Abstract
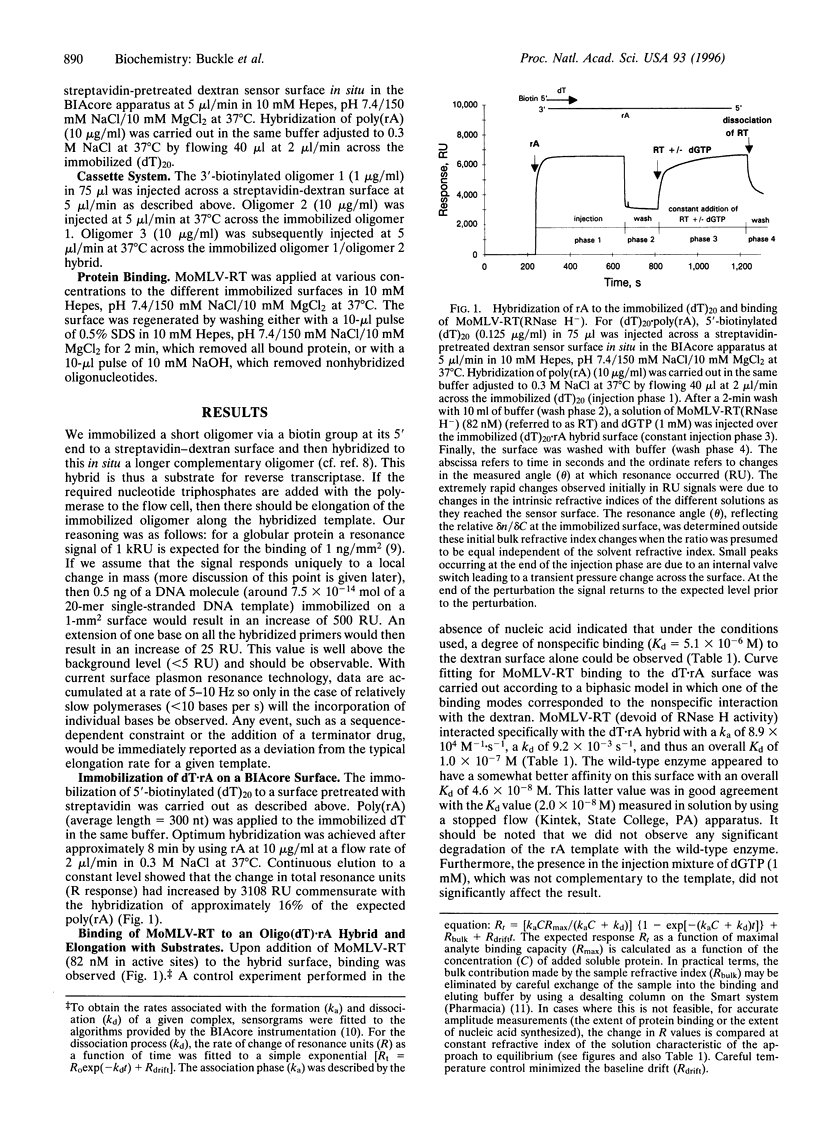
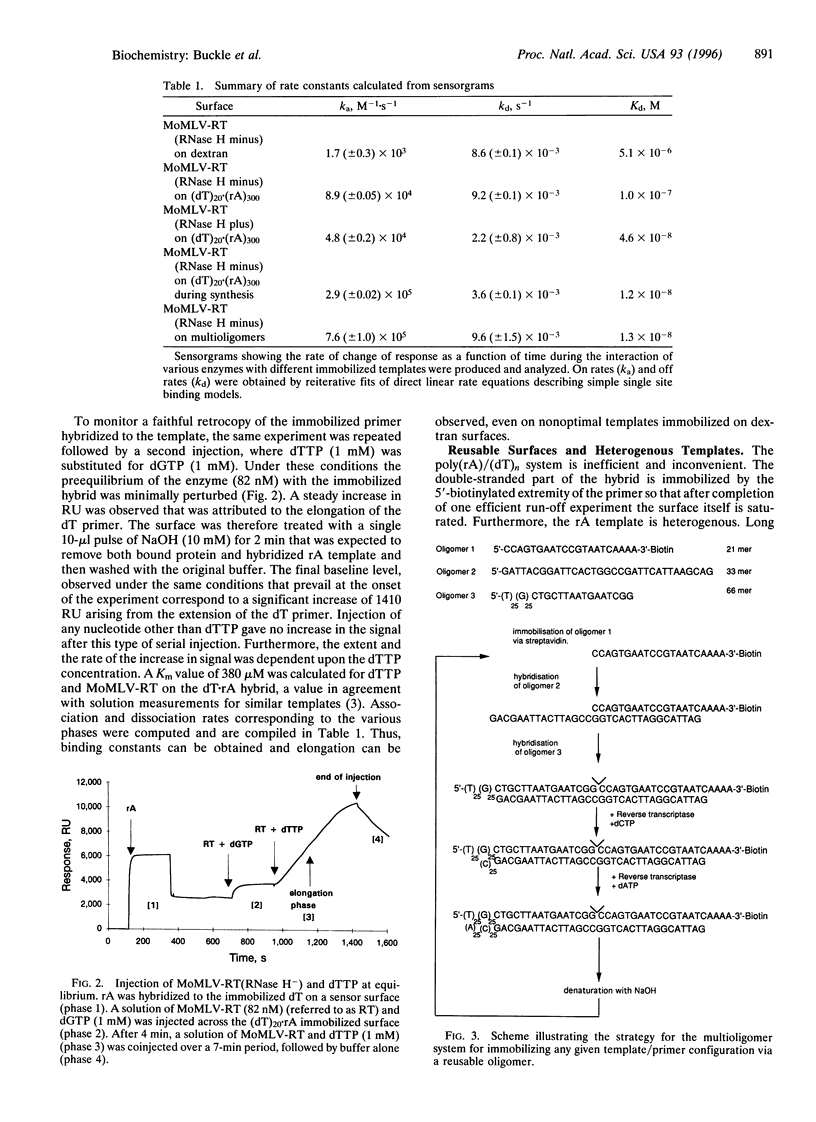
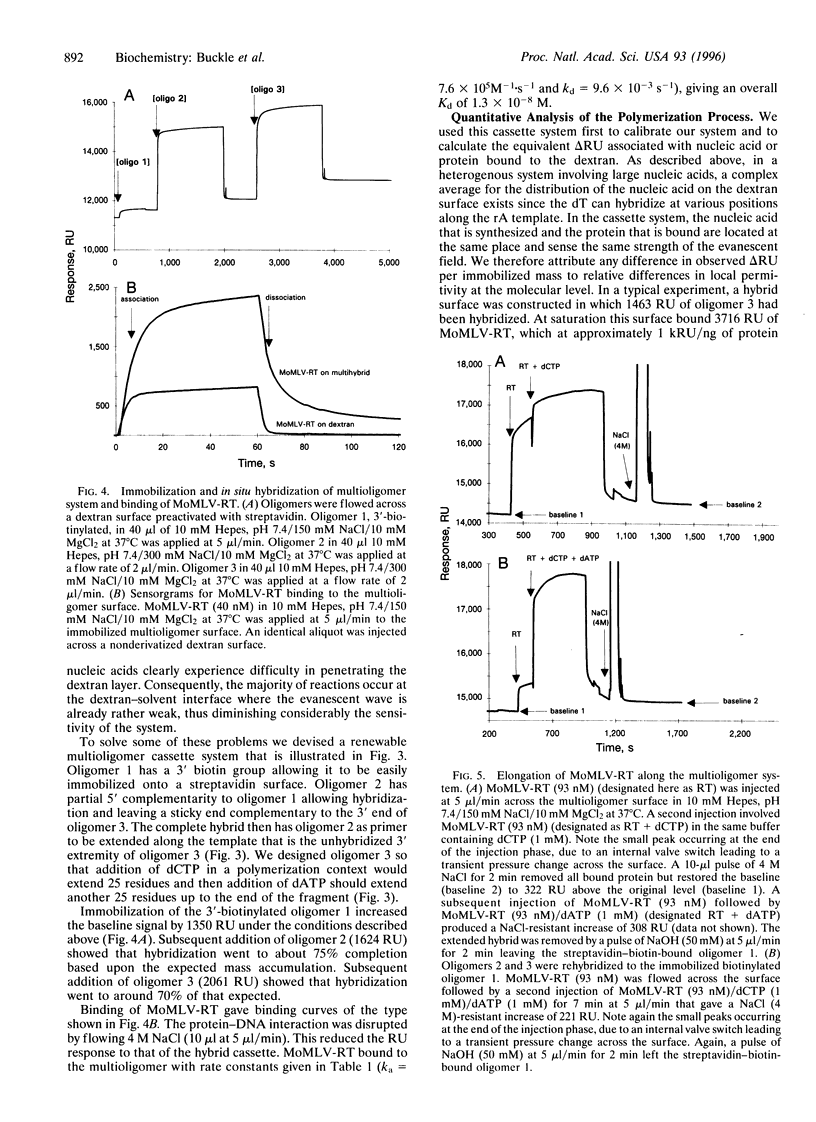
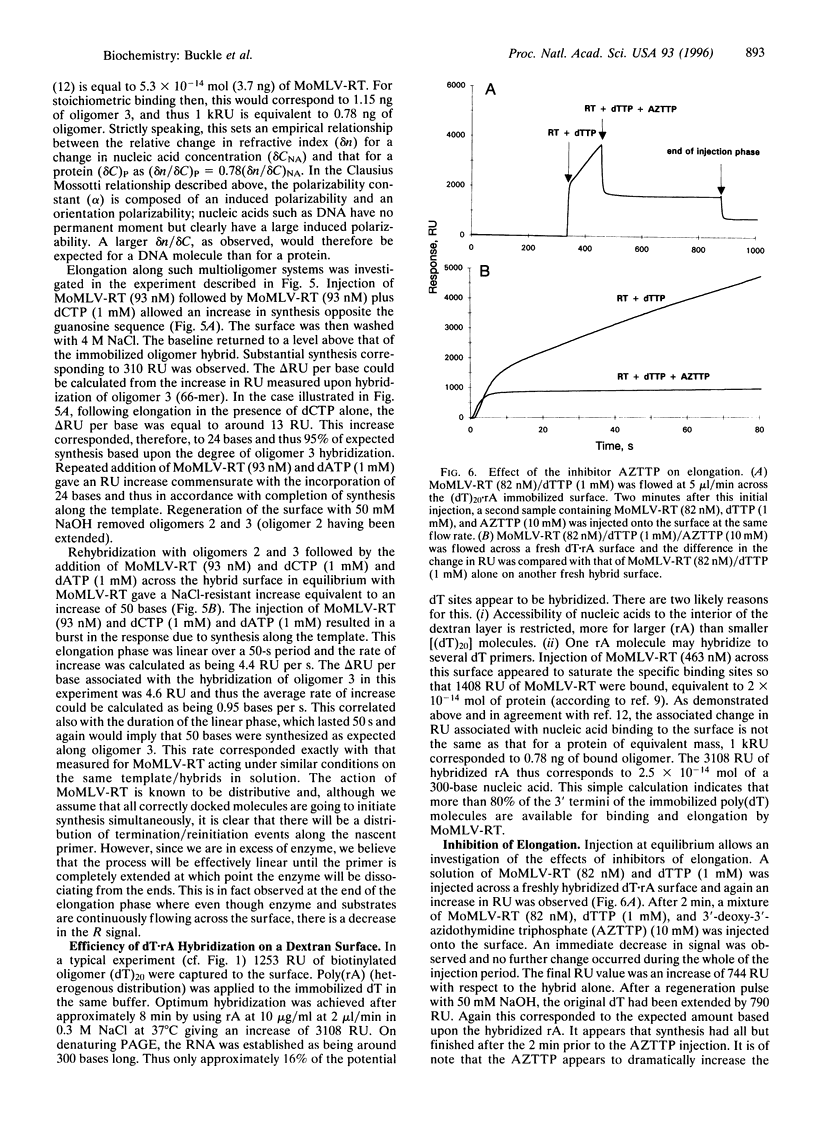
A rapid direct assay for polymerase-induced elongation along a given template is an obligate requirement for understanding the processivity of polymerization and the mode of action of drugs and inhibitors on this process. Surface plasmon resonance can be used to follow the association and the dissociation rates of a given reverse transcriptase on DNA.RNA and DNA.DNA hybrids immobilized on a biotin-streptavidin surface. The addition of nucleotides complementary to the template strand produces an increase in the local mass, as deduced from an increase in the measured signal, due to elongation of the primer strand that allows an estimation of both the extent and rate of the polymerization process. The terminator drug 3'-deoxy-3'-azidothymidine triphosphate completely abolishes the increase in signal as would be expected from an inhibition of elongation. This technique provides a sensitive assay for the affinities of different polymerases for specific templates and for the effects of terminators of the elongation process.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bondeson K., Frostell-Karlsson A., Fägerstam L., Magnusson G. Lactose repressor-operator DNA interactions: kinetic analysis by a surface plasmon resonance biosensor. Anal Biochem. 1993 Oct;214(1):245–251. doi: 10.1006/abio.1993.1484. [DOI] [PubMed] [Google Scholar]
- Fisher R. J., Fivash M., Casas-Finet J., Erickson J. W., Kondoh A., Bladen S. V., Fisher C., Watson D. K., Papas T. Real-time DNA binding measurements of the ETS1 recombinant oncoproteins reveal significant kinetic differences between the p42 and p51 isoforms. Protein Sci. 1994 Feb;3(2):257–266. doi: 10.1002/pro.5560030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H. E., McCoy J. M., Seehra J. S., Richardson C. C. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J Biol Chem. 1989 Mar 15;264(8):4669–4678. [PubMed] [Google Scholar]
- Kati W. M., Johnson K. A., Jerva L. F., Anderson K. S. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992 Dec 25;267(36):25988–25997. [PubMed] [Google Scholar]
- Malmqvist M. Biospecific interaction analysis using biosensor technology. Nature. 1993 Jan 14;361(6408):186–187. doi: 10.1038/361186a0. [DOI] [PubMed] [Google Scholar]
- Nice E., Lackmann M., Smyth F., Fabri L., Burgess A. W. Synergies between micropreparative high-performance liquid chromatography and an instrumental optical biosensor. J Chromatogr A. 1994 Feb 4;660(1-2):169–185. doi: 10.1016/0021-9673(94)85110-7. [DOI] [PubMed] [Google Scholar]
- O'Shannessy D. J., Brigham-Burke M., Soneson K. K., Hensley P., Brooks I. Determination of rate and equilibrium binding constants for macromolecular interactions using surface plasmon resonance: use of nonlinear least squares analysis methods. Anal Biochem. 1993 Aug 1;212(2):457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
- Oyama F., Kikuchi R., Crouch R. J., Uchida T. Intrinsic properties of reverse transcriptase in reverse transcription. Associated RNase H is essentially regarded as an endonuclease. J Biol Chem. 1989 Nov 5;264(31):18808–18817. [PubMed] [Google Scholar]
- Ricchetti M., Buc H. Reverse transcriptases and genomic variability: the accuracy of DNA replication is enzyme specific and sequence dependent. EMBO J. 1990 May;9(5):1583–1593. doi: 10.1002/j.1460-2075.1990.tb08278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



