract (Becton, Dickinson and Co, Sparks, MD) was used for PHB secretion studies in E. coli[38]. Overnight E. coli cultures harboring specific plasmids were inoculated from freezer stocks into 5 ml of M9 media with chloramphenicol (34 μg/ml), ampicillin (50 μg/ml), and grown in an orbital shaker table at 220 rpm at 37°C. Overnight cultures were then used to seed larger 250 ml flasks (50 ml media volume) at an initial optical density (OD600) of 0.05 at time 0 h. 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Gold Biotechnology, Inc. St. Louis, MO) was added to each flask at time 0 h. Flasks were removed at 24, and 48 h and analyzed for PHB. CFU/mL was measured at time points 0, 4, 6, 8, 12, and 24 h.
Recovery of secreted PHB
PHB granules can agglomerate together naturally and previous studies have demonstrated the use of Calcium chloride (CaCl2) to enhance this process. A study by Fidler et al. used CaCl2 to purify PHB from lysed cells by selectively aggregating PHB granules. It was observed that PHB granules fell to the bottom of the test tube after addition of CaCl2[54]. Another CaCl2 method for PHB recovery by Resch et al. used a low speed centrifugation step to further enhance PHB recovery from cell debris [55].
In this study, techniques for secreted PHB recovery were adapted from the methods outlined in [55]. At 24, and 48 h 0.01 M CaCl2 (final concentration, Avantor Performance Materials, Inc. Center Valley, PA) was added to the bacterial culture and mixed by inverting the tube several times. The tubes were then allowed to sit for 10 mins at room temperature and then centrifuged at 54 × g for 5 min. The supernatant was removed and transferred to a fresh tube and the pellet was freeze dried. The supernatant was centrifuged at 3452 × g for 10 mins and the pellet was freeze dried. The pellet from the first centrifugation contained secreted PHB with CaCl2 and the pellet from the second centrifugation contained bacterial mass and non-secreted PHB. Secretion studies and PHB analysis were conducted in triplicate.
PHB concentration determination
PHB concentrations were determined based on a NMR-GC method described in Linton et al. 2012 [53]. Briefly, samples were lyophilized after which approximately 15 mg of sample was mixed with equal volumes of sodium hypochlorite and deuterated chloroform. Samples were centrifuged and 1H NMR was carried out on the PHB fraction. PHB concentration was determined from a NMR-GC standard.
Scanning electron microscopy (SEM)
To visually show PHB secretion from E. coli, SEM was performed. SEM protocols were used as mentioned in [56]. Briefly, secreting and non-secreting strains were grown up overnight and fixed onto glass cover slips. Samples were mounted on aluminum stubs and sputter coated with 10 nm gold. SEM was carried out using a Hitachi S4000 SEM.
Statistical analysis
All growth (CFU/mL) and PHB yield studies were carried out in triplicate to show consistency of data. Statistical analysis was conducted with Statistical Analysis Software (SAS 9.3, SAS Institute Inc., Cary, NC). A two-way analysis of variance (ANOVA) with tukey post hoc comparison performed on significant results (confidence level 95%).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AR and EL conducted experiments and wrote the manuscript draft. CDM conceived and designed the experiments and wrote the manuscript. AH assisted with experiments and data analysis. RCS assisted with experimental design and manuscript revision. All authors read and approved the final manuscript.
Contributor Information
Asif Rahman, Email: asif.rahman@aggiemail.usu.edu.
Elisabeth Linton, Email: libbielinton@gmail.com.
Alex D Hatch, Email: a.hatch@aggiemail.usu.edu.
Ronald C Sims, Email: ron.sims@usu.edu.
Charles D Miller, Email: charles.miller@usu.edu.
Acknowledgements
The authors would also like to acknowledge FenAnn Shen and T.C. Shen in the Physics Department at Utah State University for assisting with the SEM. Marie K. Walsh in Nutrition, Dietetics and Food Sciences at Utah State University helped with phasin translocation studies. The authors would like to thank I. Barry Holland and Thorsten Jumpertz for the pLG575 plasmid. The authors would also like to acknowledge the gift of the primary phasin antibody from Anthony J. Sinskey and JoAnne Stubbe. This research was supported by the Utah State University Synthetic Bio-Manufacturing Center (SBC) and Utah Science Technology and Research initiative (USTAR).
References
- Philp JC, Ritchie RJ, Guy K. Biobased plastics in a bioeconomy. Trends Biotechnol. 2013;7:65–67. doi: 10.1016/j.tibtech.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Keshavarz T, Roy I. Polyhydroxyalkanoates: bioplastics with a green agenda. Curr Opin Microbiol. 2010;7:321–326. doi: 10.1016/j.mib.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Dias JML, Lemos PC, Serafim LS, Oliveira C, Eiroa M, Albuquerque MGE, Ramos AM, Oliveira R, Reis MAM. Recent advances in polyhydroxyalkanoate production by mixed aerobic cultures: from the substrate to the final product. Macromol Biosci. 2006;7:885–906. doi: 10.1002/mabi.200600112. [DOI] [PubMed] [Google Scholar]
- Madison LL, Huisman GW. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev. 1999;7:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L, Fei Q, Zhang YH, Wang XZ, Fan DD, Chang HN. Thermal properties and biodegradability studies of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) Journal of Polymers and the Environment. 2012;7:23–28. doi: 10.1007/s10924-011-0362-9. [DOI] [Google Scholar]
- Mergaert J, Anderson C, Wouters A, Swings J. Microbial degradation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in compost. Journal of Environmental Polymer Degradation. 1994;7:177–183. doi: 10.1007/BF02067443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D, Handrick R. Microbial degradation of polyhydroxyalkanoates. Annu Rev Microbiol. 2002;7:403–432. doi: 10.1146/annurev.micro.56.012302.160838. [DOI] [PubMed] [Google Scholar]
- Jendrossek D, Schirmer A, Schlegel HG. Biodegradation of polyhydroxyalkanoic acids. Appl Microbiol Biotechnol. 1996;7:451–463. doi: 10.1007/s002530050844. [DOI] [PubMed] [Google Scholar]
- Agnew DE, Pfleger BF. Synthetic biology strategies for synthesizing polyhydroxyalkanoates from unrelated carbon sources. Chemical Engineering Science. 2012. In press. Available online, ISSN 0009-2509, http://dx.doi.org/10.1016/j.ces.2012.12.023. [DOI] [PMC free article] [PubMed]
- Rehm BHA. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol. 2010;7:578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- Chanprateep S. Current trends in biodegradable polyhydroxyalkanoates. J Biosci Bioeng. 2010;7:621–632. doi: 10.1016/j.jbiosc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Akaraonye E, Keshavarz T, Roy I. Production of polyhydroxyalkanoates: the future green materials of choice. J Chem Technol Biotechnol. 2010;7:732–743. doi: 10.1002/jctb.2392. [DOI] [Google Scholar]
- Gumel AM, Annuar MSM, Chisti Y. Recent advances in the production, recovery and applications of polyhydroxyalkanoates. J Polym and the Environ. 2013;7:580–605. doi: 10.1007/s10924-012-0527-1. [DOI] [Google Scholar]
- Numata K, Doi Y. Dual biosyntheses of poly[(R)-3-hydroxybutyric acid] and silk protein for the fabrication of biofunctional bioplastic. Polymer Journal. 2011;7:642–647. doi: 10.1038/pj.2011.27. [DOI] [Google Scholar]
- Xu C, Qiu Z. Crystallization behavior and thermal property of biodegradable poly(3-hydroxybutyrate)/multi-walled carbon nanotubes nanocomposite. Polymers for Advanced Technologies. 2011;7:538–544. doi: 10.1002/pat.1540. [DOI] [Google Scholar]
- Spiekermann P, Rehm BHA, Kalscheuer R, Baumeister D, Steinbüchel A. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch Microbiol. 1999;7:73–80. doi: 10.1007/s002030050681. [DOI] [PubMed] [Google Scholar]
- Grage K, Jahns AC, Parlane N, Palanisamy R, Rasiah IA, Atwood JA, Rehm BHA. Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules. 2009;7:660–669. doi: 10.1021/bm801394s. [DOI] [PubMed] [Google Scholar]
- Rehm BHA. Polyester synthases: natural catalysts for plastics. Biochem J. 2003;7:15–33. doi: 10.1042/BJ20031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pötter M, Müller H, Reinecke F, Wieczorek R, Fricke F, Bowien B, Friedrich B, Steinbüchel A. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in ralstonia eutropha. Microbiology. 2004;7:2301–2311. doi: 10.1099/mic.0.26970-0. [DOI] [PubMed] [Google Scholar]
- Jacquel N, Lo C-W, Wei Y-H, Wu H-S, Wang SS. Isolation and purification of bacterial poly(3-hydroxyalkanoates) Biochem Eng J. 2008;7:15–27. doi: 10.1016/j.bej.2007.11.029. [DOI] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. 2007;7:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;7:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou A, Christie PJ, Fernandez RC, Palmer T, Plano GV, Pugsley AP. Secretion by numbers: protein traffic in prokaryotes. Mol Microbiol. 2006;7:308–319. doi: 10.1111/j.1365-2958.2006.05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvaux M, Hébraud M, Talon R, Henderson IR. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 2009;7:139–145. doi: 10.1016/j.tim.2009.01.004. [DOI] [PubMed] [Google Scholar]
- York GM, Stubbe J, Sinskey AJ. New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J Bacteriol. 2001;7:2394–2397. doi: 10.1128/JB.183.7.2394-2397.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbuchel A, Aerts K, Babel W, Follner C, Kiebergesell M, Madkour MH, Mayer F, Pieper-Furst U, Pries A, Valentin HE, Wieczorek R. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can J Microbiol. 1995;7:94–105. doi: 10.1139/m95-175. [DOI] [PubMed] [Google Scholar]
- Giddings G, Allison G, Brooks D, Carter A. Transgenic plants as factories for biopharmaceuticals. Nat Biotechnol. 2000;7:1151–1155. doi: 10.1038/81132. [DOI] [PubMed] [Google Scholar]
- Maehara A, Ueda S, Nakano H, Yamane T. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J Bacteriol. 1999;7:2914–2921. doi: 10.1128/jb.181.9.2914-2921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek R, Pries A, Steinbuchel A, Mayer F. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol. 1995;7:2425–2435. doi: 10.1128/jb.177.9.2425-2435.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergulhão FJM, Summers DK, Monteiro GA. Recombinant protein secretion in escherichia coli. Biotechnol Adv. 2005;7:177–202. doi: 10.1016/j.biotechadv.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Holland IB, Schmitt L, Young J. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway. Mol Membr Biol. 2005;7:29–39. doi: 10.1080/09687860500042013. [DOI] [PubMed] [Google Scholar]
- Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;7:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- Kenny B, Haigh R, Holland IB. Analysis of the haemolysin transport process through the secretion from Escherichia coli of PCM, CAT or β-galactosidase fused to the Hly C-terminal signal domain. Mol Microbiol. 1991;7:2557–2568. doi: 10.1111/j.1365-2958.1991.tb02102.x. [DOI] [PubMed] [Google Scholar]
- Mackman N, Baker K, Gray L, Haigh R, Nicaud JM, Holland IB. Release of a chimeric protein into the medium from Escherichia coli using the C-terminal secretion signal of haemolysin. EMBO J. 1987;7:2835–2841. doi: 10.1002/j.1460-2075.1987.tb02580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton E, Walsh MK, Sims RC, Miller CD. Translocation of green fluorescent protein by comparative analysis with multiple signal peptides. Biotechnol J. 2012;7:667–676. doi: 10.1002/biot.201100158. [DOI] [PubMed] [Google Scholar]
- Fernández LA, De Lorenzo V. Formation of disulphide bonds during secretion of proteins through the periplasmic-independent type I pathway. Mol Microbiol. 2001;7:332–346. doi: 10.1046/j.1365-2958.2001.02410.x. [DOI] [PubMed] [Google Scholar]
- Lee SY, Yim KS, Chang HN, Chang YK. Construction of plasmids, estimation of plasmid stability, and use of stable plasmids for the production of poly(3-hydroxybutyric acid) by recombinant Escherichia coli. J Biotechnol. 1994;7:203–211. doi: 10.1016/0168-1656(94)90183-X. [DOI] [PubMed] [Google Scholar]
- Kang Z, Wang Q, Zhang HJ, Qi QS. Construction of a stress-induced system in Escherichia coli for efficient polyhydroxyalkanoates production. Appl Microbiol Biotechnol. 2008;7:203–208. doi: 10.1007/s00253-008-1428-z. [DOI] [PubMed] [Google Scholar]
- Choi JI, Lee SY. Efficient and economical recovery of poly(3-hydroxybutyrate) from recombinant Escherichia coli by simple digestion with chemicals. Biotechnol Bioeng. 1999;7:546–553. doi: 10.1002/(SICI)1097-0290(19990305)62:5<546::AID-BIT6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Ramachander TVN, Rohini D, Belhekar A, Rawal SK. Synthesis of PHB by recombinant E. coli harboring an approximately 5 kb genomic DNA fragment from Streptomyces aureofaciens NRRL 2209. Int J Biol Macromol. 2002;7:63–69. doi: 10.1016/S0141-8130(02)00068-5. [DOI] [PubMed] [Google Scholar]
- Molitoris HP, Moss ST, De Koning GJM, Jendrossek D. Scanning electron microscopy of polyhydroxyalkanoate degradation by bacteria. Appl Microbiol Biotechnol. 1996;7:570–579. doi: 10.1007/s002530050863. [DOI] [Google Scholar]
- Peters V, Rehm BHA. In vivo monitoring of PHA granule formation using GFP-labeled PHA synthases. FEMS Microbiol Lett. 2005;7:93–100. doi: 10.1016/j.femsle.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Peters V, Becher D, Rehm BHA. The inherent property of polyhydroxyalkanoate synthase to form spherical PHA granules at the cell poles: The core region is required for polar localization. J Biotechnol. 2007;7:238–245. doi: 10.1016/j.jbiotec.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Idempotent vector design for standard assembly of biobricks. http://hdl.handle.net/1721.1/21168.
- A New biobrick assembly strategy designed for facile protein engineering. http://hdl.handle.net/1721.1/32535.
- Anderson JC, Dueber JE, Leguia M, Wu GC, Arkin AP, Keasling JD. BglBricks: a flexible standard for biological part assembly. J Biol Eng. 2010;7:1. doi: 10.1186/1754-1611-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackman N, Nicaud JM, Gray L, Holland IB. Genetical and functional organisation of the Escherichia coli haemolysin determinant 2001. Molecular and General Genetics. 1985;7:282–288. doi: 10.1007/BF00425672. [DOI] [PubMed] [Google Scholar]
- Jobling MG, Holmes RK. Construction of vectors with the p15a replicon, kanamycin resistance, inducible lacZα and pUC18 or pUC19 multiple cloning sites. Nucleic Acids Res. 1990;7:5315–5316. doi: 10.1093/nar/18.17.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Registry of standard biological parts. http://partsregistry.org/
- Sambrook J, Russel DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- York GM, Junker BH, Stubbe J, Sinskey AJ. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J Bacteriol. 2001;7:4217–4226. doi: 10.1128/JB.183.14.4217-4226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, He A, Lawrence AG, Liu P, Watson N, Sinskey AJ, Stubbe J. Analysis of transient polyhydroxybutyrate production in Wautersia eutropha H16 by quantitative Western analysis and transmission electron microscopy. J Bacteriol. 2005;7:3825–3832. doi: 10.1128/JB.187.11.3825-3832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton E, Rahman A, Viamajala S, Sims RC, Miller CD. Polyhydroxyalkanoate quantification in organic wastes and pure cultures using a single-step extraction and 1H NMR analysis. Water Sci Technol. 2012;7:1000–1006. doi: 10.2166/wst.2012.273. [DOI] [PubMed] [Google Scholar]
- Fidler S, Dennis D. Polyhydroxyalkanoate production in recombinant Escherichia coli. FEMS Microbiol Rev. 1992;7:231–235. doi: 10.1111/j.1574-6968.1992.tb05842.x. [DOI] [PubMed] [Google Scholar]
- Resch S, Gruber K, Wanner G, Slater S, Dennis D, Lubitz W. Aqueous release and purification of poly([beta]-hydroxybutyrate) from Escherichia coli. J Biotechnol. 1998;7:173–182. doi: 10.1016/S0168-1656(98)00127-8. [DOI] [PubMed] [Google Scholar]
- Mortensen T, Shen S, Shen F, Walsh MK, Sims RC, Miller CD. Investigating the effectiveness of St John’s wort herb as an antimicrobial agent against mycobacteria. Phytother Res. 2012;7:1327–1333. doi: 10.1002/ptr.3716. [DOI] [PubMed] [Google Scholar]


