Abstract
Purpose
To assess the prevalence of phosphoinositide 3-kinase (PI3K) pathway alterations in pure high-grade ductal carcinoma in situ (DCIS) and DCIS associated with invasive breast cancer (IBC), and to determine whether DCIS and adjacent IBCs harbor distinct PI3K pathway aberrations.
Experimental Design
89 cases of pure high-grade DCIS and 119 cases of high-grade DCIS associated with IBC were characterized according to estrogen receptor (ER) and HER2 status, subjected to immunohistochemical analysis of PTEN, INPP4B, phosphorylated (p)AKT and pS6 expression, and to microdissection followed by Sequenom genotyping of PIK3CA and AKT1 hotspot mutations.
Results
Alterations affecting the PI3K pathway were found in a subset of pure DCIS and DCIS adjacent to IBC. A subtype-matched comparison of pure DCIS and DCIS adjacent to IBC revealed that PIK3CA hotspot mutations and pAKT expression were significantly more prevalent in ER-positive/HER2-negative DCIS adjacent to IBC (p-values, 0.005 and 0.043, respectively), and that in ER-negative/HER2-positive cases, INPP4B loss of expression was more frequently observed in pure DCIS (p-value 0.013). No differences in the parameters analyzed were observed in a pairwise comparison of the in situ and invasive components of cases of DCIS and adjacent IBC. Analysis of the PIK3CA mutant allelic frequencies in DCIS and synchronous IBC revealed cases where PIK3CA mutations were either restricted to the DCIS or to the invasive components.
Conclusion
Molecular aberrations affecting the PI3K pathway may play a role in the progression from high-grade DCIS to IBC in a subset of cases (e.g., a subgroup of ER-positive/HER2-negative lesions).
Keywords: ductal carcinoma in situ, breast cancer, PI3K pathway, progression, evolution
INTRODUCTION
Ductal carcinoma in situ (DCIS) is a neoplastic proliferation of epithelial cells of the breast, which is separated from the breast stroma by the presence of an intact basement membrane and a discontinuous layer of myoepithelial cells(1–3). Widespread mammographic screening has led to an increase in the detection of DCIS, which now accounts for approximately 30% of new screen-detected breast cancers(4). Although DCIS has been shown to constitute a non-obligate precursor of invasive breast cancer (IBC)(5–9), with up to 40% of these lesions progressing to invasive disease if untreated, identifying which cases will either recur as in situ disease or progress to invasive breast cancer has proven challenging. Clinically useful predictors of progression from in situ to invasive disease have yet to be developed or introduced in clinical practice(2, 10, 11). In addition, the molecular mechanisms that underpin the progression from DCIS to IBC have yet to be defined(2, 3).
Previous studies based on immunohistochemistry, in situ hybridization, comparative genomic hybridization (aCGH), and microarray-based gene expression profiling have demonstrated that DCIS and IBCs are remarkably similar at the molecular level(12–22). It should be noted, however, that most of these studies have not focused on matched DCIS and IBC from the same patient. In those that have focused on synchronous DCIS and IBC, amplification of MYC(19) and FGFR1(22) has been reported to be more frequent in the invasive component(3). Furthermore, recent studies have demonstrated that intra-tumor genetic heterogeneity is a common phenomenon from the early stages of breast cancer development(23, 24), suggesting that the progression from DCIS to IBC may follow Darwinian evolutionary rules(3, 23, 24). Hence, one could posit that this biological phenomenon may constitute an evolutionary bottleneck, with the selection of non-modal populations of cancer cells harboring specific genetic aberrations in the progression from DCIS to invasive disease.
It is currently accepted that breast cancer comprises multiple entities with distinct risk factors, clinical presentation, histologic features, response to therapy, and outcomes(25). In fact, the transcriptomic differences between estrogen receptor (ER)-positive and ER-negative breast cancers are such that they are likely to constitute completely different diseases that originate in the same microanatomical structure and affect the same anatomical site(25). Despite the molecular differences between ER-positive and ER-negative disease, some molecular pathways seem to be frequently targeted by genetic aberrations in both ER-positive and ER-negative breast cancers(26). For example, the PI3K pathway, which plays pivotal roles in cell survival, proliferation, and migration(27) is altered not only in the majority of ER-positive breast cancers, but also in a large subset of ER-negative breast cancers(26, 28).
Activating mutations in PIK3CA, encoding the PI3K catalytic subunit p110α, and loss of function of the negative regulator of PI3K signaling, PTEN, have been reported in up to 35% and 13% of IBCs, respectively, and vary according to the subtype of the disease as defined by molecular subtyping, and ER and HER2 status(26, 28–30). Loss of protein expression of the putative tumor suppressor INPP4B, which inhibits PI3K signaling, has been found in 7–11% of ER-positive and 44–58% of ER-negative IBCs, in particular in basal-like breast cancer(31, 32). In addition, somatic mutations of AKT1 have been reported in 1–8% of IBCs; however, their effect on the PI3K pathway is not yet entirely understood(26, 28, 33). PIK3CA mutations have been reported in approximately 30% of DCIS(23, 34–37), and qualitative comparisons between DCIS and IBC have demonstrated that if a PIK3CA mutation is present in the DCIS, it would also be present in the invasive component in the vast majority of cases(35, 37); however, discordances have also been recorded(36). In a pilot study using semi-quantitative methods to infer the percentage of cancer cells harboring specific mutations, we have recently documented the presence of PIK3CA mutations in the modal population of samples of DCIS, which were either present in a non-modal subset of the neoplastic cells of the invasive component or entirely absent in the invasive lesion, providing another line of evidence to support the contention that progression from DCIS to invasive breast cancers may result in the selection of genetically distinct clones(3, 23).
Given the non-obligate precursor nature of DCIS, questions that are germane to our ability to develop predictors of progression include whether DCIS that does not progress to invasive cancer harbors distinct molecular aberrations as compared to those that do, and how similar synchronous DCIS and IBCs are at the molecular level. Therefore, defining these molecular differences may offer valuable insights into the mechanisms that result in the establishment of invasive disease. Given the pivotal roles played by the PI3K pathway in both ER-positive and ER-negative breast cancers, here we sought to define the prevalence of PI3K pathway alterations in a matched cohort of high-grade DCIS that did or did not progress to IBC, and to define the differences in the frequency of molecular alterations of this pathway in samples of synchronous DCIS and IBC.
MATERIALS AND METHODS
Patient and tissue samples
Following approval from the institutional review board, the breast surgical database at Memorial Sloan-Kettering Cancer Center (MSKCC) was queried for patients who underwent definitive surgical treatment for either pure DCIS or DCIS with associated IBC from 1999–2003. To maximize our ability to obtain adequate material for analysis, we restricted our query to those cases with pure DCIS (i.e., cases where the most advanced lesion found in the surgical specimen was a DCIS and who did not develop IBC in the ipsilateral breast within 5 years of follow-up) or DCIS and synchronous invasive disease in at least two available archival formalin-fixed paraffin-embedded (FFPE) blocks, and to cases in which the invasive tumor component was at least 1.0 cm in size. In addition, to minimize the impact of known confounding factors, such as histologic grade, we restricted the study to high-grade DCIS. Consecutive cases meeting these criteria were selected. Ultimately, 89 cases of pure high-grade DCIS and 119 cases of DCIS with synchronous adjacent IBC (in the same FFPE block) were available for analysis (Supplementary Table S1). All cases were reviewed by two pathologists (DG and VPA) to confirm the reported histologic features and grade. Histologic grade for in situ lesions was assessed based on nuclear pleomorphism and necrosis. High-grade DCIS corresponded to groups 2 and 3 of the Van Nuys classification for DCIS(38). Invasive lesions were graded based on nuclear pleomorphism using Black’s nuclear grading method as modified by Fisher et al.(39). Given that ER-positive and ER-negative breast cancers have been shown to have distinct repertoires of molecular aberrations(25, 26, 28), the comparisons between pure DCIS and DCIS adjacent to IBC were only performed within groups stratified according to ER and HER2 status (i.e., ER-positive/HER2-negative, ER-positive/HER2-positive, ER-negative/HER2-positive, and ER-negative/HER2-negative).
Immunohistochemistry
Immunohistochemistry (IHC) for ER, progesterone receptor (PR), HER2, PTEN, INPP4B, phosphorylated (p)AKT(Ser473), and pS6(Ser240/244) was performed on representative 5μm-thick FFPE sections containing either pure DCIS, or DCIS and adjacent IBCs, as described previously(31, 32, 40, 41). In brief, monoclonal antibodies against ER (clone 1D5, Dako, Carpinteria, CA, USA), PR (clone PgR636, Dako), HER2 (HercepTest™, Dako), and PTEN (clone 6H2.1, Dako) were diluted 1:100, against pAKT (Ser473, clone D9E, Cell Signaling, Danvers, MA, USA) 1:40, against INPP4B (clone EPR3108, Abcam, Cambridge, MA, USA) and against pS6 (Ser240/244, clone D68F8, Cell Signaling) 1:1000. ER, PR, PTEN, HER2, INPP4B and pS6 staining was performed using the Dako Autostainer Plus. pAKT staining was performed manually using an overnight primary antibody incubation at 4°C, and immunodetection with an avidin-biotin-peroxidase complex (Vectastain, Vector, Burlingame, CA, USA). All sections were counterstained with hematoxylin and reviewed by 4 observers (VPA, EGR, DG and RAS). HER2 was scored as per the HER2 American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP) guidelines for IBC(42) adapted to in situ disease, namely: 0 = no membranous expression; 1+ = weak, incomplete membrane staining in any proportion of tumor cells, or <10% of complete membrane staining; 2+ = complete membrane staining that is either non-uniform or weak in intensity, but with obvious circumferential distribution in at least 10% of cells; and 3+ = uniform intense membrane staining of >30% of invasive tumor cells(42). ER and PR were scored according to the current ASCO/CAP guidelines (i.e., >1% of nuclear staining in morphologically unequivocal neoplastic cells was considered positive)(43). For PTEN expression, normal epithelium and stroma served as an internal control, and tumor immunoreactivity was scored using a semi-quantitative system as previously described(40): score 0 = no immunoreaction; score 1 = reduced intensity of immunoreaction compared to normal epithelium; and score 2 = intensity equal to normal epithelium. For pAKT, the overall intensity score (0, 1+, 2+, 3+) was multiplied by the percentage of neoplastic cells expressing the marker (0–100%), and lesions were considered pAKT-positive if the final score was moderate (5–60) to high (61–300). For PTEN and pAKT analysis, FFPE pellets of PTEN wild-type (MCF7) and PTEN null (MDA-MB-468) cancer cell lines served as positive and negative controls, respectively. INPP4B loss of expression was defined as complete absence of expression or expression in <5% of neoplastic cells, as previously described(31). Normal breast epithelium and stroma served as internal controls. pS6 expression was evaluated using the H score system; lesions were considered positive if the final score was >100, as previously described(44).
HER2 fluorescence in situ hybridization (FISH)
HER2 amplification assessed by HercepTest™ was confirmed by FISH analysis using probes for HER2 (ERBB2, BAC clones RP11-94L15 and CTD-3211L18, Red-dUTP labeled) and CEP17 (plasmid clone p17H8, Green-dUTP labeled) (Abbott Molecular, Des Plaines, IL, USA). Following the ASCO/CAP guidelines(42), cases were considered amplified if the HER2/CEP17 ratio was greater than 2.2, equivocal between 2.2 and 1.8, and not amplified if the ratio was less than 1.8.
Microdissection and DNA extraction
Eight representative 10μm-thick sections were cut from each case and manually microdissected with a sterile scalpel under a stereomicroscope to ensure a tumor cell content of >70%. DNA of DCIS and IBC components were extracted separately using the QuickGene™ DNA tissue kit (Fujifilm, Singapore) as previously described(41). DNA quantification was performed with Quant-iT™ Picogreen® (Invitrogen, Life Technologies, Grand Island, NY, USA).
Mutation detection
DNA samples extracted from microdissected pure DCIS and from each component of the cases with adjacent DCIS and IBC were subjected to Sequenom™ MassARRAY™ (Sequenom, San Diego, CA, USA) analysis to detect PIK3CA hotspot (H1047R, E542K, E545K or N345K) and AKT1 (E17K) mutations, as previously described(41, 45). The multiplexed assays were designed using the Assay Design 3.1 Sequenom software. In brief, pre-PCR amplification (15ng gDNA) using the same primers as for Sequenom was performed before the iPLEX Gold genotyping assay, and 7nl of the purified primer extension reaction was loaded on a matrix pad of a SpectroCHIP (Sequenom) for analysis and measured by laser desorption/ionization of time-of-flight mass spectrometry. The prevalence of mutant alleles was estimated by calculating the ratio of the area of the raw spectra of the mutant allele to its wild-type, as previously described(23).
Statistical analysis
The association between PI3K pathway aberrations, type (DCIS vs. IBC), and ER and HER2 status was assessed using Fisher’s exact and Chi-Square tests for categorical data, and the two-tailed Student’s t-test for comparison of mean values. 95% confidence intervals were adopted, and p-values <0.05 were considered significant. Statistical analyses were performed using SAS software, version 9.1 (SAS Institute, Cary, NC, USA).
RESULTS
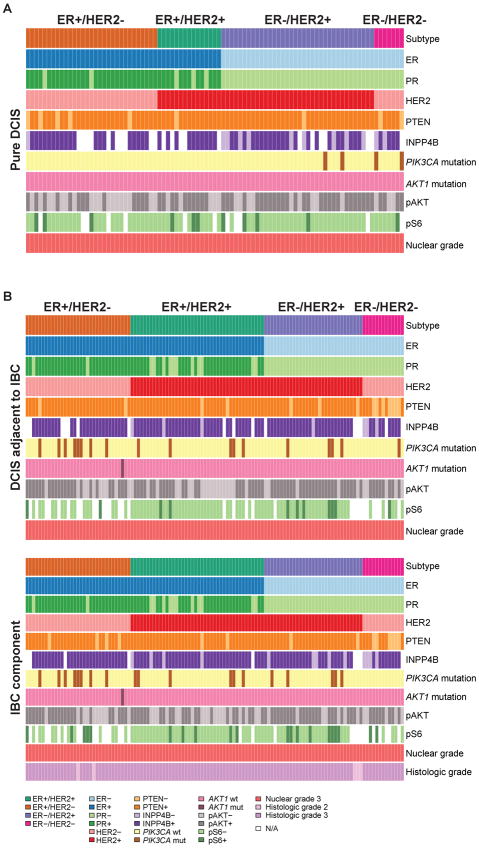
The clinical and pathologic characteristics of the 89 patients with pure high-grade DCIS were similar to those of the 119 patients with DCIS adjacent to invasive cancer (Supplementary Table S1). In brief, out of 89 cases of pure DCIS, 31 (35%) were ER-positive/HER2-negative, 15 (17%) ER-positive/HER2-positive, 36 (40%) ER-negative/HER2-positive, and 7 (8%) ER-negative/HER2-negative, whereas out of 119 cases of DCIS associated with invasive cancer, 33 (28%) were ER-positive/HER2-negative, 42 (35%) ER-positive/HER2-positive, 31 (26%) ER-negative/HER2-positive, and 13 (11%) ER-negative/HER2-negative. All DCIS and IBC cases included in this study had nuclear grade 3; the IBC components of the 119 cases with adjacent DCIS and IBC were of histologic grade 3 (n=114; 96%) and grade 2 (n=5; 4%) (Figure 1; Supplementary Table S1). In cases of DCIS adjacent to IBC, there was a perfect concordance in the ER, PR, and HER2 status of the in situ and invasive components of each case (Supplementary Table S1).
Figure 1. Summary of the histological, immunohistochemical and genetic characteristics of cases included in this study.
A) Pure high-grade DCIS and B) DCIS adjacent to invasive breast cancer and their matched invasive component.
DCIS, ductal carcinoma in situ; ER, estrogen receptor; IBC, invasive breast cancer; mut, mutant; N/A, not available; PR, progesterone receptor; wt, wild-type
PI3K pathway alterations differ in subtypes of pure DCIS and DCIS adjacent to IBC
Previous studies have reported PI3K pathway alterations in IBC and DCIS(23, 26, 28, 34–37), and have demonstrated that the mechanisms resulting in PI3K pathway activation vary according to the subtype of IBC(26, 28, 31). We posited that different subtypes of DCIS would differ in the prevalence of PTEN and INPP4B loss of expression, PIK3CA and AKT1 hotspot mutations, and pAKT and pS6 expression. Consistent with previous observations(23, 34–37), alterations of the PI3K pathway were frequently found in both pure DCIS and DCIS adjacent to IBC (Table 1; Figure 1; Supplementary Table S2; Supplementary Figure S1). In pure DCIS, significant differences in both the prevalence of PIK3CA hotspot mutations and loss of INPP4B expression were observed according to the subtypes, with 0%, 0%, 5% and 28% of ER-positive/HER2-negative, ER-positive/HER2-positive, ER-negative/HER2-positive, and ER-negative/HER2-negative lesions harboring PIK3CA mutations, respectively (4×2 Fisher’s exact test, p-value=0.0220; Table 1), and 5%, 0%, 30% and 34% of ER-positive/HER2-negative, ER-positive/HER2-positive, ER-negative/HER2-positive, and ER-negative/HER2-negative lesions, showing loss of INPP4B expression, respectively (4×2 Fisher’s exact test, p-value=0.027; Table 1). Differences in the prevalence of pAKT expression were also found in pure DCIS, with 42%, 67%, 75%, and 57% of ER-positive/HER2-negative, ER-positive/HER2-positive, ER-negative/HER2-positive, and ER-negative/HER2-negative lesions displaying pAKT expression, respectively (4×2 Fisher’s exact test, p-value=0.0427; Table 1). Of note, the only PIK3CA mutation observed in pure DCIS was the oncogenic H1047R kinase domain mutation (Supplementary Table S3). No significant differences in the prevalence of PTEN loss of expression, AKT1 mutations or pS6 expression were observed.
Table 1.
PI3K pathway alterations in pure high-grade DCIS and high-grade DCIS adjacent to IBC.
| ER-positive/HER2-negative | ER-positive/HER2-positive | ER-negative/HER2-positive | ER-negative/HER2-negative | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pure DCIS | DCIS adj IBC* | p-value | Pure DCIS | DCIS adj IBC | p-value | Pure DCIS | DCIS adj IBC | p-value | Pure DCIS* | DCIS adj IBC* | p-value | |
| PTEN loss | 5/31 (16%) | 2/33 (6%) | 0.250 | 1/15 (7%) | 2/42 (5%) | 1.000 | 1/36 (3%) | 3/31 (10%) | 0.329 | 2/7 (28%) | 7/13 (54%) | 0.374 |
| INPP4B loss | 1/21 (5%) | 0/26 (0%) | 0.447 | 0/10 (0%) | 3/39 (8%) | 1.000 | 9/30 (30%) | 1/27 (4%) | 0.013 | 2/6 (33%) | 3/11 (27%) | 1.000 |
| PIK3CA mut | 0/31 (0%) | 8/33 (24%) | 0.005 | 0/15 (0%) | 5/42 (12%) | 0.311 | 2/36 (5%) | 4/31 (13%) | 0.404 | 2/7 (28%) | 1/13 (8%) | 0.270 |
| AKT1 mut | 0/31 (0%) | 1/33 (3%) | 1.000 | 0/15 (0%) | 0/42 (0%) | 1.000 | 0/36 (0%) | 0/31 (0%) | 1.000 | 0/7 (0%) | 0/13 (0%) | 1.000 |
| pAKT pos | 13/31 (42%) | 23/33 (70%) | 0.043 | 10/15 (67%) | 23/42 (55%) | 0.547 | 27/36 (75%) | 26/31 (84%) | 0.548 | 4/7 (57%) | 8/13 (61%) | 1.000 |
| pS6 pos | 3/26 (11%) | 2/17 (12%) | 1.000 | 3/13 (23%) | 2/38 (5%) | 0.098 | 4/32 (12%) | 6/26 (23%) | 0.319 | 1/6 (17%) | 1/6 (17%) | 1.000 |
Adj, adjacent to; DCIS, ductal carcinoma in situ; ER, estrogen receptor; IBC, invasive breast cancer; mut, mutant; pos, positive
One sample had concurrent PTEN loss and PIK3CA mutation.
In contrast, in high-grade DCIS adjacent to IBC, significant differences between subtypes, as defined by ER and HER2 status, were found in relation to PTEN and INPP4B loss of expression (4×2 Fisher’s exact test, p-value<0.001 and p-value=0.0336, respectively; Table 1; Supplementary Table S2). No other significant differences in the prevalence of pAKT and pS6 expression, and PIK3CA or AKT1 mutations were observed. In fact, AKT1 mutations, albeit previously reported in a subset of DCIS (4%) and adjacent IBC(35), were shown to be remarkably rare in our study (0.5% of all lesions analyzed), suggesting that mutations affecting this gene may not constitute an important driver of high-grade DCIS.
Consistent with previous studies, which have demonstrated that PIK3CA mutations and PTEN loss are generally mutually exclusive in IBCs(26, 28), here we demonstrate that alterations affecting these genes were largely mutually exclusive in all subtypes of both pure DCIS and DCIS adjacent to IBC. In fact, PTEN loss of expression and PIK3CA mutations were concurrently found only in one pure DCIS of ER-negative/HER2-negative phenotype, and in two DCIS adjacent to IBC, one ER-positive/HER2-negative, and another ER-negative/HER2-negative (Figures 1 and 2). We also observed that INPP4B loss of expression was preferentially found in cases lacking PTEN loss of expression and/or PIK3CA hotspot mutations. In fact, concurrent INPP4B and PTEN loss of expression was found only in two pure DCIS and two DCIS adjacent to IBC, whereas concurrent INPP4B loss of expression and PIK3CA hotspot mutations were found only in one case of pure DCIS (Figure 1).
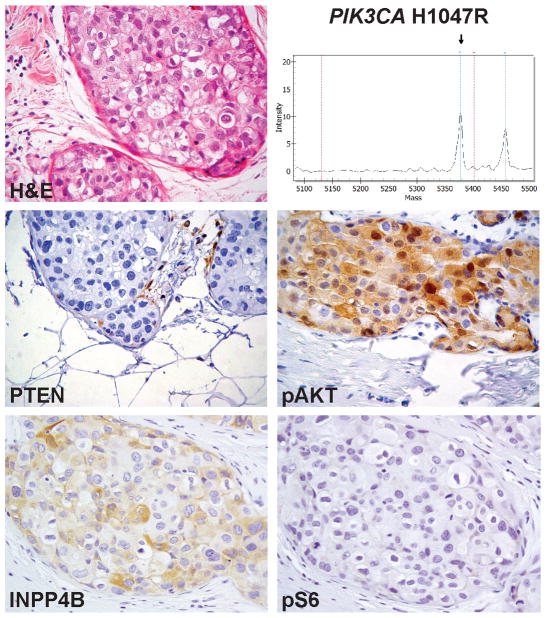
Figure 2. PIK3CA mutations and PTEN loss of expression may not be mutually exclusive in breast cancer.
Representative micrograph of a pure DCIS concurrently harboring a H1047R PIK3CA mutation and PTEN loss of expression as defined by immunohistochemistry (case D094). Note that INPP4B and pAKT were expressed at moderate-to-high levels, whilst pS6 expression was absent.
H&E, hematoxylin and eosin. Magnification of micrographs: 200x
Comparative analysis of the cases of pure DCIS and DCIS adjacent to IBC matched according to nuclear grade and subtype revealed remarkable similarities in the prevalence of PTEN and INPP4B loss of expression, presence of PIK3CA hotspot mutations, AKT1 mutations, and pAKT and pS6 expression (Table 1). In fact, significant differences were only observed in the group of ER-positive/HER2-negative lesions, where significantly higher frequencies of PIK3CA hotspot mutations and pAKT expression were found in DCIS adjacent to IBC than in pure DCIS (Fisher’s exact test p-values: 0.005 and 0.043, respectively; Table 1), and in ER-negative/HER2-positive lesions, where a significantly higher prevalence of INPP4B loss of expression was found in pure DCIS than in DCIS adjacent to IBC (30% vs. 4%, respectively; Fisher’s exact test p-value=0.013; Table 1). No significant differences were found between pure DCIS and DCIS adjacent to IBC in ER-positive/HER2-positive and ER-negative/HER2-negative lesions.
These observations demonstrate that in a way akin to IBC, different subtypes of DCIS, as defined by ER and HER2 status, have different patterns of PI3K pathway alterations, and that this pathway is altered in similar ways in pure DCIS and DCIS adjacent to IBC. Although PIK3CA mutations and pAKT expression were more frequently found in DCIS adjacent to IBC than in pure ER-positive/HER2-negative DCIS, in ER-negative/HER2-positive lesions, a significantly higher frequency of loss of INPP4B expression was found in pure DCIS than in DCIS adjacent to IBC. Taken together, our findings suggest that alterations in the PI3K pathway may play a role in the progression from in situ to invasive disease in a subset of ER-positive/HER2-negative DCIS.
PI3K pathway alterations are largely maintained in the progression from DCIS to IBC
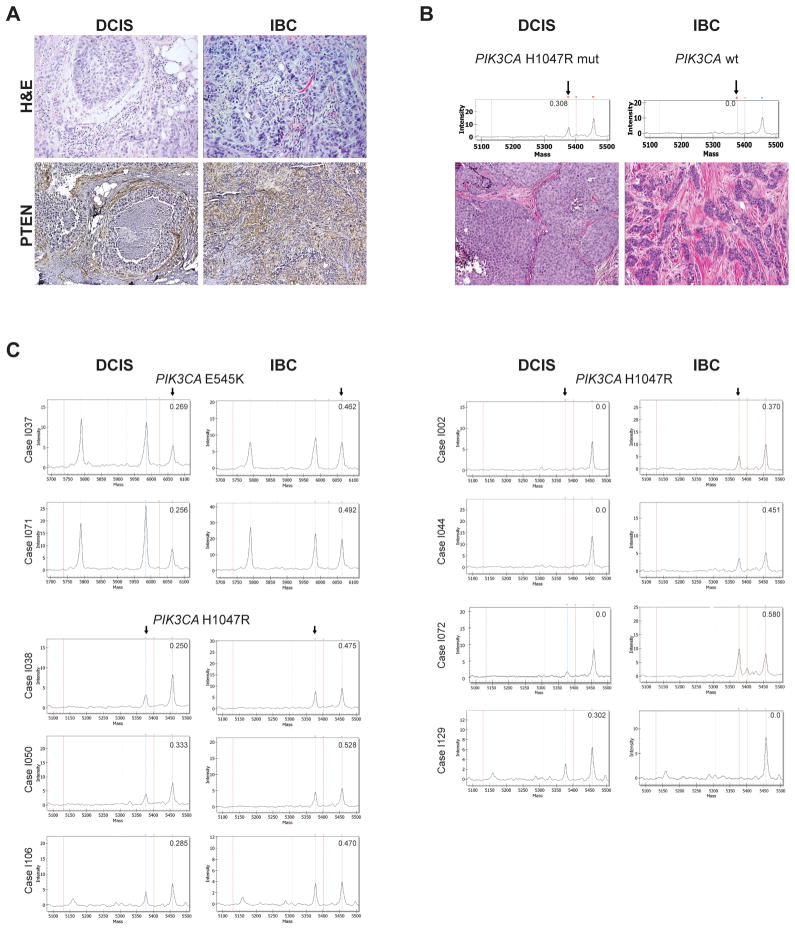
Despite the qualitative similarities between synchronous DCIS and IBC, recent pair-wise comparisons of these lesions have demonstrated that DCIS and IBC may differ by the presence of specific genetic aberrations(3, 23, 24). We sought to determine whether PTEN and INPP4B loss of expression, PIK3CA and AKT1 hotspot mutations, and pAKT and pS6 expression would differ between the in situ and invasive components of cases of synchronous DCIS and IBC. Although differences in PTEN loss of expression, presence of PIK3CA mutations, and pAKT and pS6 expression were observed between matched DCIS and IBC, these changes were not unidirectional (Figure 1). For example, out of the 8 cases with differences in PTEN loss of expression, 4 showed PTEN loss in the IBC component but not in the DCIS, whereas in the remaining 4 cases, PTEN was expressed in the invasive component but absent in the DCIS (Figures 1 and 3A). Changes in INPP4B loss of expression, on the other hand, were unidirectional; in the progression from DCIS to IBC, six cases displayed loss of INPP4B expression, five of which were ER-negative lesions (Figure 1). Consistent with previous observations(23, 34–37), no significant qualitative differences were observed between the DCIS and invasive components of the cases analyzed (Table 2; Supplementary Table S2). When stratified according to ER and HER2 status, again, the DCIS and invasive components of each case were remarkably similar in regards to PTEN and INPP4B loss of expression, presence of PIK3CA and AKT1 mutations, and pAKT and pS6 expression (Table 2).
Figure 3. PI3K pathway alterations may be discordant between the in situ and adjacent invasive components of breast cancer.
In A, representative micrographs of a DCIS and its synchronous, adjacent invasive breast cancer where PTEN loss of expression was detected in the in situ but not in the invasive component (case I063). In B, representative micrographs of a DCIS and its synchronous, adjacent invasive breast cancer where mutant PIK3CA was detected in the modal population of the DCIS cells, but absent in the invasive component (case I068). In C, representative Sequenom MassARRAY plots of cases with discordant PIK3CA mutation status between the in situ and invasive components of cases of DCIS adjacent to invasive breast cancer. The numbers in each plot refer to the PIK3CA mutant allele frequency. In B and C, arrows highlight the mutant allele peak in the Sequenom MassARRAY plots.
DCIS, ductal carcinoma in situ; H&E, hematoxylin and eosin; IBC, invasive breast cancer.
Table 2.
PI3K pathway alterations in high-grade DCIS component and in matched adjacent IBC component.
| ER-positive/HER2-negative | ER-positive/HER2-positive | ER-negative/HER2-positive | ER-negative/HER2-negative | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DCIS* | IBC* | p-value | DCIS | IBC | p-value | DCIS | IBC | p-value | DCIS* | IBC | p-value | |
| PTEN loss | 2/33 (6%) | 4/33 (12%) | 0.672 | 2/42 (5%) | 2/42 (5%) | 1.000 | 3/31 (10%) | 2/31 (6%) | 1.000 | 7/13 (54%) | 6/13 (46%) | 1.000 |
| INPP4B loss | 0/26 (0%) | 0/26 (0%) | 1.000 | 3/39 (8%) | 4/39 (10%) | 1.000 | 1/27 (4%) | 5/27 (18%) | 0.192 | 3/11 (27%) | 4/11 (36%) | 1.000 |
| PIK3CA mut | 8/33 (24%) | 8/33 (24%) | 1.000 | 5/42 (12%) | 6/42 (14%) | 1.000 | 4/31 (13%) | 5/31 (16%) | 1.000 | 1/13 (8%) | 0/13 (0%) | 1.000 |
| AKT1 mut | 1/33 (3%) | 1/33 (3%) | 1.000 | 0/42 (0%) | 0/42 (0%) | 1.000 | 0/31 (0%) | 0/31 (0%) | 1.000 | 0/13 (0%) | 0/13 (0%) | 1.000 |
| pAKT pos | 23/33 (70%) | 22/33 (67%) | 1.000 | 23/42 (55%) | 23/42 (55%) | 1.000 | 26/31 (84%) | 24/31 (77%) | 0.749 | 8/13 (61%) | 8/13 (61%) | 1.000 |
| pS6 pos | 2/16 (12%) | 5/16 (31%) | 0.394 | 2/37 (5%) | 5/37 (13%) | 0.430 | 6/26 (23%) | 8/26 (31%) | 0.755 | 0/5 (0%) | 1/5 (20%) | 1.000 |
DCIS, ductal carcinoma in situ; ER, estrogen receptor; IBC, invasive breast cancer; mut, mutant; pos, positive
One sample had concurrent PTEN loss and PIK3CA mutation.
Changes in PIK3CA mutation status in the progression from DCIS to IBC
Although PIK3CA mutations have been reported at similar frequencies in DCIS and IBC(23, 34–37), recent studies have described changes in the PIK3CA status in the progression from in situ to invasive disease(23, 36). Here we have observed that in 3 cases, the H1047R PIK3CA mutation was present in the invasive component but not detectable in the synchronous DCIS areas, and in 5 additional cases, the PIK3CA mutation was present in a non-modal population of the DCIS cells (PIK3CA mutant allele frequencies ranging from 25%–33.3%), but likely present in the modal population of the IBC (PIK3CA mutant allele frequencies ranging from 46.2%–52.8%; Table 3; Figures 1 and 3C). In two additional cases, the H1047R PIK3CA mutation was restricted to the DCIS component but absent in the IBC (Figures 1, 3B and 3C, Table 3).
Table 3.
Cases with discordant PIK3CA mutant frequencies in the high-grade DCIS and synchronous IBC components.
| Case ID | Component | ER | PR | HER2 | PIK3CA mutation | Allele | WT frequency | Mutation frequency | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | I037 | DCIS | + | + | + | E545K | A | 0.731 | 0.269 |
| IBC | + | + | + | E545K | A | 0.538 | 0.462 | ||
| 2 | I071 | DCIS | − | − | + | E545K | A | 0.744 | 0.256 |
| IBC | − | − | + | E545K | A | 0.508 | 0.492 | ||
| 3 | I038 | DCIS | + | + | + | H1047R | G | 0.750 | 0.250 |
| IBC | + | + | + | H1047R | G | 0.525 | 0.475 | ||
| 4 | I050 | DCIS | − | − | + | H1047R | G | 0.667 | 0.333 |
| IBC | − | − | + | H1047R | G | 0.472 | 0.528 | ||
| 5 | I106 | DCIS | + | + | − | H1047R | G | 0.715 | 0.285 |
| IBC | + | + | − | H1047R | G | 0.530 | 0.470 | ||
| 6 | I002 | DCIS | + | + | + | none | 1.000 | 0 | |
| IBC | + | + | + | H1047R | G | 0.630 | 0.370 | ||
| 7 | I044 | DCIS | − | − | + | none | 1.000 | 0 | |
| IBC | − | − | + | H1047R | G | 0.549 | 0.451 | ||
| 8 | I072 | DCIS | − | − | + | none | 1.000 | 0 | |
| IBC | − | − | + | H1047R | G | 0.344 | 0.580 | ||
| 9 | I068 | DCIS | − | − | + | H1047R | G | 0.692 | 0.308 |
| IBC | − | − | + | none | 1.000 | 0 | |||
| 10 | I129 | DCIS | − | − | − | H1047R | G | 0.698 | 0.302 |
| IBC | − | − | − | none | 1.000 | 0 |
DCIS, ductal carcinoma in situ; ER, estrogen receptor; IBC, invasive breast cancer; PR, progesterone receptor; WT, wild-type
Taken together, our results demonstrate the existence of intra-tumor genetic heterogeneity in DCIS and suggest that in the progression from DCIS to IBC, subclones of neoplastic cells harboring specific repertoires of genetic aberrations may be selected. Furthermore, our data support the contention that although PIK3CA mutations may play a role from the early stages of breast tumorigenesis, their role as driver of the progression from in situ to invasive disease is less clear, given that examples of both PIK3CA wild-type DCIS adjacent to PIK3CA mutant IBC and of PIK3CA wild-type IBC adjacent to PIK3CA mutant DCIS were observed.
DISCUSSION
Here we demonstrate that both pure high-grade DCIS and high-grade DCIS adjacent to IBC often harbor molecular alterations that result in activation of the PI3K pathway, and that in a way akin to IBCs, different subtypes of DCIS, as defined by ER and HER2 status, display different patterns of alterations affecting genes in the PI3K canonical pathway. We have also confirmed previous observations demonstrating that synchronous DCIS and IBCs display remarkably similar patterns of alterations of this pathway; however, we also provide direct evidence of the existence of intra-tumor genetic heterogeneity in DCIS and that the PIK3CA mutation status may change in the progression from in situ to invasive disease.
PI3K pathway alterations have previously been documented in breast cancer(23, 26, 28, 31, 32, 34–37). Here we not only confirmed that a subset of DCIS do harbor PIK3CA mutations, but also provided an integrative analysis combining an assessment of the most common mechanisms of activation of this pathway, and an immunohistochemical assessment of the PI3K pathway activity employing pAKT and pS6 as surrogates of activation of this pathway. In both pure DCIS and DCIS adjacent to IBC, we have observed that the presence of PTEN and INPP4B loss of expression and/or mutations in PIK3CA or AKT1 varied significantly according to subtype (Table 1). Importantly, the observation that PIK3CA hotspot mutations and pAKT expression were significantly more frequent in ER-positive/HER2-negative DCIS adjacent to IBC than in pure DCIS is consistent with the notion that PI3K pathway activation may impart increased risk of or association with invasive progression in this disease subtype. Additional studies to test this hypothesis are warranted.
Using a subtype-matched approach, we have observed a significantly higher prevalence of PIK3CA mutations and pAKT activity in high-grade ER-positive/HER2-negative DCIS adjacent to IBC than in pure DCIS. In high-grade ER-negative/HER2-positive lesions, however, INPP4B loss of expression, an event that can potentially activate the PI3K pathway, was more frequent in pure DCIS than in high-grade DCIS adjacent to IBC. These observations demonstrate that loss of PTEN and INPP4B expression, and mutations affecting PIK3CA and AKT1 are present in a subset of both pure high-grade DCIS and high-grade DCIS adjacent to IBC, providing additional evidence to support the role of this pathway in the early stages of breast cancer development.
In pure high-grade DCIS, PIK3CA mutations were relatively infrequent and were not found in ER-positive lesions; on the other hand, 5% of ER-negative/HER2-positive and 28% of the ER-negative/HER2-negative high-grade DCIS harbored the H1047R PIK3CA mutation. In high-grade DCIS adjacent to IBC, however, PIK3CA mutations were present in all subtypes, ranging from 8% in ER-negative/HER2-negative lesions to 24% in ER-positive/HER2-positive disease. Out of all ER-positive DCIS analyzed in this study, only 11% harbored PIK3CA hotspot mutations. Albeit at first glance at variance with the notion that PIK3CA mutations are more frequently found in ER-positive IBCs, these seemingly unexpected findings can be reconciled by the fact that we have focused on high-grade ER-positive lesions, which have been reported to less frequently harbor PIK3CA mutations than low-grade ER-positive IBCs (24%-49%)(26, 36, 46), low-grade pure DCIS (34%)(36), and early precursors of low-grade forms of DCIS (54%)(47). The high prevalence of PIK3CA mutations in non-obligate precursors of low-grade DCIS (e.g., columnar cell lesions) and low-grade DCIS, in conjunction with the low frequencies of PIK3CA mutations in high-grade ER-positive DCIS analyzed in this study are consistent with the notion that the molecular pathways involved in the development and progression of low- and high-grade DCIS are likely distinct(1), and that in high-grade DCIS, PIK3CA mutations may only play a role in a minority of cases.
Our analysis of PI3K pathway activation using pAKT and pS6 as surrogate markers suggested that activation of this pathway is more frequent than alterations of PTEN, INPP4B, PIK3CA, and AKT1 in pure high-grade DCIS and high-grade DCIS adjacent to IBC. In all subtypes, pAKT expression was more prevalent than pS6 expression in both pure high DCIS and DCIS adjacent to IBC (Table 1). Concurrent expression of pAKT and pS6 was observed in 13% and 38% of pure DCIS and DCIS adjacent to IBC, respectively. The vast majority of cases (69% of pure DCIS and 75% of DCIS adjacent to IBC) harboring PTEN or INPP4B loss of expression, or PIK3CA or AKT1 mutations, displayed pAKT expression, whereas pS6 expression was found in 6% and 21% of pure DCIS and DCIS adjacent to IBC harboring these molecular aberrations. The lack of pAKT and pS6 expression in cases with alterations in these genes may stem from the fact that we have i) assessed pAKT and pS6 expression in surgical specimens and that previous analyses have shown that their immunohistochemical assessment is affected by pre-analytical variables, and its expression levels are significantly lower in surgical specimens than in core biopsies(48, 49); and ii) employed antibodies that recognize only a few phosphorylation sites of AKT (i.e. Ser473) and S6 (i.e. Ser240/244). Although our study may have underestimated the prevalence of PI3K pathway activation in DCIS, our results do demonstrate that mechanisms other than PTEN or INPP4B loss of expression, and PIK3CA or AKT1 mutations may result in activation of this pathway in DCIS, and warrant further studies investigating the causes of PI3K pathway activation in these lesions.
Recent studies based on aCGH, FISH, and Sequenom analysis, or on multi-probe FISH analysis of synchronous DCIS and IBC, have demonstrated that from a qualitative standpoint, DCIS and IBC samples from a given patient have strikingly similar genomic profiles(23, 24). These studies, however, have revealed not only intra-lesion genetic heterogeneity but also differences in the prevalence of amplifications affecting specific loci and in the prevalence of mutations between the DCIS and invasive samples. In Hernandez et al.(23), in three of 13 cases of synchronous DCIS and IBC harboring PIK3CA mutations, these mutations were either restricted to the DCIS component (n=2) or the frequency of the PIK3CA mutant allele was decreased in the IBC when compared to the DCIS component(23). Our results confirm and expand on previous observations, given that in two cases, PIK3CA mutations were present in the DCIS but absent in the IBC component, whereas in three cases, mutations affecting this gene were found in the invasive component but not in the DCIS. In addition, differences in the frequencies of the mutant allele varied from the DCIS to the invasive component in five cases. Taken together, these observations are consistent with a model where DCIS is composed of a mosaic of tumor cells that, in addition to the founder genetic aberrations, harbor private mutations, and that clonal selection is likely to take place in the progression from in situ to invasive disease(3). These results provide another line of evidence to suggest that PIK3CA mutations may play a role in the progression from in situ to invasive disease in a small subset of cases.
Our study has several limitations. First, the retrospective identification of tissue specimens and the selection criteria that included samples with at least two FFPE blocks available may limit the generalizability of the results; however, we have assembled a large cohort of carefully analyzed cases and our findings should be considered as exploratory and hypothesis generating. Second, the sample size of some of the subtypes of DCIS investigated in this study (i.e. ER-negative/HER2-negative DCIS) is small; hence, we cannot rule out type II or β-errors in the comparative analyses performed in this subgroup. Third, given the evidence to suggest that low-and high-grade DCIS and IBCs are likely to evolve through distinct pathways (reviewed in(1)), we have focused only on the subset of high-grade lesions; hence, our conclusions should be considered relevant only to high-grade disease. Fourth, the surrogate markers employed to determine activation of the PI3K pathway, pAKT and pS6 immunohistochemistry, have been shown to be affected by pre-analytical parameters, including delayed fixation and the type of specimen(48, 49). Although the frequency of PI3K pathway activation may have been underestimated in this study due to the use of surgical specimens, given that all tissues were collected during the same time frame at a single institution, the reduction in pAKT and pS6 expression driven by pre-analytical parameters should be equally prevalent among all groups. Finally, we have only investigated a limited number of PIK3CA hotspot mutations; hence, we may not have captured all cases harboring activating mutations in this gene. It should be noted, however, that in IBC the PIK3CA mutations included in the Sequenom MassARRAY (i.e., H1047R, E542K, E545K or N345K) assay employed here account for 87% of all mutant cases reported by The Cancer Genome Atlas(26).
In conclusion, here we demonstrate that PTEN and INPP4B loss of expression, PIK3CA hotspot mutations, and AKT1 mutations are found in a subset of pure high-grade DCIS and high-grade DCIS adjacent to IBC, that the prevalence of alterations affecting these genes vary more according to the ER/HER2 subtype of DCIS than to its association with synchronous IBC. PTEN loss of expression was infrequent in subtypes other than high-grade ER-negative/HER2-negative DCIS, INPP4B loss of expression was preferentially found in ER-negative/HER2-positive and ER-negative/HER2-negative DCIS, PIK3CA mutations were relatively uncommon in all subtypes of high-grade in situ disease (0%-28%), and AKT1 mutations were only found in 0.5% of all lesions analyzed, yet activation of the PI3K pathway, as defined by pAKT and/or pS6 expression, was shown to be a more pervasive biological phenomenon, possibly driven by genetic (e.g., HER2 gene amplification) or epigenetic alterations other than those surveyed in our study. Our findings also demonstrate the qualitative similarities in ER, PR, and HER2 status, and PI3K pathway alterations between DCIS and synchronous invasive IBCs, suggesting that the overall phenotype of a breast cancer is likely to be determined early in tumorigenesis. Intra-tumor genetic heterogeneity and selection of genetically distinct clones in the progression from in situ to invasive disease, however, were documented in a subset cases, and qualitative and quantitative differences in the presence and percentage of PIK3CA mutant alleles between matched DCIS and IBC. Our findings provide additional evidence to demonstrate the importance of the PI3K pathway in breast cancer and that PI3K pathway aberrations may be associated with a higher risk of progression in a subset of lesions; however, its role in mediating the progression from in situ to invasive disease appears to be more limited.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Ductal carcinoma in situ (DCIS) is considered a non-obligate precursor of invasive breast cancer (IBC). The introduction of mammography screening has resulted in a dramatic increase in the incidence of DCIS. The majority of women with DCIS will not develop IBC, yet we currently lack effective tools to predict which lesions are most likely to progress and, as such, current treatment recommendations are based on the notion that every DCIS has the potential to progress to IBC over time. Germane to the development of biomarkers to differentiate between DCIS that will or will not progress to IBC is the characterization of the mechanisms that drive progression. Although the PI3K pathway plays a pivotal role in breast cancer, here we demonstrate that alterations in key components of this pathway may play a role in the progression from high-grade DCIS to IBC only in a subset of cases.
Acknowledgments
Financial support: This study was funded in part by the Walsh Family Fund, the Cary Grossman Breast Surgery Research Fund, and NIH/NCI Cancer Center Support Grant P30 CA008748.
The authors acknowledge V Barbashina and JP Catalano for the collection of the archived FFPE samples, and M Leversha for the assessment of HER2 by FISH. They also gratefully acknowledge H Liyanage and A Heguy for assistance with the Sequenom MassArray analysis.
Footnotes
Conflict of interest: The authors have no conflict of interest to declare.
AUTHOR CONTRIBUTIONS
Conception and design: R.A. Sakr, T.A. King, S. Chandarlapaty, N. Rosen
Development of methodology: R.A. Sakr, T.A. King, S. Chandarlapaty
Acquisition of data (acquired and managed patients, provided facilities, etc.): R.A. Sakr, V.P. Andrade, E. Guerini-Rocco, D. Giri
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): R.A. Sakr, T.A. King, S. Chandarlapaty, B. Weigelt, C.K.Y. Ng, C.F. Cowell, J.S. Reis-Filho
Writing, review and/or revision of the manuscript: R.A. Sakr, B. Weigelt, T.A. King, S. Chandarlapaty, J.S. Reis-Filho
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): R.A. Sakr, T.A. King, B. Weigelt, J.S. Reis-Filho
Study supervision: R.A. Sakr, T.A. King
References
- 1.Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, Marchio C, Reis-Filho JS. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology. 2010;57:171–92. doi: 10.1111/j.1365-2559.2010.03568.x. [DOI] [PubMed] [Google Scholar]
- 2.Polyak K. Molecular markers for the diagnosis and management of ductal carcinoma in situ. J Natl Cancer Inst Monogr. 2010;2010:210–3. doi: 10.1093/jncimonographs/lgq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowell CF, Weigelt B, Sakr RA, Ng CK, Hicks J, King TA, et al. Progression from ductal carcinoma in situ to invasive breast cancer: Revisited. Mol Oncol. 2013;7:859–69. doi: 10.1016/j.molonc.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Society AC. Cancer Facts and Figures 2013. Atlanta, Ga: American Cancer Society; 2013. [Google Scholar]
- 5.Wellings SR, Jensen HM. On the origin and progression of ductal carcinoma in the human breast. J Natl Cancer Inst. 1973;50:1111–8. doi: 10.1093/jnci/50.5.1111. [DOI] [PubMed] [Google Scholar]
- 6.Buerger H, Otterbach F, Simon R, Poremba C, Diallo R, Decker T, et al. Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. J Pathol. 1999;187:396–402. doi: 10.1002/(SICI)1096-9896(199903)187:4<396::AID-PATH286>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Buerger H, Otterbach F, Simon R, Schafer KL, Poremba C, Diallo R, et al. Different genetic pathways in the evolution of invasive breast cancer are associated with distinct morphological subtypes. J Pathol. 1999;189:521–6. doi: 10.1002/(SICI)1096-9896(199912)189:4<521::AID-PATH472>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 8.Burkhardt L, Grob TJ, Hermann I, Burandt E, Choschzick M, Janicke F, et al. Gene amplification in ductal carcinoma in situ of the breast. Breast Cancer Res Treat. 2010;123:757–65. doi: 10.1007/s10549-009-0675-8. [DOI] [PubMed] [Google Scholar]
- 9.O’Connell P, Pekkel V, Fuqua SA, Osborne CK, Clark GM, Allred DC. Analysis of loss of heterozygosity in 399 premalignant breast lesions at 15 genetic loci. J Natl Cancer Inst. 1998;90:697–703. doi: 10.1093/jnci/90.9.697. [DOI] [PubMed] [Google Scholar]
- 10.Solin LJ, Gray R, Baehner FL, Butler SM, Hughes LL, Yoshizawa C, et al. A multigene expression assay to predict local recurrence risk for ductal carcinoma in situ of the breast. J Natl Cancer Inst. 2013;105:701–10. doi: 10.1093/jnci/djt067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerlikowske K, Molinaro AM, Gauthier ML, Berman HK, Waldman F, Bennington J, et al. Biomarker expression and risk of subsequent tumors after initial ductal carcinoma in situ diagnosis. J Natl Cancer Inst. 2010;102:627–37. doi: 10.1093/jnci/djq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, Niu Y, Wang X, Wei L, Lu S. Genetic changes at specific stages of breast cancer progression detected by comparative genomic hybridization. J Mol Med (Berl) 2009;87:145–52. doi: 10.1007/s00109-008-0408-1. [DOI] [PubMed] [Google Scholar]
- 13.Liao S, Desouki MM, Gaile DP, Shepherd L, Nowak NJ, Conroy J, et al. Differential copy number aberrations in novel candidate genes associated with progression from in situ to invasive ductal carcinoma of the breast. Genes Chromosomes Cancer. 2012;51:1067–78. doi: 10.1002/gcc.21991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065–78. doi: 10.1158/0008-5472.CAN-05-4083. [DOI] [PubMed] [Google Scholar]
- 15.Consortium EP, Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moelans CB, de Wegers RA, Monsuurs HN, Maess AH, van Diest PJ. Molecular differences between ductal carcinoma in situ and adjacent invasive breast carcinoma: a multiplex ligation-dependent probe amplification study. Cell Oncol (Dordr) 2011;34:475–82. doi: 10.1007/s13402-011-0043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–75. [PubMed] [Google Scholar]
- 18.Johnson CE, Gorringe KL, Thompson ER, Opeskin K, Boyle SE, Wang Y, et al. Identification of copy number alterations associated with the progression of DCIS to invasive ductal carcinoma. Breast Cancer Res Treat. 2012;133:889–98. doi: 10.1007/s10549-011-1835-1. [DOI] [PubMed] [Google Scholar]
- 19.Robanus-Maandag EC, Bosch CA, Kristel PM, Hart AA, Faneyte IF, Nederlof PM, et al. Association of C-MYC amplification with progression from the in situ to the invasive stage in C-MYC-amplified breast carcinomas. J Pathol. 2003;201:75–82. doi: 10.1002/path.1385. [DOI] [PubMed] [Google Scholar]
- 20.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:5974–9. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vincent-Salomon A, Lucchesi C, Gruel N, Raynal V, Pierron G, Goudefroye R, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res. 2008;14:1956–65. doi: 10.1158/1078-0432.CCR-07-1465. [DOI] [PubMed] [Google Scholar]
- 22.Jang MH, Kim EJ, Choi Y, Lee HE, Kim YJ, Kim JH, et al. FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Res. 2012;14:R115. doi: 10.1186/bcr3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez L, Wilkerson PM, Lambros MB, Campion-Flora A, Rodrigues DN, Gauthier A, et al. Genomic and mutational profiling of ductal carcinomas in situ and matched adjacent invasive breast cancers reveals intra-tumour genetic heterogeneity and clonal selection. J Pathol. 2012;227:42–52. doi: 10.1002/path.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heselmeyer-Haddad K, Berroa Garcia LY, Bradley A, Ortiz-Melendez C, Lee WJ, Christensen R, et al. Single-cell genetic analysis of ductal carcinoma in situ and invasive breast cancer reveals enormous tumor heterogeneity yet conserved genomic imbalances and gain of MYC during progression. Am J Pathol. 2012;181:1807–22. doi: 10.1016/j.ajpath.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet. 2011;378:1812–23. doi: 10.1016/S0140-6736(11)61539-0. [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–83. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–91. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marty B, Maire V, Gravier E, Rigaill G, Vincent-Salomon A, Kappler M, et al. Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast Cancer Res. 2008;10:R101. doi: 10.1186/bcr2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones N, Bonnet F, Sfar S, Lafitte M, Lafon D, Sierankowski G, et al. Comprehensive analysis of PTEN status in breast carcinomas. Int J Cancer. 2013;133:323–34. doi: 10.1002/ijc.28021. [DOI] [PubMed] [Google Scholar]
- 31.Fedele CG, Ooms LM, Ho M, Vieusseux J, O’Toole SA, Millar EK, et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc Natl Acad Sci U S A. 2010;107:22231–6. doi: 10.1073/pnas.1015245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–25. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–44. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 34.Troxell ML, Levine J, Beadling C, Warrick A, Dunlap J, Presnell A, et al. High prevalence of PIK3CA/AKT pathway mutations in papillary neoplasms of the breast. Mod Pathol. 2010;23:27–37. doi: 10.1038/modpathol.2009.142. [DOI] [PubMed] [Google Scholar]
- 35.Dunlap J, Le C, Shukla A, Patterson J, Presnell A, Heinrich MC, et al. Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res Treat. 2010;120:409–18. doi: 10.1007/s10549-009-0406-1. [DOI] [PubMed] [Google Scholar]
- 36.Miron A, Varadi M, Carrasco D, Li H, Luongo L, Kim HJ, et al. PIK3CA mutations in in situ and invasive breast carcinomas. Cancer Res. 2010;70:5674–8. doi: 10.1158/0008-5472.CAN-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalinsky K, Heguy A, Bhanot UK, Patil S, Moynahan ME. PIK3CA mutations rarely demonstrate genotypic intratumoral heterogeneity and are selected for in breast cancer progression. Breast Cancer Res Treat. 2011;129:635–43. doi: 10.1007/s10549-011-1601-4. [DOI] [PubMed] [Google Scholar]
- 38.Silverstein MJ, Poller DN, Waisman JR, Colburn WJ, Barth A, Gierson ED, et al. Prognostic classification of breast ductal carcinoma-in-situ. Lancet. 1995;345:1154–7. doi: 10.1016/s0140-6736(95)90982-6. [DOI] [PubMed] [Google Scholar]
- 39.Fisher ER, Gregorio RM, Fisher B, Redmond C, Vellios F, Sommers SC. The pathology of invasive breast cancer. A syllabus derived from findings of the National Surgical Adjuvant Breast Project (protocol no. 4) Cancer. 1975;36:1–85. doi: 10.1002/1097-0142(197507)36:1<1::aid-cncr2820360102>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 40.Sakr RA, Barbashina V, Morrogh M, Chandarlapaty S, Andrade VP, Arroyo CD, et al. Protocol for PTEN expression by immunohistochemistry in formalin-fixed paraffin-embedded human breast carcinoma. Appl Immunohistochem Mol Morphol. 2010;18:371–4. doi: 10.1097/PAI.0b013e3181d50bd5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandarlapaty S, Sakr RA, Giri D, Patil S, Heguy A, Morrow M, et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res. 2012;18:6784–91. doi: 10.1158/1078-0432.CCR-12-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–45. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 43.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–95. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adamo B, Deal AM, Burrows E, Geradts J, Hamilton E, Blackwell KL, et al. Phosphatidylinositol 3-kinase pathway activation in breast cancer brain metastases. Breast Cancer Res. 2011;13:R125. doi: 10.1186/bcr3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duprez R, Wilkerson PM, Lacroix-Triki M, Lambros MB, Mackay A, Hern RA, et al. Immunophenotypic and genomic characterization of papillary carcinomas of the breast. J Pathol. 2012;226:427–41. doi: 10.1002/path.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–59. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 47.Troxell ML, Brunner AL, Neff T, Warrick A, Beadling C, Montgomery K, et al. Phosphatidylinositol-3-kinase pathway mutations are common in breast columnar cell lesions. Mod Pathol. 2012;25:930–7. doi: 10.1038/modpathol.2012.55. [DOI] [PubMed] [Google Scholar]
- 48.Pinhel IF, Macneill FA, Hills MJ, Salter J, Detre S, A’Hern R, et al. Extreme loss of immunoreactive p-Akt and p-Erk1/2 during routine fixation of primary breast cancer. Breast Cancer Res. 2010;12:R76. doi: 10.1186/bcr2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meric-Bernstam F, Akcakanat A, Chen H, Sahin A, Tarco E, Carkaci S, et al. Effect of biospecimen variables on proteomic biomarker assessment in breast cancer. Cancer Res. 2012;72:P1-07-6. doi: 10.1158/1078-0432.CCR-13-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.