Abstract
The common CHRNA5 mis-sense coding single nucleotide polymorphism (SNP) rs16969968:G>A (D398N) has been shown repeatedly to confer risk for heavy smoking in individuals who carry the ‘A’ allele (encoding the 398N amino acid). The mis-sense SNP has a minor allele frequency (MAF) of ~40% in European-Americans, but only ~7% in African-Americans (http://www.ncbi.nlm.nih.gov/projects/SNP/). We reasoned that there might be other mis-sense variants among African-Americans that could confer the heavy smoking phenotype (defined here as ≥20 cigarettes per day), perhaps in a similar manner to that of the D398N polymorphism in Europeans. As such, we re-sequenced 250 African-American heavy smokers, most of whom were homozygous ‘G’ at rs16969968:G>A (MAF of 9.6% within the population). Although many novel coding SNPs were not observed, we report an interesting, although rare (perhaps personal) variant in CHRNA5 that could result in nonsense-mediated decay of the aberrant transcript.
Keywords: mutation, nicotinic acetylcholine receptor, nAChR, nicotine dependence
Introduction
Numerous genome-wide studies and meta-analyses conducted in multiple ethnic groups have identified genes thought to be responsible for smoking related measures and behaviors, including smoking quantity (cigarettes smoked per day: CPD) and the Fagerström Test for Nicotine Dependence (FTND) (Bierut et al., 2007; Berrettini et al., 2008; Vink et al., 2009; TAG 2010; Thorgeirsson et al., 2010; Liu et al., 2010; Li et al., 2006; Li et al., 2008; David et al., 2012; Chen et al., 2012; Rice et al., 2012). In addition, targeted association studies have identified the CHRNA5-A3-B4 gene cluster as being associated with nicotine dependence in various ethnic populations (Saccone et al., 2007, 2009, 2009b; Li et al., 2010, 2010b). The missense polymorphism rs16969968:G>A in CHRNA5 is thought to confer risk for heavy smoking in people of European ancestry as it encodes an amino acid change (D398N) that leads to hypo-functionality of (α4β2)2α5 nAChRs (Bierut et al, 2008; Kuryatov et al., 2011; Tammimäki et al, 2012). However, rs16969968:G>A has a low minor allele frequency (MAF) in populations other than those of European descent (Bierut et al., 2008) and the linkage disequilibrium block that includes this SNP extends upstream and downstream of the CHRNA5 gene itself (Saccone et al, 2010). We hypothesized that African-American heavy smokers might have different common (MAF>1%) or rare (MAF<1%) variants in CHRNA5 that would associate with the nicotine addiction phenotype, as does rs16969968:G>A in Europeans.
Previous re-sequencing studies have been done to assess the contribution of rare variants in the CHRNA5/A3/B4 gene cluster to the etiologies of various disorders, such as megacystis-microcolon-hypoperistalsis syndrome (MMIHS – A3/B4) (Lev-Lehman et al, 2001), autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE – A5/A3/B4) (Duga et al, 2001) and sporadic amyotrophic lateral sclerosis (SALS – A3/B4) (Sabatelli et al, 2009). Whereas these prior studies were mainly on people of European descent, Rana et al (2009) re-sequenced the CHRNA5/A3/B4 genes in 34 Caucasian-American and 30 African-American individuals seeking to discover natural variation in these genes that might explain heritable autonomic traits. More recent CHRNA5/A3/B4 re-sequencing efforts have studied the relationship between common and rare variants in these genes and their association with the Fagerström Test for Nicotine Dependence (Wessel et al, 2010; Haller et al, 2012). Whereas Wessel et al (2010) re-sequenced European-Americans, re-sequencing by Haller et al (2012) included 400 European-Americans and 352 African-Americans; half of each sequenced population being nicotine dependent cases or smoking controls. Here, we re-sequenced the coding regions of the CHRNA5 gene (GenBank: NM_000745.3) in an independent population of 250 African-American heavy smokers (defined as people who smoked 20 or more cigarettes per day). In most cases, rare variants (MAF<1%) in CHRNA5 are unlikely to explain the heavy smoking phenotype. Nonetheless, we identified a rare frame-shift variant that might result in nonsense mediated decay (NMD) of the aberrant transcript.
Materials and Methods
Ethics Statement
The NIMH and NIDA Genetics Initiative samples were collected by consortia (see Acknowledgments) with written informed consent and were purchased from the Rutgers University Cell & DNA Repository (RUCDR) before use in this study.
Study Subjects
We screened the NIMH Genetics Initiative African-American control population for their self-report of maximal CPD smoked (questionnaire subsection M3B). Using a 20 CPD cut-off for inclusion in the CHRNA5 re-sequencing project yielded 250 African-American heavy smokers. The African-American cocaine (N=487) and opiate (N=325) addicted populations from the NIDA Genetics Initiative were obtained from the RUCDR. These de-identified genomic DNA (gDNA) samples were used to determine the frequency of the exon 2 deletion in a larger, independent African-American population. Ethnicity was by self-report.
CHRNA5 re-sequencing and genotyping
Amplicons of CHRNA5 exons (1–6) were verified by agarose gel electrophoresis prior to Sanger sequencing. Sequencher 4.9 (GeneCodes Corporation, Ann Arbor, MI, USA) auto-called common polymorphisms and rare polymorphisms were called using the ‘call secondary peaks’ command with secondary peak height set to 45%. Initial quality control was by visual inspection of chromatogram peaks with additional verification done by sequencing the opposite strands of amplicons, by sequencing a second amplicon from the same individual or by TaqMan® allelic discrimination assays (Life Technologies, Grand Island, NY, USA). The frame-shift deletion in CHRNA5 exon 2 was verified by TA-cloning of amplicons into pCRII-TOPO vector (Life Technologies), transforming E. coli, purifying plasmid DNA and fully sequencing both alleles. TaqMan® assays used were: C_1502678_10 (rs2036527:G>A), C_26000428_20 (rs16969968:G>A) and custom assays for the exon 2 frame-shift deletion, rs80087508:A>G and rs79109919:T>A. TaqMan® assays were cycled in a 9700 PCR machine under standard parameters in 384 well plates with 4 ng gDNA per 5 μl reaction, post-read into SDS 2.2.2 on a 7900HT and final genotypes called automatically using TaqMan® Genotyper (Life Technologies) prior to export into Excel for recoding and analysis using Haploview 4.2 (Barrett et al., 2005).
Haplotype analysis
Haploview 4.2 (Barrett et al., 2005) was used to build haplotypes, calculate linkage disequilibrium (LD) and generate statistics (expectation maximization) on single markers and haplotypes determined by genotypes at various loci within CHRNA5 in the re-sequenced African-American population (Figure 2), and to calculate LD between SNPs using version 3, release 27 of the CEU+TSI (European descent), CHB+JPT (Asian descent) and ASW (African-American) HapMap data sets for the region encompassing CHRNA5.
Figure 2. Haplotype and Linkage Disequilibrium Structure of Variants in CHRNA5 of African-American Heavy Smokers.
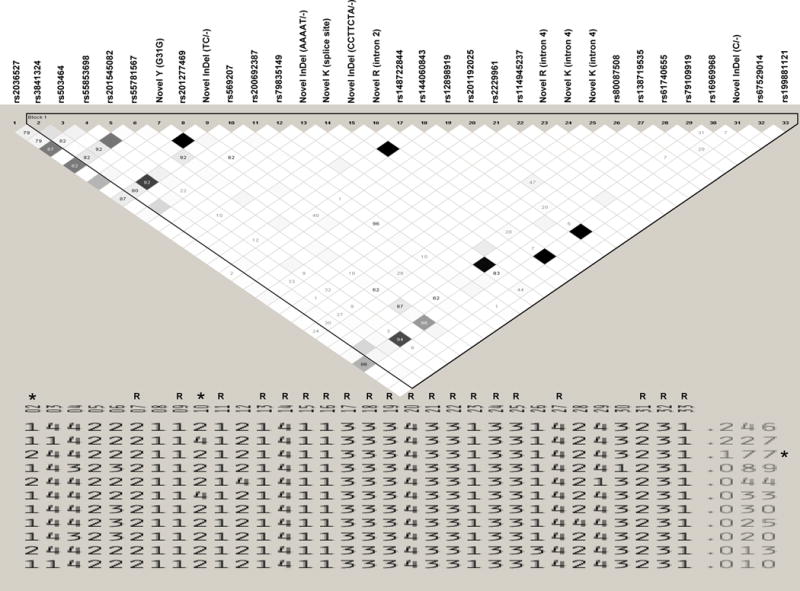
The linkage disequilibrium (LD) and haplotype block structures containing the indicated SNPs are shown. Analysis was done using Haploview 4.2 (Barrett et al., 2005) using a solid spine of LD. The color scheme of the LD plot is for r2 whereby white indicates r2=0 and black indicates r2=1. Numbers within the shaded squares are those for D′. Haplotypes with frequencies (indicated to the right of the haplotypes) of less than 1% in the population are not shown (see Supplemental Figure S1 for all other haplotypes). Unless otherwise indicated, SNPs are coded in the traditional manner whereby A=1, C=2, G=3 and T=4. An “R” above a SNP number indicates that it is a “rare” variant which may appear monomorphic, but see Supplemental Figure S1. Values for rs2036527:G>A (SNP #1) are not included in the haplotypes as it resides in a different block than the other SNPs examined. Values for rs3841324:GGGCGGGGCCAGAGGGAAATAG>- (SNP #2), which is a 22 bp deletion in the CHRNA5 promoter (Buckland et al., 2005), have been set to “1” for “normal” or “2” for “deletion”. SNP #8 (rs201277469) is coded such that “no insertion”=1 and “T-insertion”=4. SNP #9 (Novel InDel (intron 1)) is coded such that “TC”=1 and “deletion of TC”=2. The 5 bp deletion variant in CHRNA5 exon 2 (SNP #13, coded “1” for “no deletion” and “2” for “deletion”) falls within a rare haplotype (f=0.003) containing the normal promoter (SNP #2) that is not shown in Figure 2, but see Supplemental Figure S1. SNP #15 (Novel InDel (intron 2)) is coded such that “CCTTCTA”=1 and “deletion of CCTTCTA”=2. SNP #31 (Novel InDel (intron 5)) is coded such that “C”=2 and “deletion of C”=1. The asterisks above the markers (SNP #2 and SNP #10) or to the right of the haplotype (f=0.177) indicate that the marker or haplotype showed a nominal trend (p<0.1) toward statistical significance in our exploratory analysis based on CPD.
Results
Sequencing results are given in Table 1. One novel rare variant, a 5 bp deletion in exon 2 (Figure 1), was identified in CHRNA5 of one African-American heavy smoker. The resultant frame-shift causes premature stop codons in exons 3 and 4 (Figure 1), which, in theory, would cause the aberrant transcript to undergo NMD. The individual smoked 30 CPD, is homozygous ‘G’ at rs16969968:G>A for D398N, and has neither the promoter deletion rs3841324:GGGCGGGGCCAGAGGGAAATAG>- nor the rs79109919:T>A (L363Q) or rs80087508:A>G (K167R) mis-sense variants. Additionally, this individual is homozygous ‘G’ at rs2036527:G>A; the only SNP to achieve genome-wide significance in a meta-analysis of African-American populations based on CPD (David et al., 2012). No other sequenced or genotyped African-American individuals had the exon 2 deletion, suggesting it is either very rare among African-Americans or is a personal variant (Table 1). A ‘frame-shift’ variant at this position in CHRNA5 exon 2 is noted in one African-American in the National Heart Lung and Blood Institute Exome Sequencing Project (NHLBI_ESP) (Exome Variant Server, 2012). Given the rarity of this variant, it is entirely possible, but not necessarily the case, that both studies re-sequenced the same individual with this polymorphism.
Table 1.
Novel and Known Simple Nucleotide Variations (SNV) Identified in CHRNA5 of African-American Heavy Smokers.d
| Marker# (Figure 2) | SNV ID | SNV Locationa | Gene Region | SNV Polymorphism | Minor Allele Freqs. (Minor alleles/Total alleles) | Type of Change | Amino Acid Change | PolyPhen2 |
|---|---|---|---|---|---|---|---|---|
| 1 | rs2036527 | Chr15:78851615 | distal promoter | G>A | 0.243 (116/478) | |||
| 2 | rs3841324 | Chr15:78857813–78857834 | proximal promoter | GGGCGGGGCCAGAGGGAAATAG>- | 0.25 (125/500) | |||
| 3 | rs503464 | Chr15:78857896 | 5′UTR | T>A | 0.251 (102/406) | |||
| 4 | rs55853698 | Chr15:78857939 | 5′UTR | T>G | 0.133 (54/406) | |||
| 5 | rs201545082 | Chr15:78857943 | 5′UTR | C>G | 0.0025 (1/406) | |||
| 6 | rs55781567 | Chr15:78857986 | 5′UTR | C>G | 0.222 (90/406) | |||
| 7 | Novel SNP | Chr15:78858154 | exon 1 | C>T | 0.0025 (1/406) | synonymous | G31G | N/A |
| 8 | rs201277469 | Chr15:78858281–78858282 | intron 1 | ->T-ins | 0.081 (40/492) | |||
| N/Ac | rs10709956 | Chr15:78872835 | intron 1 | A>- | NDc | |||
| N/A | rs621849 | Chr15:78872861 | intron 1 | A>G | ND | |||
| 9 | Novel Mutation | Chr15:78873006–78873007 | intron 1 | TC>- | 0.002 (1/476) | |||
| 10 | rs569207 | Chr15:78873119 | intron 1 | C>T | 0.246 (117/476) | |||
| 11 | rs200692387 | Chr15:78873147 | intron 1 | A>T | 0.002 (1/476) | polypyrimidine tract | ||
| 12 | rs79835149 | Chr15:78873220 | exon2 | C>T | 0.050 (24/476) | synonymous | Y58Y | N/A |
| 13 | Novel Mutation | Chr15:78873257–78873261 | exon 2 | AAAAT>- | 0.002 (1/484)b | frame-shift | N/A | N/A |
| 14 | Novel SNP | Chr15:78873311 | intron 2 | T>G | 0.002 (1/484) | splice site | ||
| 15 | Novel Mutation | Chr15:78873319–78873325 | intron 2 | CCTTCTA>- | 0.008 (4/484) | |||
| 16 | Novel SNP | Chr15:78878907 | intron 2 | A>G | 0.002 (1/490) | |||
| 17 | rs148722844 | Chr15:78879017 | exon 3 | G>A | 0.002 (1/490) | non-synonymous | V97I | Probably damaging |
| 18 | rs144060843 | Chr15:78879066 | intron 3 | G>T | 0.008 (4/490) | |||
| 19 | rs12898919 | Chr15:78880577 | intron 3 | G>C | 0.004 (2/488) | |||
| 20 | rs201192025 | Chr15:78880650 | intron 3 | T>C | 0.002 (1/488) | polypyrimidine tract | ||
| 21 | rs2229961 | Chr15:78880752 | exon 4 | G>A | 0.004 (2/488) | non-synonymous | V134I | Probably damaging |
| 22 | rs114945237 | Chr15:78880887 | intron 4 | G>A | 0.006 (3/488) | |||
| 23 | Novel SNP | Chr15:78880907 | intron 4 | A>G | 0.002 (1/488) | |||
| 24 | Novel SNP | Chr15:78880919 | intron 4 | G>T | 0.004 (2/488) | |||
| 25 | Novel SNP | Chr15:78880925 | intron 4 | G>T | 0.002 (1/488) | |||
| 26 | rs80087508 | Chr15:78882233 | exon 5 | A>G | 0.025 (10/398) | non-synonymous | K167R | Probably damaging |
| 27 | rs138719535 | Chr15:78882529 | exon 5 | T>A | 0.002 (1/482) | non-synonymous | F266I | Probably damaging |
| 28 | rs61740655 | Chr15:78882726 | exon 5 | C>T | 0.029 (14/488) | synonymous | T331T | N/A |
| 29 | rs79109919 | Chr15:78882821 | exon 5 | T>A | 0.05 (24/478) | non-synonymous | L363Q | Probably damaging |
| 30 | rs16969968 | Chr15:78882925 | exon 5 | G>A | 0.096 (46/478) | non-synonymous | D398N | Benign |
| 31 | Novel Mutation | Chr15:78885285 | intron 5 | C/- | 0.008 (4/474) | |||
| 32 | rs67529014 | Chr15:78885369 | intron 5 | G>C | 0.004 (2/474) | |||
| 33 | rs199881121 | Chr15:78885677 | 3′UTR | A>G | 0.002 (1/474) |
SNV locations are based upon human genome build NCBI/GRCh37.p5.
May be personal SNV - mutant allele not seen in larger AA populations genotyped (0/974 cocaine addicted and 0/650 opiate addicted)
N/A=not applicable; ND = not determinable
Due to amplification failures and/or poor sequencing of some samples, total alleles analyzed varies from 398 to 500.
Figure 1. African-American individual with a 5 bp deletion in CHRNA5 exon 2.

(A) The original sequence trace of African-American individual with the 5 bp (1 and 2/3 codon) deletion is shown. (B) The theoretical translation of the mutant allele of CHRNA5 is shown. The ‘AAAAT’ deletion mutation causes a frame-shift leading to stop codons (indicated by stars) in exons 3 and 4 of the spliced transcript. Thus, it is possible the aberrant transcript undergoes non-sense mediated decay (NMD). This frame-shift deletion variant was also found in the NHLBI_ESP. We have submitted the variant to dbSNP.
Other CHRNA5 rare variants identified in our African-American heavy smoker population (Table 1), included a novel synonymous C>T SNP in exon 1 (G31G), known rare variants in exons 3, 4 and 5 (rs148722844:G>A, V97I, rs2229961:G>A, V134I and rs138719535:T>A, F266I, respectively), and more common known variants in exon 2 (rs79831749:C>T, Y58Y) and exon 5 (rs80087508:A>G, K167R; rs61740655:C>T, T331T; rs79109919:T>A, L363Q and rs16969968:G>A, D398N). Like Haller et al. (2012), no novel polymorphisms were identified in the coding region of CHRNA5 exon 6. Whereas most SNPs detected in our study population were also detected by Haller et al. (2012), they did not report detection of the rare synonymous G31G or non-synonymous V97I SNPs in their African-American study population. The V97I variant was detected in one African-American individual in the NHLBI_ESP re-sequencing study. The individual in our study with the V97I variant also has the L363Q variant, but not the V134I, K167R, F266I or D398N variants. Valine 97 in α5 protein is invariant among species and isoleucine substitution is predicted to be “probably damaging” by PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/). Similar PolyPhen2 predictions are made for V134I, K167R, F266I and L363Q (Table 1). It remains unknown if this individual is a compound heterozygote or if both damaging variants occur on the same CHRNA5 allele.
We did an exploratory analysis in which individuals who smoked CPD<25 (N=130) or CPD≥25 (N=120), the defined “breakpoint” (whereby specificity improves from 65% (CPD=20) to 90% (CPD≥25)) for DSM-IV nicotine dependence in Europeans (Berrettini et al., 2008), were categorized as nicotine dependent ‘controls’ or ‘cases’, respectively. Even with very limited power, a nominal trend toward statistical significance was observed for two markers: rs3841324:GGGCGGGGCCAGAGGGAAATAG>- (X2=2.782, p=0.0953) and rs569207:C>T (X2=3.713, p=0.054). One haplotype (f=0.177) showed a nominal trend toward statistical significance (X2=2.954, p=0.0856) (Figure 2). No other single marker or haplotype showed a trend (p<0.1) toward statistically significant association with the CPD phenotype.
Discussion
Chrna5 knockout mice self-administer more nicotine at high doses than their wild-type littermates (Fowler et al., 2011). Similar to heterozygous Chrna5 mutant mice, the individual with the deletion in exon 2 (Figure 1) may be haplo-insufficient for α5 receptor expression due to NMD of the allele. Thus, without compensatory up-regulation of the other CHRNA5 allele (see below), this heavy smoker may have fewer α5-containing receptors than normal individuals, being able to administer more nicotine for reward, without the aversive consequences of higher nicotine doses.
Functional polymorphisms within regulatory, intronic or untranslated regions that affect α5 receptor expression levels may modulate nicotine dependence (Wang et al., 2009, 2009b; Buckland et al., 2005). Indeed, rs2036527:G>A, located at -6246 from the transcription start site of CHRNA5, obtained genome-wide statistical significance in a meta-analysis of African-American GWAS (David et al., 2012) and a meta-analysis of European GWAS identified the CHRNA5 5′UTR SNP rs55853698:T>G as having the highest statistical significance for the CPD phenotype (Liu et al., 2010). The minor allele (‘G’) of rs55853698:T>G is in moderate LD (D′=0.872, r2=0.52) with the rs2036527:G>A minor allele (‘A’) in our African-American heavy smoker population (Figure 2). Thus, rs2036527:G>A may be a proxy for greater expression of CHRNA5 in African-Americans (David et al., 2012) and other ethnic populations where LD between rs2036527:G>A and rs55853698:T>G is perfect (1kGenome_CEU: D′=1.0, r2=1.0).
A CHRNA5 promoter study, that excluded rs2036527:G>A, found significantly more reporter activity was obtained from the T-G than the T-C (rs55853698:T>G-55781567:C>G) 5′UTR haplotype (Doyle et al., 2011). Additionally, individuals with the minor allele (‘T’) at CHRNA5 intronic SNP rs588765:C>T had 2–3-fold higher CHRNA5 mRNA expression (Wang et al., 2009b). The individual with the exon 2 frame-shift is homozygous ‘T’ at rs55853698:T>G, but heterozygous at rs55781567:C>G in the CHRNA5 5′UTR, and is heterozygous at intronic SNP rs588765:C>T raising the possibility of higher expression from one of the CHRNA5 alleles. If the exon 2 frame-shift occurs on a background allele that is rs55853698:T-rs55781567:C-rs588765:C, then we cannot exclude the possibility that, although the aberrant allele may undergo NMD, this individual has a normal level of α5 protein expression from higher mRNA expression from the other CHRNA5 allele that is rs55853698:T-rs55781567:G-rs588765:T. Therefore, the role of the exon 2 deletion in mediating this individual’s heavy smoking remains unclear.
According to HapMap, rs2036527:G>A is in almost perfect LD (D′=0.979, r2=0.902) with rs16969968:G>A in Europeans, but these SNPs are in moderate to low LD, respectively, in Asians (D′=0.872, r2=0.673) and African-Americans (D′=0.87, r2=0.218). The LD between rs2036527:G>A and rs16969968:G>A was slightly higher in our African-American heavy smoker population (Fig. 2, D′=0.965, r2=0.302) than found in HapMap and the MAF of rs16969968:G>A was higher in our African-American heavy smoker population (9.6%) than in either the 1000 genomes ASW (7%) or NHLBI_ESP African-American (6%) populations. This suggests that, similar to European and Asian populations (Berrettini et al., 2008; Chen et al., 2012), at least one hypo-functional α5 allele (Bierut et al, 2008; Kuryatov et al., 2011; Tammimäki et al, 2012) might be contributing to heavy smoking in a subset of our re-sequenced African-American population.
Similar to our results (Table 1), Haller et al. (2012) found few novel mis-sense variants in CHRNA5 that were more prevalent in the nicotine dependent (FTND≥4) than non-dependent (FTND≤1) African-American Collaborative Genetic Study of Nicotine Dependence sample (N=352). Likewise, whereas the NHLBI_ESP identified both known common and novel rare mis-sense CHRNA5 variants in African-Americans, the rare variants were extremely uncommon (MAF«1%). Two exceptions – rs2229961:G>A (V134I) and rs76766434:C>T (R401C) – occurred at slightly higher frequencies, but still had MAF<1% (Exome Variant Server, 2012). We identified 2 individuals in our heavy smoker population that were heterozygous at rs2229961:G>A, but did not find heterozygosity at rs76766434:C>T in any sequenced individual (Table 1). Haller et al. (2012) identified SNPs in CHRNB4 (rs12914008:A>G and rs61737499:G>A) and in CHRNA3 (rs8192475:C>T) that decreased risk for nicotine dependence in both Europeans and African-Americans, but did not find association of rare variants in CHRNB3, CHRNA6 or CHRNA5 with nicotine dependence in either population. Therefore, additional common or rare variants in CHRNA5 were not found in our African-American heavy smoker population likely because SNPs in other cholinergic receptor subunits predict risk for nicotine intake levels among different ethnicities (Thorgeirsson et al., 2010; Haller et al., 2012; Saccone et al., 2009b; Chen et al., 2012; Rice et al., 2012) and polymorphisms within other genes, such as CYP2A6, a risk locus for European and Asian population smoking phenotypes (TAG 2010; Thorgeirsson et al., 2010; Liu et al., 2010; Kumasaka et al., 2012), may influence smoking status by affecting nicotine metabolism.
This work is an extension of that by Rana et al. (2009) and Haller et al. (2012) who re-sequenced CHRNA5 (and other) genes in African-Americans. Similar to their results, we find few novel variants in CHRNA5, and those that we did find are mostly rare in the African-American population (Table 1). Nonetheless, we describe a novel frame-shift mutation that likely results in nonsense-mediated decay of the transcript from the aberrant allele (Figure 1). Several limitations of our study should be acknowledged: We re-sequenced mainly the coding (exonic) portions of the CHRNA5 gene, which, although some intronic variants were found, excluded large portions of CHRNA5 that might have functional relevance to CHRNA5 expression levels. The boundaries of the region associated with nicotine dependence stretches both upstream and downstream of CHRNA5 (Saccone et al, 2010), incorporating other genes (i.e. CHRNA3/B4) that we have not re-sequenced in this study, but that others reported to contain variants that are protective for nicotine dependence (Haller et al, 2012). Finally, we conclude that, in African-Americans, variants (common or rare) in genes other than CHRNA5 most likely contribute to the nicotine dependent phenotype, either independently or in combination with variants in CHRNA5. The functional significance, on CHRNA5 expression or protein function, of the variants found herein must be determined in future studies.
Supplementary Material
Shown are all possible haplotypes predicted by Haploview 4.2 (Barrett et al, 2005) for common and rare variants in CHRNA5 among the re-sequenced African-American population of heavy smokers. SNPs are color coded such that blue indicates the major allele and red indicates the minor allele. See Table 1 for further details regarding the identity of each SNP number and the major and minor alleles.
Acknowledgments
The authors would like to thank the Nucleic acid and PCR core of the Children’s Hospital of Philadelphia for performing the Sanger sequencing. The authors also would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010). The authors thank all study participants without whom these studies could not have been done.
We would also like to acknowledge NIDA’s Center for Genetic Studies in conjunction with Washington University and Rutgers University Cell & DNA Repository for providing DNA samples collected from the following studies and investigators: Opioid Samples: Addictions: Genotypes, Polymorphisms and Function, Mary Jeanne Kreek, M.D.; Genetics of Opioid Dependence, Joel Gelernter, M.D., Kathleen Brady, M.D., Ph.D., Henry Kranzler, M.D., Roger Weiss, M.D.; Opioid Dependence, Wade Berrettini, M.D., Ph.D. Cocaine Samples: An Introduction to the Family Study of Cocaine Dependence, Laura Bierut, M.D.; Genetics of Cocaine Induced Psychosis, Joseph F Cubells, M.D., Ph.D. We would also like to acknowledge the NIDA Clinical Trials Network (CTN) and the University of Pennsylvania CTN node (George Woody, M.D., grant: 2U10DA013043) for support and provision of additional DNA samples from opioid addicted individuals. The NIMH control subjects were collected by the NIMH Schizophrenia Genetics Initiative ‘Molecular Genetics of Schizophrenia II′ (MGS-2) collaboration. The investigators and co-investigators are: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, M.D.; Emory University School of Medicine, Atlanta, GA, MH59587, Farooq Amin, M.D. (PI); Louisiana State University Health Sciences Center; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA, MH60870, William Byerley, M.D. (PI); Washington University, St. Louis, MO, U01,MH060879, C. Robert Cloninger, M.D. (PI); University of Iowa, Iowa, IA, MH59566, Raymond Crowe, M.D. (PI), Donald Black, M.D.; University of Colorado, Denver, CO, MH059565, Robert Freedman, M.D. (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson M.D. (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, M.D. (PI); Mt. Sinai School of Medicine, New York, NY, MH59586, Jeremy Silverman, Ph.D. (PI).
Funding: This work was supported by a grant (R01-DA025201) from the National Institute on Drug Addiction at the National Institutes of Health to WHB and by a grant (K08-MH080372) from the National Institute of Mental Health at the National Institutes of Health to FWL.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland PR, Hoogendoorn B, Coleman SL, Guy CA, Smith SK, O’Donovan MC. Strong bias in the location of functional promoter polymorphisms. Hum Mutat. 2005;26:214–223. doi: 10.1002/humu.20207. [DOI] [PubMed] [Google Scholar]
- Chen LS, Saccone NL, Culverhouse RC, Bracci PM, Chen CH, Dueker N, et al. Smoking and genetic risk variation across populations of European, Asian, and African American ancestry—a meta-analysis of chromosome 15q25. Genet Epidemiol. 2012;36:340–351. doi: 10.1002/gepi.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David SP, Hamidovic A, Chen GK, Bergen AW, Wessel J, Kasberger JL, et al. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry. 2012;2:e119. doi: 10.1038/tp.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle GA, Wang MJ, Chou AD, Oleynick JU, Arnold SE, Buono RJ, et al. In vitro and ex vivo analysis of CHRNA3 and CHRNA5 haplotype expression. PLoS One. 2011;6:e23373. doi: 10.1371/journal.pone.0023373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duga S, Soldà G, Asselta R, Bonati MT, Dalprà L, Malcovati M, Tenchini ML. Characterization of the genomic structure of the human neuronal nicotinic acetylcholine receptor CHRNA5/A3/B4 gene cluster and identification of novel intragenic polymorphisms. J Hum Genet. 2001;46(11):640–8. doi: 10.1007/s100380170015. [DOI] [PubMed] [Google Scholar]
- Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP) Seattle, WA: (URL: http://evs.gs.washington.edu/EVS/) [date (November, 2012) accessed] [Google Scholar]
- Fowler CD, Lu Q, Johnson PM, Marks MJ, Kenny PJ. Habenular α5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471:597–601. doi: 10.1038/nature09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller G, Druley T, Vallania FL, Mitra RD, Li P, Akk G, et al. Rare missense variants in CHRNB4 are associated with reduced risk of nicotine dependence. Hum Mol Genet. 2012;21:647–655. doi: 10.1093/hmg/ddr498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumasaka N, Aoki M, Okada Y, Takahashi A, Ozaki K, Mushiroda T, et al. Haplotypes with copy number and single nucleotide polymorphisms in CYP2A6 locus are associated with smoking quantity in a Japanese population. PLoS One. 2012;7:e44507. doi: 10.1371/journal.pone.0044507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryatov A, Berrettini W, Lindstrom J. Acetylcholine receptor (AChR) α5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (α4β2)2α5 AChR function. Mol Pharmacol. 2011;79:119–125. doi: 10.1124/mol.110.066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Lehman E, Bercovich D, Xu W, Stockton DW, Beaudet AL. Characterization of the human beta4 nAChR gene and polymorphisms in CHRNA3 and CHRNB4. J Hum Genet. 2001;46(7):362–6. doi: 10.1007/PL00010921. [DOI] [PubMed] [Google Scholar]
- Li MD, Ma JZ, Payne TJ, Lou XY, Zhang D, Dupont RT, et al. Genome-wide linkage scan for nicotine dependence in European Americans and its converging results with African Americans in the Mid-South Tobacco Family sample. Mol Psychiatry. 2008;13:407–416. doi: 10.1038/sj.mp.4002038. [DOI] [PubMed] [Google Scholar]
- Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT, et al. A genomewide search finds major susceptibility loci for nicotine dependence on chromosome 10 in African Americans. Am J Hum Genet. 2006;79:745–751. doi: 10.1086/508208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Xu Q, Lou XY, Payne TJ, Niu T, Ma JZ. Association and interaction analysis of variants in CHRNA5/CHRNA3/CHRNB4 gene cluster with nicotine dependence in African and European Americans. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:745–756. doi: 10.1002/ajmg.b.31043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Yoon D, Lee JY, Han BG, Niu T, Payne TJ, et al. Associations of variants in CHRNA5/A3/B4 gene cluster with smoking behaviors in a Korean population. PLoS One. 2010b;5:e12183. doi: 10.1371/journal.pone.0012183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Wellcome Trust Case Control Consortium Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana BK, Wessel J, Mahboubi V, Rao F, Haeller J, Gayen JR, Eskin E, Valle AM, Das M, Mahata SK, Taupenot L, Stridsberg M, Talley TT, Ziegler MG, Smith DW, Schork NJ, O’Connor DT, Taylor P. Natural variation within the neuronal nicotinic acetylcholine receptor cluster on human chromosome 15q24: influence on heritable autonomic traits in twin pairs. J Pharmacol Exp Ther. 2009 Nov;331(2):419–28. doi: 10.1124/jpet.109.157271. Epub 2009 Aug 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JP, Hartz SM, Agrawal A, Almasy L, Bennett S, Breslau N, et al. GENEVA Consortium CHRNB3 is more strongly associated with Fagerström test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction. 2012;107:2019–2028. doi: 10.1111/j.1360-0443.2012.03922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli M, Eusebi F, Al-Chalabi A, Conte A, Madia F, Luigetti M, Mancuso I, Limatola C, Trettel F, Sobrero F, Di Angelantonio S, Grassi F, Di Castro A, Moriconi C, Fucile S, Lattante S, Marangi G, Murdolo M, Orteschi D, Del Grande A, Tonali P, Neri G, Zollino M. Rare missense variants of neuronal nicotinic acetylcholine receptor altering receptor function are associated with sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2009 Oct 15;18(20):3997–4006. doi: 10.1093/hmg/ddp339. Epub 2009 Jul 23. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, Giegling I, Han S, Han Y, Keskitalo-Vuokko K, Kong X, Landi MT, Ma JZ, Short SE, Stephens SH, Stevens VL, Sun L, Wang Y, Wenzlaff AS, Aggen SH, Breslau N, Broderick P, Chatterjee N, Chen J, Heath AC, Heliövaara M, Hoft NR, Hunter DJ, Jensen MK, Martin NG, Montgomery GW, Niu T, Payne TJ, Peltonen L, Pergadia ML, Rice JP, Sherva R, Spitz MR, Sun J, Wang JC, Weiss RB, Wheeler W, Witt SH, Yang BZ, Caporaso NE, Ehringer MA, Eisen T, Gapstur SM, Gelernter J, Houlston R, Kaprio J, Kendler KS, Kraft P, Leppert MF, Li MD, Madden PA, Nöthen MM, Pillai S, Rietschel M, Rujescu D, Schwartz A, Amos CI, Bierut LJ. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010 Aug 5;6(8) doi: 10.1371/journal.pgen.1001053. doi:pii:e1001053.10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009b;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammimäki A, Herder P, Li P, Esch C, Laughlin JR, Akk G, Stitzel JA. Impact of human D398N single nucleotide polymorphism on intracellular calcium response mediated by α3β4α5 nicotinic acetylcholine receptors. Neuropharmacology. 2012 Nov;63(6):1002–11. doi: 10.1016/j.neuropharm.2012.07.022. Epub 2012 Jul 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tobacco and Genetics (TAG) Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Smit AB, de Geus EJ, Sullivan P, Willemsen G, Hottenga JJ, et al. Genome-wide association study of smoking initiation and current smoking. Am J Hum Genet. 2009;84:367–379. doi: 10.1016/j.ajhg.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, Budde JP, et al. COGEND collaborators and GELCC collaborators Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009;18:3125–3135. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009b;14:501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel J, McDonald SM, Hinds DA, Stokowski RP, Javitz HS, Kennemer M, Krasnow R, Dirks W, Hardin J, Pitts SJ, Michel M, Jack L, Ballinger DG, McClure JB, Swan GE, Bergen AW. Resequencing of nicotinic acetylcholine receptor genes and association of common and rare variants with the Fagerström test for nicotine dependence. Neuropsychopharmacology. 2010 Nov;35(12):2392–402. doi: 10.1038/npp.2010.120. Epub 2010 Aug 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shown are all possible haplotypes predicted by Haploview 4.2 (Barrett et al, 2005) for common and rare variants in CHRNA5 among the re-sequenced African-American population of heavy smokers. SNPs are color coded such that blue indicates the major allele and red indicates the minor allele. See Table 1 for further details regarding the identity of each SNP number and the major and minor alleles.


