Introduction
Copy number variations in human population and disease genetics
A copy number variation (CNV) arises when the number of copies of a segment of a chromosome, ranging from a few hundred base pairs (bps) to megabases (Mbs), differs from the expected number of copies (e.g., two copies for autosomes and X chromosomes in females) due to duplication or deletion. CNVs are a major source of genomic diversity in human populations (Redon et al., 2006). Moreover, common or rare CNVs have been associated with genetic susceptibility for many diseases including various cancers, autoimmune disorders, schizophrenia, and autism (see Merikangas and colleagues’ review (Merikangas, Corvin, & Gallagher, 2009)). One recent large study in the Wellcome Trust Case Control Consortium of 16,000 cases (2,000 cases for each of eight different complex diseases) and a shared set of 3,000 controls identified and replicated three loci with CNV associations with disease: IRGM for Crohn's disease, HLA for Crohn's disease, rheumatoid arthritis and type 1 diabetes, and TSPAN8 for type 2 diabetes (Wellcome Trust Case Control et al., 2010). While it is important to note that most common CNVs have demonstrated little or no contribution to disease risk, CNVs have explained the elusive genetic effects in some complex diseases like rheumatoid arthritis, and as such remain one of many viable categories of genomic variation to be explored for possible genetic contributions to disease.
Much work has already been done in defining copy number variations throughout the genome, and this has led to an explosion of bioinformatics resources. The Human Genome Structural Variation Project website curated by Eichler and colleagues at the University of Washington Department of Genome Sciences provides a detailed map of CNVs and large structural variants (Kidd et al., 2008; http://hgsv.washington.edu/). The Copy Number Variation (CNV) Project (http://www.sanger.ac.uk/research/areas/humangenetics/cnv/) from the Wellcome Trust Sanger Institute curates CNVs identified through a variety of genotyping and hybridization approaches and provides extensive information of known CNV- phenotype associations (Bochukova et al., 2010; Conrad et al., 2010). The Center for Human and Clinical Genetics at Leiden University Medical Center maintains a comprehensive list of genetic variation databases (http://www.humgen.nl/SNP_databases.html), including CNV databases.
Detecting copy number variations
A variety of technologies are available to detect CNVs such as fluorescence in situ hybridization (FISH), Array-comparative genomic hybridization (aCGH) (see Unit 4.14), genome-wide single nucleotide polymorphism (SNP) arrays (see also Unit 8.13), and most recently, high-throughput sequencing. These methods have their unique advantages and limitations in cost, equipment needs, size resolution, and sensitivity.
High-throughput, high-density genotyping technologies used in genome-wide association studies such as Illumina BeadArrays enable detection of CNVs. These technologies are based on hybridizations with SNP marker probes designed specifically for particular genomic locations (see Unit 2.9). These array platforms typically target biallelic SNPs. For each SNP, an array platform includes two types of hybridization probes specific to two types of known alleles, usually coded as A and B, and the SNP genotype can be determined by the ratios of the hybridization intensities for A and B probes (Figure 1a). CNVs such as duplications and deletions increase or decrease the total measured intensities; moreover, for large CNVs that span multiple SNPs, intensity ratios have patterns distinct from normal disomic genomic regions (Figure 1b). Computational methods such as PennCNV (Wang et al., 2007), QuantiSNP (Colella et al., 2007), or R/CNVtools (Barnes et al., 2008) have been developed that make full use of these properties to detect common or rare CNVs using hybridization intensities and allele frequencies from SNP markers.
Figure 1.
(a) Calling SNP genotypes by the ratio of probe intensities (allele frequencies) on hybridization arrays. (b) Examples where copy number variations alter total intensities and allele frequencies.
Outline
In this unit we present three basic protocols that: (1) apply PennCNV (Wang et al., 2007) to Illumina SNP array data to detect CNVs, and perform quality assessment; (2) use R to perform association testing of common CNVs; and (3) use PLINK (Purcell et al., 2007) to perform burden tests to find associations with rare or non-overlapping CNVs. We also include a support protocol to visualize CNVs using the UCSC Genome Browser. These protocols assume the reader is familiar with using Linux-based operating systems and software, and has experience using PLINK (Purcell et al., 2007) to analyze GWAS data. Note that some additional terminology is discussed in the commentary section.
Basic Protocol I
Title: Detect CNVs from Illumina Whole-Genome Genotyping array data using PennCNV.
Introduction
In this protocol we describe using PennCNV (Wang et al., 2007) to analyze genotyping data obtained from the Illumina Human660-Quad v1 SNP array to detect CNVs. With minor adjustment these methods can be applied to data collected from other genotyping arrays. Quality control measures of the data can be divided into two phases: 1) at SNP genotyping, including removing failed probes, removing individuals based on call rate, population structure, Hardy-Weinberg Equilibrium (see Units 1.19 and 1.22); and 2) at CNV calling, including removing individuals with highly variable signal intensity data.
Materials List
Signal intensity data - LRR (Log R Ratio) and BAF (B Allele Frequency) - of each individual and each probe.
Additional input files for PennCNV as described in its manual: PFB (Population Frequency of B allele), HMM, and GCModel files.
Linux environment with PennCNV installed. We assume the user has PennCNV installed or has the knowledge on how to obtain and install the software; more information is available on the PennCNV website (http://www.openbioinformatics.org/penncnv/penncnv_installation.html).
Steps and Annotations
Generate a signal intensity file by the export function provided in Illumina GenomeStudio or BeadStudio. The following fields are required: SNP information (rs ID is required while chromosome and location are optional), and LRR and BAF values for each sample. The PennCNV website (http://www.openbioinformatics.org/penncnv/penncnv_input.html) provides step-by-step instructions. Assume the file name is lrr_baf.txt.
Remove probes that can not be uniquely mapped to the genome. Although Illumina selects SNPs that can be uniquely mapped to the reference genome when an array was designed, this might not be the case for some of the SNPs when a newer reference genome assembly is released. One may detect this when the SNP location is mapped to the newer reference genome assembly using UCSC Genome Browser's liftover tool or NCBI's genome-remapping service. For each version of dbSNP, UCSC Genome Browser's provides of a list of SNPs that are mapped to multiple loci. Users can use such file to filter out such potentially problematic SNPs.
Choose proper PFB and GCmodel files. The PennCNV package comes with a number of PFB files for different genotyping arrays. The PFB file describes which B-allele frequencies to use for all markers on the array platform, and the GCmodel file specifies parameters for adjustment of the differences in GC-content (e.g. waviness) across the genome (Diskin et al., 2008) . Additional PFB and GCmodel files occasionally become available on the PennCNV website. The coordinates used in these files and the GCmodel file are, however, based on the hg18 reference genome. One can use UCSC Genome Browser's liftover tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver) to convert them to the desired version of genome assembly. We named the hg19 version of the PFB and GCmodle files for Illumina Human660 array hh660.hg19.pfb and hh660.hg19.gcmodel respectively.
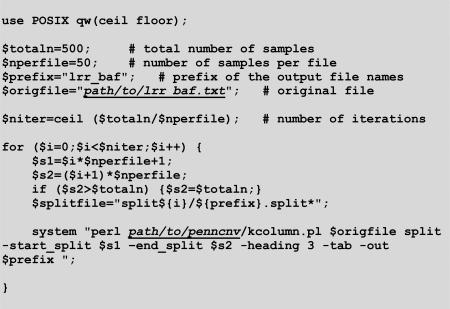
If there are many samples to process, it is more efficient to split the intensity file by sample into smaller files such that they can be processed by PennCNV in parallel. For instance, in the example below we use kcolumn.pl of the PennCNV package to split an intensity file lrr_baf.txt into 50 samples per file. Output files are named incrementally as lrr_baf.split1, lrr_baf.split2, and so on.
-
5
Run PennCNV. Here is an example of the command used to identify CNVs. This command processes the intensity file lrr_baf.split1 and generates a log file lrr_baf_1.log, which contains summary statistics that provide information on the quality of input data, and an output file lrr_baf_1.rawcn that contains called CNVs. The user should edit and run this command for every split file from step 4.

The above perl script reads the three model files (hhall.hmm which comes with PennCNV, and the two files hh660.hg19.pfb, and hh660.hg19.gcmodel we described earlier), and calls CNVs using the intensity file lrr_baf1 Result The user should change the path to the files in the command above according to where these reside on the user's computer system. This command processes the intensity file lrr_baf.split1 and generates a log file, which contains summary statistics that inform on the quality of input data, and an output file that contains called CNVs.
-
6
Run PennCNV's filter_cnv.pl to generate a quality control summary for each sample. The output file, lrr_baf_1_qcpass_default.rawcn, contains CNVs that pass QC.

-
7
Review the quality control summary file to exclude samples of poor quality. PennCNV computes the following statistics for each sample: LRR_SD (standard deviation of LRRs), BAF_drift (measuring departure of the BAF from the expected values), WF (waviness factor, the amount of dispersion in signal intensity, which is a good indicator for DNA quality after accounting for GC content (Diskin et al., 2008)), and NumCNV (number of called-CNVs). PennCNV uses the following exclusion criteria by default: LRR_SD > 0.3, BAF_drift > 0.01, WF > 0.05. Samples meeting any of these three criteria will be excluded. For more stringent quality control, use lower exclusion thresholds. Our experience indicates that NumCNV is sensitive to the platform being used, so we recommend checking the distribution across all samples and exclude outliers.
-
8
Review each .rawcnv file and check the number of SNPs spanning each called CNV. The .rawcnv file is a tab-separated text file that can be easily opened using Excel or processed using R. Typically a minimum of three SNPs is used as a filter. However, our experience suggests that a minimum of 10 SNPs produces results with reasonable sensitivity.
-
9
Remove called CNVs in certain genomic regions: (1) HLA regions, (2) genomic regions that are near the centromeres and the telomeres (PennCNV recommends a 1Mb neighborhood). The PennCNV website provides physical locations (in hg18) of these regions (see http://www.openbioinformatics.org/penncnv/penncnv_faq.html#ig).
Basic Protocol 2
Use of R to perform association tests for common CNVs
In this protocol we demonstrate using logistic regression, which was implemented with the glm() function (generalized linear models) in R to find CNV associations with disease. All input and output files are tab-delimited with headers. To make the files a manageable size while simplifying the input process to R, we divide the variants into one file per chromosome, and each file contains both phenotype and genotype data.
Materials List
Output file from the PennCNV software (see Basic Protocol I) that contains all called CNVs.
Individuals’ case/control status and phenotypes or factors that may confound the relation between CNVs and the disease state. Theses pieces of information are equivalent to those of PLINK FAM and covariate files.
Linux environment with R installed. We assume the user has installed R or has the knowledge on how to obtain and install the software from the R website (http://www.r-project.org/). Comprehensive documentation is available there.
Make a phenotype file that can be read by R as a data frame or a matrix (e.g. using the read.table() command). This file includes case/control status and covariates, which can include individual phenotype data such as sex and age and sample characteristics such as tissue type and batch.
Generate Copy Number Polymorphic Regions (CNPRs). Predicted CNVs from the same genomic locus can have various start and end points across individuals. To make the analysis simpler, divide the genome into CNPRs. Each individual is then assigned a Copy Number (CN) state for each CNPR according to the CNV predicted in that region, with CN=2 if no CNV is predicted. Details on constructing CNPRs are illustrated in Figure 2. The drawback to this approach is that the origin of a CNV cannot be readily interpreted because one CNV may span multiple CNPRs. We provide an R script (penncnv2cnpr.r, which can be downloaded from the journal website as supplementary material) that takes PennCNV-predicted CNVs pooled from multiple individuals, creates CNPRs and breaks each called CNV into corresponding CNPRs.
Encode CN state with deletion (DEL) and duplication (DUP) variables. CNVs can be the results of deletion or duplication. We use two variables to code for copy number. Without further information, the copy number derived from genotyping array data does not provide allele specific copy number. For instance, a typical copy number of two at one locus, in fact, can indicate a single duplication of one allele and single deletion of the other (i.e., uniparental disomy). Such events are probably rare, but without dual coding of deletion and duplication, it would not be treated as a CNV. Thus, for one individual in one CNPR, if no CNV is predicted, both DEL and DUP are 0. If CN=1, then DEL=1 and DUP=0. If CN=0, then DEL=2 and DUP=0; DEL = 0 and DUP = 1 for CN=3.
Divide the data into smaller subsets (e.g., one dataset per chromosome) for regression analysis. Analyzing large datasets in R can be computationally intensive, and so dividing the data into smaller files can reduce the computational demand on hardware.
R commands for association analysis.
Figure 2.
A section of a chromosome to demonstrate how Copy Number Polymorphic Regions (CNPRs) are constructed. In this example, PennCNV has been run to call CNVs from SNP array data of six individuals (indiv1 through 6). All called CNVs from all individuals were pooled together. All non-redundant end points of the CNVs become break points that would be used to partition the chromosome. A pair of break points form a CNPR. Every CNV is then decomposed into multiple consecutive CNPRs. Red: Copy Number (CN) = 1; Blue: CN=3; Black: CN=4. Based on the the type of a CNV (CN=1 or CN=3) one individual has in a CNPR (CN=2 if no CNV was called), a matrix can be generated.
We use logistic regression for testing association of copy number and disease status. Logistic regression can be performed in R using the glm() function with the family parameter set to “binomial”. The following are examples of R commands for with and without covariates at a given CNPR:

-
6
Results of the regression are stored in nocovar and wcovar. One can then use the summary() function in R (see below) to generate human-readable summaries. DX is a vector of integers denoting affection status of every individual, where 0 and 1 code for control and case, respectively. DEL and DUP are vectors of non-negative integers. Copy Number (CN) at a diploid locus is denoted by one number in DEL and another in DUP. If CN = 2, DEL = 0 and DUP = 0. If CN > 2, DEL = 0 and DUP = CN - 2. If CN < 2, DUP = 0 and DEL = 2 - CN. AGE is a vector of numbers denoting age at exam or age of onset. SEX is a “factor” data type. It can be a vector of integers (e.g. 1 and 2 for male and female respectively), or characters (M/F or male/female). Age and sex are included on the right-hand side to adjust for the effects of age and sex on disease risk. The data set is a dataframe composed of columns DX, DEL, DUP, AGE, and SEX at a given CNPR,
-
7
Interpret the glm output.
The R summary() function provides a detailed view of the regression results. An example is shown below. summary(nocov)$coefficients displays the coefficient and significance information in four columns (“Estimate” “Std. Error” “z value” “Pr(>|z|)”) for each predictor term. Note that as the regression was carried out once per CNPR, the P-value obtained here should be corrected for multiple tests if multiple CNPRs were tested. In the example below, deletions reduce the disease risk and duplications slightly elevate the disease risk (sign of the z value), but neither is statistically significant (P=0.709 and 0.933 respectively).
Basic Protocol 3
Use of PLINK to perform burden tests for rare or non-overlapping CNVs
This protocol allows for testing if there is significant difference in the frequency and total length of CNVs in case and control subjects without requiring all CNVs to span the same genomic region. The test can be performed either genome-wide or over specific genomic regions (e.g. a list of candidate genes). Plink uses permutation to compute empirical P-values.
Materials
Output file from the PennCNV software (see Basic Protocol I) that contains all called CNVs
PLINK FAM file from the Genome-Wide Association Study (GWAS) SNP data. An optional file describing user-specified genomic regions for burden tests. For example, a file containing the coordinates of all known genes on the human genome. Each row specifies one genomic region (chromosome, start, and end positions)
Linux environment with PLINK installed. We assume the user has installed PLINK or has the knowledge on how to obtain and install it (http://pngu.mgh.harvard.edu/~purcell/plink/). Comprehensive documentation is available there.
Convert the PennCNV output file into the PLINK CNV file format. In this format, each row is a called CNV for a subject (specified by the Family ID and Individual ID fields), the physical locations of its breakpoints, the number of copies, and two optional fields (confidence score, number of SNP markers) for filtering. See http://pngu.mgh.harvard.edu/~purcell/plink/cnv.shtml#format for details on the file format. Such conversion can be done using tools such as Excel, R or generic text editors, or computer programs written in Linux, Perl, Python, C, etc. We named the converted file mydata.cnv as the input CNV file for the PLINK commands demonstrated below.
Create a map file for the input CNV data. PLINK CNV burden analysis program requires a map file that describes the start and stop locations (base pairs) for all input CNVs. The following command takes mydata.cnv as input and saves the map information in the file mydata_cnv.map.
![]()
-
3
Perform a genome-wide burden test using the formatted CNV data and the newly created MAP file.
![]()
The function randomly shuffles the case/control status of all subjects 10,000 times to compute P-values empirically for the hypothesis that the burden of CNVs is different between case and control subjects. PLINK will generate two files: mydata.summary contains CNV frequencies by case/control status, and mydata.summary.mperm contains P-values from permutations.
-
4Numerous options can be added to the PLINK command shown above.
- Both “--cnv-dup” or “--cnv-del” options limit the test to only duplications or deletions respectively.
- The options “--cnv-intersect region.txt --cnv-test-region” limit the test to given genomic regions and only those CNVs intersecting with such regions are included. The option “--cnv-overlap 0.6” further specifies that the overlapping covers at least 60% of the intersecting CNV.
- Similarly,“--cnv-exclude regions.list” excludes those CNVs intersecting specified genomic regions. The format of ‘regions.list’ is the same as that of item 3 in the Materials List. This option is useful to avoid regions known for generating spurious CNV calls.
- It is also possible to filter CNVs based on frequency. For instance, to focus on rare CNVs, “--cnv-freq-exclude-above 10” excludes CNVs that are present in at least 10 subjects (or 1% frequency in a 1,000-sample dataset).
Support Protocol
Visually inspect CNVs on the UCSC Genome Browser
This protocol describes steps to format called CNVs in the Browser Extended Data (BED) format, specify red/blue color schemes for copy numbers, and upload data to the UCSC Genome Browser for visualization.
Materials
Output file from the PennCNV software (see Basic Protocol I) that contains all called CNVs.
A web browser that is compatible with the UCSC Genome Browser.
Format CNV files into the BED format. A BED file is a tab-delimited file that represents genomic features, such as genes or CNVs as integer intervals one interval per line, and describes how these intervals to be displayed on the UCSC browser as a custom track (see UNIT 18.6). Only the first three fields -chromosome/scaffold name, start and end positions - describing the genomic location are required but the optional fields, such as name, strand etc., and the “track line” make the visualization more informative. Please refer to http://genome.ucsc.edu/FAQ/FAQformat.html#format1 for more details. For this protocol, we put seven fields and a track line in one BED file. An example may look like this:

![]()
Although the track line appears as two lines, it is in fact one single line. Both of the last two fields being identical to the second one, i.e. the start position, makes the bars representing the CNVs thinner so as to accommodate more CNVs in one given space. For a small number of items, a generic text editor or Excel is sufficient to do the conversion manually. For a large number, however, a program is usually needed to do it efficiently and correctly. The following is an example Perl script that reads a PennCNV output file and converts it into a BED file. The script gives the contrast between deletions and duplications by assigning a strand status to each CNV (‘+’ when CN < 2 and ‘-’ when CN > 2), and using the “colorByStrand” attribute in the track line. The two colors for the two strands are specified by RGB color codes and divided by a space. To visualize the contrast between cases and controls, then the coding for strand should be used to encode disease status instead, and thus duplications and deletions should be separated into two tracks.
-
2Upload to UCSC Genome Browser.
- Open the web browser and connect to the UCSC Genome Browser (http://genome.ucsc.edu/). Click on “Genomes” at the top-left of the home page.
- Select the Human genome using the drop-down lists. It is important to choose the version of the genome assembly (hg18/NCBI release 36, or hg19/GRCh 37) that corresponds to the coordinate system used in your CNV data.
- Click on the “add custom tracks” button right below the drop-down lists. You can then choose to copy-paste the BED file generated in the previous step into the “Paste URLs or data” text field, or click the “Choose File” button and then upload the BED file. Click the Submit button.
- If the input BED file is correct, you will be brought to a new page with title “Manage Custom Tracks”. You should see a table that lists custom tracks you have uploaded. Your BED file should become available as a track with the name you specified. Click on “go to genome browser” to view the uploaded data.
Commentary
Background Information
Terminology for SNP genotyping array platform data types
Illumina and Affymetrix Genome-wide SNP genotyping platforms use designed probes that specifically hybridize with the genomic DNA flanking chosen SNPs. Although it is possible that two kinds of nucleotides (biallelic), three, or even four are observed at these SNPs in the general population, by design, the probes on a commercial genotyping array only detect two alleles. In general, the two alleles are labeled A and B. The A/B designation is manufacturer-specific but should be consistent across platforms from the same manufacturer; the reader should consult the company for designation rules and/or annotations. A scanner is used to measure the fluorescence intensity of hybridized A and B probes for each SNP on the array: these data are referred as the raw intensities of the A and B alleles (RA and RB respectively). SNP genotypes are determined by comparing A and B intensities: a genotype of A/A is called when A fluorescence intensity is strong and B allele intensity is low; B/B is called when B fluorescence intensity is strong and A allele intensity is low; and A/B is called if the two intensities are similar and of an intermediate level of intensity (e.g. Figure 1a). The following two derived measures are informative about the copy number status.
The log R ratio (LRR) is the log2-transformed value of the normalized intensity of the SNP, (RA + RB)/Rexpected, where Rexpected is an interpolation generated by GenomeStudio (http://www.illumina.com/documents/products/technotes/technote_cnv_algorithms.pdf). LRR indicates the relative abundance of the genomic DNA around the SNP and is expected to correlate with copy number status.
The B allele frequency (BAF) of a SNP reflects the relative abundance of B allele intensity; it is an adjusted value generated by GenomeStudio, assuming three canonical clusters (A/A: 0.0, A/B: 0.5, B/B: 1). Please refer to the Illumina technical note on algorithms for detecting CNVs (http://www.illumina.com/documents/products/technotes/technote_cnv_algorithms.pdf) for the precise definition. Normally BAF is close to 0, 0.5, or 1 for autosomal loci, and one expects to observe BAF close to 0 and 1 but not 0.5 for SNPs in single deletions (copy number=1). Similarly, one expects BAFs for SNPs in single duplications (copy number=3) to be around 0, 0.33, 0.67, or 1. In more recent products, Illumina and Affymetrix have introduced CNV-specific probes. These probes do not have distinct alleles and only return intensity information. Therefore LRR is available but BAF is not available for these probes.
Statistical approaches for CNV calling
SNP arrays call SNP genotypes with good robustness since after proper normalization A and B allele intensities typically group into three distinct clusters in a dataset with large number of samples (e.g. Figure 1a). Observed intensities from individual SNPs are quite noisy for calling CNVs, requiring aggregation of information across multiple SNPs/samples to improve the detection accuracy. Depending on how information is aggregated, we can classify CNV calling algorithms into three types of approaches:
1. Aggregate along chromosomes
The first approach aggregates information across adjacent SNPs to improve the specificity for CNV detection. These algorithms are based on the observation that a CNV spanning multiple probes will have similar effects perturbing their BAF and LRR values. PennCNV (Wang et al., 2007) and QuantiSNP (Colella et al., 2007)} developed a Hidden Markov Model algorithm to generate smoothed copy number calls using LRR and BAF as input. The cnvPartition algorithm developed by Illumina (http://www.illumina.com/documents/products/technotes/technote_cnv_algorithms.pdf) detects breakpoints as sudden changes in BAF and LRR values and assigns copy number values to the partitioned genomic regions.
2. Aggregate across samples
The second approach aggregates information across multiple samples to make CNV calls for a single SNP. For example, CNVtools {(Barnes et al., 2008)} fits LRR values from multiple samples using a Gaussian mixture for each probe, creating discrete intensity clusters that correspond to different copy numbers at the same time.
3. Hybrid approaches
Other computer programs combine both concepts by aggregating information from multiple SNPs and multiple samples. The Birdseye component of the Birdsuite software {(Korn et al., 2008)}implements a Hidden Markov model for aggregating adjacent SNPs. The software then searches for genomic regions showing correlated intensity patterns across samples for better specificity.
Critical Parameters
Points to consider before beginning experiments
1. Confounding factors
Since all calling algorithms rely on the intensity of SNP probes to detect CNVs, confounding factors arise easily and care must be taken to eliminate them. Different genotyping platforms have different probe designs and usually lead to different sensitivities for CNV detection. Different labs or different technicians may perform DNA extraction differently. Our experience also indicates when DNAs are extracted from different tissue types (e.g. DNA from whole blood versus from cell lines versus from frozen brain), they have considerable difference in the distribution of called CNVs genome-wide. We also found that such difference is minimized when we limit our analysis to large CNVs (which require many more SNPs), indicating the tissue specificity effect may be artifacts due to DNA extraction. If such confounding factors cannot be avoided in the study design, one can introduce indicator variables (e.g. DNA source tissue, batch index) in the statistical analysis to help control for confounding. We also recommend the analysis to focus on large CNVs (e.g. 10 SNPs or longer for PennCNV).
2. Special genomic regions
Repeat-rich regions such as telomeres and centromeres are highly copy-number polymorphic, and may cause problems in the analysis. Regions that contain nearly identical duplicate sequences may also contain segmental duplications that look like low copy number duplications in CNV analysis. The HLA Major Histocompatibility Complex (MHC) region on chromosome 6 is also highly polymorphic, and may lead to false positive calls either because of cell-line artifacts or by difference in the specificity of SNP probe hybridization on genotyping arrays, indirectly affecting CNV detection. These physical locations of these regions can be downloaded either from the PennCNV website (need to be converted to coordinates in the correct version of the reference genome) or the UCSC Genome Browser.
3. Threshold parameters for calling a CNV
Many CNV calling programs do not assign a confidence score to individual CNV calls, and users of these programs often set additional criteria to exclude unreliable calls. For instance, PennCNV recommends analyzing CNVs spanning at least three SNPs. This is can be done by the “-numsnp 3” option when invoking PennCNV, or one can filter the output file by checking the ‘numsnp’ field. PennCNV also allows users to set length threshold; e.g., only output CNVs 50Kbp or longer (using the “-length 50k” option or checking the length field in the output file). We recommend excluding CNVs spanning fewer than 10 SNPs. It is important to note that these two criteria assume probes are uniformly distributed on chromosomes, which is not always the case for many platforms. For example, many Illumina platforms contain CNV-specific probes densely located in known copy number polymorphic genomic regions. SNP arrays lack probes in centromeres, which span millions of bases in length. It is critical to take notice of the fact that the wide variation in probe distribution can inflate the number of SNPs supporting a called CNV or the length of the CNV.
Appendix
Internet Resources
PennCNV website: http://www.openbioinformatics.org/penncnv/ Users can download the PennCNV source code, compile, and install on their own computers. The website also contains a wealth of information including program manual, annotation files, tutorials for the PennCNV software, and other useful tips such as visualization and quality control recommendations.
R website: http://www.r-project.org/ R is a free program for statistical computing and visualization. Users can download the compiled R package for their specific computing platforms. The website also lists URLs to the Comprehensive R Archive Network (CRAN). CRAN hosts user-contributed packages that provide additional analysis capabilities.
Illumina GenomeStudio Website: http://www.illumina.com/software/genomestudio_software.ilmn The website contains instructions and FAQs for the GenomeStudio software which is required to export SNP intensities from Illumina Chip projects for CNV calling. Illumina customers can obtain the software for free.
PLINK website: http://pngu.mgh.harvard.edu/~purcell/plink/ PLINK is developed by Shaun Purcell at Harvard University. The free, open-source program is widely used by the research community to process and analyze genome-wide association studies (GWAS). Users can download the source code or obtain pre-compiled binaries for installation from this website. This website also contains very detailed instructions on how to use the program.
UCSC Genome Browser: http://genome.ucsc.edu/ Users can go to UCSC Genome Browser to download genomic annotations, or visualize CNV calls on the reference genome as outlined in the Alternative Protocol.
List of Genetic variation databases: http://www.humgen.nl/SNP_databases.html The Center for Human and Clinical Genetics at Leiden University Medical Center maintains a comprehensive list of genetic variation databases, including CNV databases.
The Human Genome Structural Variation Project: http://hgsv.washington.edu/ This website, maintained by the Eichler lab at the University of Washington, provides a detailed map of CNVs and large structural variants.
The Copy Number Variation (CNV) Project: http://www.sanger.ac.uk/research/areas/humangenetics/cnv/ The database is maintained by the Wellcome Trust Sanger Institute. It hosts CNVs identified through a variety of genotyping and hybridization approaches and provides extensive information of known CNV/phenotype associations.
The Database of Genomic Variants: http://projects.tcag.ca/variation/project.html This database is maintained by the University of Toronto Centre for Applied Genomics. The database is a comprehensive catalog of structural variants in the human genome by collecting published reports on healthy controls in the literature. It can be used as controls in studies to correlate CNVs with diseases and traits.
Literature Cited
- Barnes C, Plagnol V, Fitzgerald T, Redon R, Marchini J, Clayton D, Hurles ME. A robust statistical method for case-control association testing with copy number variation. Nat Genet. 2008;40(10):1245–1252. doi: 10.1038/ng.206. doi: 10.1038/ng.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, Saeed S, Hamilton-Shield J, Clayton-Smith J, O'Rahilly S, Hurles ME, Farooqi IS. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463(7281):666–670. doi: 10.1038/nature08689. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35(6):2013–2025. doi: 10.1093/nar/gkm076. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Wellcome Trust Case Control Consortium. Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464(7289):704–712. doi: 10.1038/nature08516. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin SJ, Li M, Hou C, Yang S, Glessner J, Hakonarson H, Bucan M, Maris JM, Wang K. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36(19):e126. doi: 10.1093/nar/gkn556. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tüzün E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56–64. doi: 10.1038/nature06862. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, Lee C, Nizzari MM, Gabriel SB, Purcell S, Daly MJ, Altshuler D. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40(10):1253–1260. doi: 10.1038/ng.237. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas AK, Corvin AP, Gallagher L. Copy-number variants in neurodevelopmental disorders: promises and challenges. Trends Genet. 2009;25(12):536–544. doi: 10.1016/j.tig.2009.10.006. doi: 10.1016/j.tig.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17(11):1665–1674. doi: 10.1101/gr.6861907. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control, Consortium. Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, Holmes C, Marchini JL, Stirrups K, Tobin MD, Wain LV, Yau C, Aerts J, Ahmad T, Andrews TD, Arbury H, Attwood A, Auton A, Ball SG, Balmforth AJ, Barrett JC, Barroso I, Barton A, Bennett AJ, Bhaskar S, Blaszczyk K, Bowes J, Brand OJ, Braund PS, Bredin F, Breen G, Brown MJ, Bruce IN, Bull J, Burren OS, Burton J, Byrnes J, Caesar S, Clee CM, Coffey AJ, Connell JM, Cooper JD, Dominiczak AF, Downes K, Drummond HE, Dudakia D, Dunham A, Ebbs B, Eccles D, Edkins S, Edwards C, Elliot A, Emery P, Evans DM, Evans G, Eyre S, Farmer A, Ferrier IN, Feuk L, Fitzgerald T, Flynn E, Forbes A, Forty L, Franklyn JA, Freathy RM, Gibbs P, Gilbert P, Gokumen O, Gordon-Smith K, Gray E, Green E, Groves CJ, Grozeva D, Gwilliam R, Hall A, Hammond N, Hardy M, Harrison P, Hassanali N, Hebaishi H, Hines S, Hinks A, Hitman GA, Hocking L, Howard E, Howard P, Howson JM, Hughes D, Hunt S, Isaacs JD, Jain M, Jewell DP, Johnson T, Jolley JD, Jones IR, Jones LA, Kirov G, Langford CF, Lango-Allen H, Lathrop GM, Lee J, Lee KL, Lees C, Lewis K, Lindgren CM, Maisuria-Armer M, Maller J, Mansfield J, Martin P, Massey DC, McArdle WL, McGuffin P, McLay KE, Mentzer A, Mimmack ML, Morgan AE, Morris AP, Mowat C, Myers S, Newman W, Nimmo ER, O'Donovan MC, Onipinla A, Onyiah I, Ovington NR, Owen MJ, Palin K, Parnell K, Pernet D, Perry JR, Phillips A, Pinto D, Prescott NJ, Prokopenko I, Quail MA, Rafelt S, Rayner NW, Redon R, Reid DM, Renwick, Ring SM, Robertson N, Russell E, St Clair D, Sambrook JG, Sanderson JD, Schuilenburg H, Scott CE, Scott R, Seal S, Shaw-Hawkins S, Shields BM, Simmonds MJ, Smyth DJ, Somaskantharajah E, Spanova K, Steer S, Stephens J, Stevens HE, Stone MA, Su Z, Symmons DP, Thompson JR, Thomson W, Travers ME, Turnbull C, Valsesia A, Walker M, Walker NM, Wallace C, Warren-Perry M, Watkins NA, Webster J, Weedon MN, Wilson AG, Woodburn M, Wordsworth BP, Young AH, Zeggini E, Carter NP, Frayling TM, Lee C, McVean G, Munroe PB, Palotie A, Sawcer SJ, Scherer SW, Strachan DP, Tyler-Smith C, Brown MA, Burton PR, Caulfield MJ, Compston A, Farrall M, Gough SC, Hall AS, Hattersley AT, Hill AV, Mathew CG, Pembrey M, Satsangi J, Stratton MR, Worthington J, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand W, Parkes M, Rahman N, Todd JA, Samani NJ, Donnelly P. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464(7289):713–720. doi: 10.1038/nature08979. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]