Abstract
Interleukin 18 (interferon gamma-inducing factor) (IL18) is an important proinflammatory cytokine that belongs to the IL1 family. This study investigated whether IL18 single-nucleotide polymorphisms (SNPs) are associated with the susceptibility to alopecia areata (AA) in a Korean population. Two hundred thirty-three AA patients and 243 healthy control subjects were recruited. One promoter SNP (rs187238, −137G/C) and exonic SNP (rs549908, Ser35Ser) in IL18 were genotyped using direct sequencing. SNPStats, SPSS 18.0, and Haploview version 4.2 programs were used to evaluate genetic data. Multiple logistic regression models were used to determine odds ratios, 95% confidence intervals, and P values. Tested 2 SNPs (rs187238 and rs549908) were associated with the development of AA (rs187238, P=0.002 in a codominant model 1, P=0.0048 in a dominant model, P=0.02 in a log-additive model, P=0.023 in allele distribution; rs549908, P=0.003 in a codominant model 1, P=0.0052 in a dominant model, P=0.016 in a log-additive model, P=0.015 in allele distribution). Our data suggest that the IL18 may be a risk factor for AA susceptibility.
Introduction
Alopecia areata (AA) is a common, chronic, and inflammatory disease. It is considered to be a tissue-specific autoimmune disease. The AA patients appear to develop antibodies to their own hair follicle structures, which suppress their hair growth (Kos and Conlon 2009). AA is found within ∼1%–2% of the general population (Safavi and others 1995; Forstbauer and others 2012). The age of onset of AA may occur at any age. However, it commonly occurs among people aged 4–5 and 15–40 years; it rarely occurs in patients over 60 years of age. The incidence rate is slightly higher in females than males, although the severity of the condition is not different between males and females.
Despite many researches about AA, the exact cause of AA is still unknown. It is thought that genetic and environmental factors likely contributed to the immune dysregulation leading to the final pathways of AA. Environmental factors, including infections, stress factors, toxins, and diet, have been suggested; it is not sure (Dudda-Subramanya and others 2007). In addition, cytomegalovirus infection and Helicobacter pylori were considered as etiology of AA. However, it is not clear, as well.
The exact mechanism causing AA is not fully elucidated. However, it is thought that hair follicles are damaged by the infiltration of CD4- and CD8-positive T cells, which disrupt an individual's hair growth (Gilhar and others 2007). Human leucocyte antigen (HLA) molecules and various inflammatory proteins, such as interleukin-1 (IL1) and autoimmune regulator, are considered to be involved in the pathogenesis of AA (Megiorni and others 2012).
It is also shown that the individual genetic background is an important role for the development of AA. A genetic basis is the strong evidence for the pathogenesis of AA. Among the studies of AA, evidence for a genetic etiology in AA is largely based on the increased incidence rate among family members of AA patients. Ten percent to 42% of AA patients report a positive family history of at least one first-degree relative (Muller and Winkelmann 1963). A family history of AA is higher in those patients who suffered early onset of AA than in late onset of AA—37% in patients who had their first patch before 30 years of age, while 7.1% if disease initiation occurred after 30 years of age (Colombe and others 1995; Dudda-Subramanya and others 2007). Furthermore, the estimated lifetime risk of AA in children of patients who suffer AA is 6%, considerably higher compared with the risk in the general population (Dudda-Subramanya and others 2007). The increased frequency of AA in people, who have been affected by their own family members, suggests that genetic factors may play an important role in the genesis of AA. Moreover, they are likely to be associated with an autoimmune etiology as a cause of AA.
Interleukin 18 (interferon gamma-inducing factor) (IL18) is an important proinflammatory cytokine that belongs to the IL1 family produced by a wide range of immune cells, such as monocytes, activated macrophages, and Kupffer cells (Dinarello 1999). IL18 can promote T helper type 1 (Th1)-mediated immune response in synergy with IL12 by producing interferon gamma (IFN-γ). In addition, IL18 can induce a type 2 response in the relative absence of IL12 by inducing the production of Th2 cytokines such as IL4 and IL13 (Yoshimoto and others 1999). In addition, the levels of IL18 in plasma are significantly more elevated in rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) patients compared with healthy controls (Wong and others 2002; Matsui and others 2003). Thus, IL18 may have a potentially pathological role in autoimmunity, such as RA and SLE.
In this study, we investigated 1 promoter single-nucleotide polymorphism (SNP) (rs187238, −137G/C) and exonic SNP (rs549908, Ser35Ser) in the IL18 gene to examine their associations with the development of AA in the Korean population. We also assessed the relationships between 2 SNPs of IL18 and the clinical pathologic features using a case–control approach.
Materials and Methods
Patients and controls
AA patients and control subjects treated at the Kyung Hee University Hospital at Gang-dong (Kyung Hee East West Neo Medical Center) were selected. A serologic work-up, including an anemia study, venereal disease research laboratory test, antinuclear antibodies, thyroid function test, and androgenic hormones, including testosterone, estradiol, luteinizing hormone, and follicle-stimulating hormone, was carried out for patients. AA patients were diagnosed by clinical features and physical examination, including the pull test and microscopic analysis of hairs by a dermatologist. Diagnosis of AA was confirmed by a skin biopsy in some cases. A careful family history was taken from each patient and the concerned individual's general health, including the existence of previous AA, triggering factors, presence of autoimmune disease, atopy, and family history of AA.
Two hundred and forty-three healthy controls (102 males and 141 females, average age at survey: 32.6 years) were included (Table 1). The healthy control subjects were recruited after they had been determined as physically healthy in a general health checkup program. Healthy controls did not have any severe diseases or symptoms. Informed consent was obtained from each subject, and the study was approved by the Institutional Review Board of Kyung Hee University Hospital at Gang-dong.
Table 1.
Demographic and Clinical Characteristics of Alopecia Areata Patients and the Control Subjects
| AA | Control | |
|---|---|---|
| Number of subjects | 233 | 243 |
| Male/female | 106/127 | 102/141 |
| Age (mean±SD) | 28.6±13.3 | 32.6±8.9 |
| First onset age | ||
| <30 years | 154 | |
| ≥30 years | 79 | |
| Family Hx | ||
| Presence | 215 | |
| Absence | 18 | |
| Type | ||
| Patch | 185 | |
| Totalis or universalis | 48 | |
| Involvement of nail | ||
| Presence | 37 | |
| Absence | 196 | |
| Involvement of body hair | ||
| Presence | 39 | |
| Absence | 194 | |
AA, alopecia areata; N, number of subjects; SD, standard deviation.
SNP selection and genotyping
For the selection among IL18 SNPs, we searched for SNPs on the promoter and exon in the IL18 gene in the SNP database of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/SNP, BUILD 137). SNPs with <10% minor allele frequency (MAF), <0.1 heterozygosity, unknown heterozygosity, and unknown genotype frequencies in Asian populations were excluded. Among the exonic SNPs of IL18 gene, all SNPs on the promoter and exon region in the IL18 gene except 2 SNPs (rs187238, −137G/C and rs549908, Ser35Ser) had an unknown heterozygosity or MAF below 0.1.
Peripheral blood samples from each subject were collected in EDTA-coated tubes and stored in a −20°C freezer. Genomic DNA was prepared from peripheral blood using a genomic DNA isolation reagent kit (High Pure PCR template preparation kit; Roche) and SNP genotyping was determined by direct sequencing. Polymerase chain reactions (PCRs) were performed using the primers for 2 exonic SNPs (rs187238, −137G/C, sense primer: 5′-GTGACCAGGAAGTCTGAAAAT-3′, anti-sense primer: 5′-CGACTGCCTGGACAGTCAGC-3′; rs549908, Ser35Ser, sense primer: 5′ GTGTGCCTTTGAGAGTAGGTT-3′, anti-sense primer: 5′-TGACTAGCTACTTCTTCCCA-3′). The PCR products were sequenced by an ABI PRISM 3730XL analyzer (PE Applied Biosystems). Sequencing data were analyzed using SeqManII software (DNASTAR).
Statistical analysis
Hardy–Weinberg equilibrium (HWE) was estimated using SNPStats (http://bioinfo.iconcologia.net/index.php?module=Snpstats) in the control group. Linkage disequilibrium (LD) block and haplotypes were calculated using Haploview version 4.2 (Daly Lab, Inc.). Multiple logistic regression models were applied by described genetic models in a previous study (Lewis 2002; Cho and others 2012, 2013) and to obtain odds ratios (ORs), 95% confidence intervals (CIs), and p-values. Data analysis was performed using SPSS 18.0 (SPSS, Inc.). P<0.05 was considered significant. To the multiple tests, the Bonferroni's correction was applied.
Results
Two hundred thirty-three AA patients and 243 healthy control subjects were genotyped to examine whether IL18 polymorphisms were associated with AA. Genotype distributions of promoter SNP and exon SNP in this study were in Hardy–Weinberg equilibrium in control subjects (rs187238, −137G/C, P=0.90; rs549908, Ser35Ser, P=1.0, data not shown). Table 2 shows the genotype and allele distributions of tested 2 SNPs in the AA group and control group. The genetic analysis between AA and control subjects was performed by the logistic regression analysis with adjusting the age and sex. rs187238 in IL18 showed significant differences between AA and control subjects [codominant model 1 (G/G versus G/C), OR=0.49, 95% CI=0.31–0.78, P=0.024; dominant model (G/G and G/C versus C/C), OR=0.54, 95% CI=0.34–0.83, P=0.0048; log-additive model (G/G versus G/C versus C/C), OR=0.63, 95% CI=0.42–0.94, P=0.02, respectively]. In addition, rs549908 showed a significant association between AA and control subjects [codominant model 1 (A/A versus A/C), OR=0.51, 95% CI=0.32–0.80, P=0.003; dominant model (A/A and A/C versus C/C), OR=0.54, 95% CI=0.35–0.84, P=0.0052; log-additive model (A/A versus A/C versus C/C), OR=0.62, 95% CI=0.42–0.92, P=0.016, respectively] (Table 2). In allele distribution analysis, the allele of rs187238 was associated with AA (OR=0.64, 95% CI=0.43–0.94, P=0.023) (Table 2). The allele of rs549908 also showed an association with AA (OR=0.62, 95% CI=0.42–0.91, P=0.016).
Table 2.
Frequencies of Genotype and Allele of IL18 in Control and Patients with Alopecia Areata
| SNP | Type | Control n (%) | AA n (%) | Model | OR (95% CI) | P | Pc |
|---|---|---|---|---|---|---|---|
| rs187238 | G/G | 173 (71.24) | 191 (824) | Codominant1 | 0.49 (0.31–0.78) | 0.002 | 0.004 |
| −137G/C | G/C | 67 (27.64) | 37 (15.94) | Codominant2 | 1.56 (0.36–6.77) | 0.56 | 1.00 |
| C/C | 3 (1.24) | 5 (2.14) | Dominant | 0.54 (0.34–0.83) | 0.0048 | 0.0096 | |
| Recessive | 1.81 (0.42–7.85) | 0.42 | 0.84 | ||||
| Log-additive | 0.63 (0.42–0.94) | 0.02 | 0.04 | ||||
| G | 413 (85.0) | 419 (89.9) | 1 | ||||
| C | 73 (15.0) | 47 (10.1) | 0.64 (0.43–0.94) | 0.023 | 0.046 | ||
| rs549908 | A/A | 173 (71.24) | 190 (81.94) | Codominant1 | 0.51 (0.32–0.80) | 0.003 | 0.006 |
| Ser35Ser | A/C | 65 (26.84) | 37 (15.94) | Codominant2 | 0.98 (0.27–3.52) | 0.97 | 1.00 |
| C/C | 5 (2.14) | 5 (2.24) | Dominant | 0.54 (0.35–0.84) | 0.0052 | 0.0104 | |
| Recessive | 1.13 (0.32–4.06) | 0.85 | 1.00 | ||||
| Log-additive | 0.62 (0.42–0.92) | 0.016 | 0.032 | ||||
| A | 411 (84.6) | 417 (89.9) | 1 | ||||
| C | 75 (15.4) | 47 (10.1) | 0.62 (0.42–0.91) | 0.015 | 0.03 |
The P values were calculated from logistic regression analyses adjusting sex and age. Bold numbers mean significance association.
The Pc values were calculated using Bonferroni's correction.
SNP, singe-nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Two SNPs (rs187238 and rs549908) of IL18 were analyzed for LD and haplotypes using Haploview 4.2. The LD block was composed of rs187238 and rs549908 (D′=0.981 and r2=0.935) (Fig. 1). There were 2 haplotypes in the LD block (haplotype GA, frequency=0.869; CC, frequency=0.124) (Table 3). Distributions of these haplotypes (AG and CC) in the LD block were associated with AA (haplotype GA, χ2=5.471, P=0.0193; haplotype CC, χ2=5.362, P=0.0206) (Table 3). After Bonferroni's correction, these significant associations still remained (P<0.05) (Tables 2 and 3).
FIG. 1.
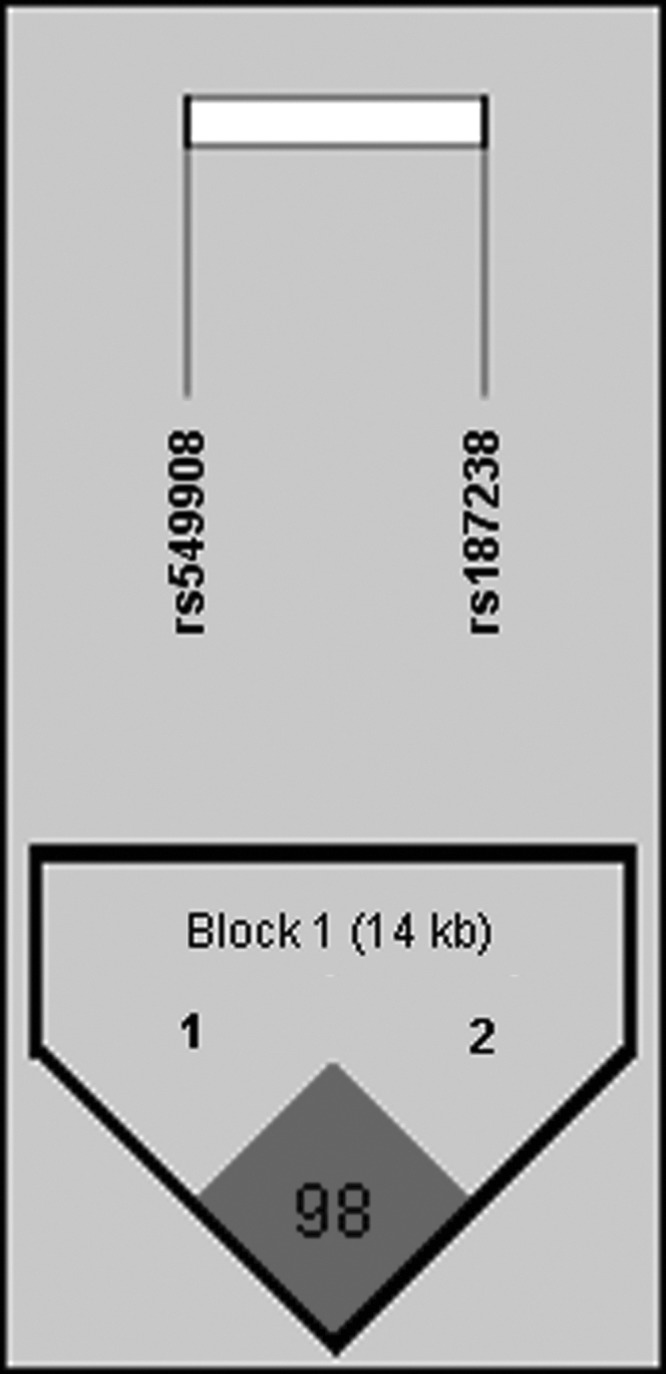
Linkage disequilibrium block between 2 single-nucleotide polymorphisms of IL18 gene. Block consists of rs549908 and rs187238.
Table 3.
Frequencies of Haplotypes in Alopecia Areata and Control Subjects
| Control | AA | |||||||
|---|---|---|---|---|---|---|---|---|
| Haplotype | Frequency | + | − | + | − | χ2 | P | Pc |
| GA | 0.869 | 410 | 76 | 417 | 49 | 5.471 | 0.0193 | 0.039 |
| CC | 0.124 | 72 | 414 | 46 | 420 | 5.362 | 0.0206 | 0.041 |
The Pc values were calculated using Bonferroni's correction. Bold numbers mean significance association.
Next, we assessed the relationship between IL18 SNPs and the clinicopathologic features (onset age, family history, type of AA, involvement of nail, and involvement of body hair) of AA. However, we could not observe a significant association with IL18 (P>0.05, data not shown).
Discussion
Although the cause of AA is poorly understood, in an attempt to determine the genetic basis of AA, a number of association studies for suspected genes have been performed from the autoimmune etiology point of view. In a previous genome-wide association study, Petukhova and others (2010) identified that cytotoxic T lymphocyte-associated antigen 4 (CTLA4), IL2/IL21, IL2 receptor A (IL2RA; CD25), Ikaros family zinc finger 4 (IKZF4), and HLA are associated with susceptibility loci for AA. They suggested that genetic factors may contribute together to induce and promote immune dysregulation in pathogenesis for AA. In addition, protein tyrosine phosphatase, nonreceptor type 22 (lymphoid) (PTPN22), and interleukin-1 receptor antagonist (IL1RN) suggested an association with AA in a case–control association study. PTPN22 plays an important role in the physiology of hair follicle (Alzolibani and others 2012). IL1RN mediates inflammatory responses and influences hair growth regulation and macrophage activated factor (Alzolibani and others 2012).
IL18 was initially described as an IFN-γ-inducing factor in T cells and natural killer cells. It is a pleiotropic cytokine involved in the regulation of innate and acquired immune responses. IL18 plays a key role in autoimmune diseases by controlling either the Th1- or Th2-type immune response (Boraschi and Dinarello 2006). IL18 induces the productions of TNF-α, granulocyte/macrophage colony-stimulating factor, and IFN-γ and increases the cytotoxic effects of NK and T cells in autoimmune diseases (Amerio and others 2002).
Despite the important and novel role of IL18 in immunomodulation, no studies have been conducted to determine associations between the IL18 gene polymorphisms and the development of AA. The objective of the present study was to assess this possible association. This is the first study on the association between IL18 and AA in the Korean population. We found that the tested 2 SNPs (rs187238 and rs549908) in the IL18 gene were associated with susceptibility to AA. Due to the important and novel role of IL18, the IL18 gene itself has been subjected to scrutiny, the aim of discovering variants may impact on disease susceptibility and/or progression. IL18 is located on 11q22.2-22.3. Several studies were performed to sequence the IL18 gene; no nonsynonymous variants have been found (Thompson and Humphries 2007). Instead, polymorphisms on the promoter region and synonymous polymorphisms on exon have been investigated and reported as associations with autoimmune diseases, such as RA, SLE, and atrophy (Lin and others 2008; Ying and others 2011; Chen and others 2012; Messaoudi and others 2012; Song and others 2012). Song and others investigated that the 3 functional IL18 promoter polymorphisms (rs1946518, −607 C/A), (rs187238, −137 G/C), and (rs360719, −1297 C/T) contributed to susceptibility of SLE in ethnically different populations. The study showed that the IL18 −607 C/A and −1297 C/T polymorphism were associated with the development of SLE in Europeans, and the IL18-137 G/C polymorphism was associated with SLE in Asians (Song and others 2012). In this study, we also found the association between the promoter SNP (rs187238, −137 G/C) and AA. The G allele distribution (89.9%) of rs187238 SNP in the AA group was higher compared with (85.0%) the control group. To investigate the binding site of transcription factors for the G and C alleles of rs187238 (−137 G/C), we used the online program AliBaba2.1 (www.gene-regulation.com/pub/programs/alibaba2). The G allele in rs187238 has the C/EBPbeta and/or NF-1 transcription factor binding site, whereas the C allele has a NF-kappaB transcription factor binding site. Previous study has determined that the promoter SNP affects the IL18 promoter activity, its expression, IL18, and IFN-γ by changing its transcription activity (Giedraitis and others 2001). Lee and others (2010) observed that expression of IL18 in serum was significantly higher in the AA patients than in the control subjects (Lee and others 2010). Furthermore, the A allele frequency (89.9) of the rs549908 SNP in the AA group was higher compared with (84.6%) the control group. Several studies were performed correlating the rs549908 SNP with certain diseases, such as spastic cerebral palsy (Hollegaard and others 2013), pulmonary tuberculosis (Han and others 2011), and SLE (Warchol and others 2009). It is suggested that tested SNPs in IL18 may be associated with susceptibility to AA.
In conclusion, our results suggest that 2 SNPs (rs187238 and rs549908) in IL18 may be a risk factor for the development of AA in the Korean population. The first study is very meaningful and could be the foundation for further related studies. Some limitations existed in our study. Replication studies will be confirmed to determine the association between the IL18 and AA.
Author Disclosure Statement
No competing financial interests exist.
References
- Alzolibani AA, Zari S, Ahmed AA. 2012. Epidemiologic and genetic characteristics of alopecia areata (part 2). Acta Dermatovenerol Alp Panonica Adriat 21(1):15–19 [PubMed] [Google Scholar]
- Amerio P, Frezzolini A, Abeni D, Teofoli P, Girardelli CR, De Pita O, Puddu P. 2002. Increased IL-18 in patients with systemic lupus erythematosus: relations with Th-1, Th-2, pro-inflammatory cytokines and disease activity. IL-18 is a marker of disease activity but does not correlate with pro-inflammatory cytokines. Clin Exp Rheumatol 20(4):535–538 [PubMed] [Google Scholar]
- Boraschi D, Dinarello CA. 2006. IL-18 in autoimmunity: review. Eur Cytokine Netw 17(4):224–252 [PubMed] [Google Scholar]
- Chen S, Jiang F, Ren J, Liu J, Meng W. 2012. Association of IL-18 polymorphisms with rheumatoid arthritis and systemic lupus erythematosus in Asian populations: a meta-analysis. BMC Med Genet 15(13):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho AR, Lee SM, Kang WS, Kim SK, Chung JH. 2013. Assessment between dopamine receptor D2 (DRD2) polymorphisms and schizophrenia in Korean population. Clin Psychopharmacol Neurosci 10(2):88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JM, Choe BK, Zheng R, Chon JM, Yoo SD, Kim CJ, Gwak G-H, Yim S-V. 2012. Association between CD40 promoter polymorphism (rs1800686, −508 C/T) and ischemic stroke with hypertension in a Korean population. Mol Cell Toxicol 8:257–262 [Google Scholar]
- Colombe BW, Price VH, Khoury EL, Garovoy MR, Lou CD. 1995. HLA class II antigen associations help to define two types of alopecia areata. J Am Acad Dermatol 33(5 Pt 1):757–764 [PubMed] [Google Scholar]
- Dinarello CA. 1999. IL-18: A TH1-inducing, proinflammatory cytokine and new member of the IL-1 family. J Allergy Clin Immunol 103(1 Pt 1):11–24 [DOI] [PubMed] [Google Scholar]
- Dudda-Subramanya R, Alexis AF, Siu K, Sinha AA. 2007. Alopecia areata: genetic complexity underlies clinical heterogeneity. Eur J Dermatol 17(5):367–374 [DOI] [PubMed] [Google Scholar]
- Forstbauer LM, Brockschmidt FF, Moskvina V, Herold C, Redler S, Herzog A, Hillmer AM, Meesters C, Heilmann S, Albert F, Alblas M, Hanneken S, Eigelshoven S, Giehl KA, Jagielska D, Blume-Peytavi U, Garcia Bartels N, Kuhn J, Hennies HC, Goebeler M, Jung A, Peitsch WK, Kortum AK, Moll I, Kruse R, Lutz G, Wolff H, Blaumeiser B, Bohm M, Kirov G, Becker T, Nothen MM, Betz RC. 2012. Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for alopecia areata. Eur J Hum Genet 20(3):326–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedraitis V, He B, Huang WX, Hillert J. 2001. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol 112(1–2):146–152 [DOI] [PubMed] [Google Scholar]
- Gilhar A, Paus R, Kalish RS. 2007. Lymphocytes, neuropeptides, and genes involved in alopecia areata. J Clin Invest 117(8):2019–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Yue J, Lian YY, Zhao YL, Wang HX, Liu LR. 2011. Relationship between single nucleotide polymorphism of interleukin-18 and susceptibility to pulmonary tuberculosis in the Chinese Han population. Microbiol Immunol 55(6):388–393 [DOI] [PubMed] [Google Scholar]
- Hollegaard MV, Skogstrand K, Thorsen P, Norgaard-Pedersen B, Hougaard DM, Grove J. 2013. Joint analysis of SNPs and proteins identifies regulatory IL18 gene variations decreasing the chance of spastic cerebral palsy. Hum Mutat 34(1):143–148 [DOI] [PubMed] [Google Scholar]
- Kos L, Conlon J. 2009. An update on alopecia areata. Curr Opin Pediatr 21(4):475–480 [DOI] [PubMed] [Google Scholar]
- Lee D, Hong SK, Park SW, Hur DY, Shon JH, Shin JG, Hwang SW, Sung HS. 2010. Serum levels of IL-18 and sIL-2R in patients with alopecia areata receiving combined therapy with oral cyclosporine and steroids. Exp Dermatol 19(2):145–147 [DOI] [PubMed] [Google Scholar]
- Lewis CM. 2002. Genetic association studies: design, analysis and interpretation. Brief Bioinform 3(2):146–153 [DOI] [PubMed] [Google Scholar]
- Lin YJ, Wan L, Sheu JJ, Huang CM, Lin CW, Lan YC, Lai CH, Hung CH, Tsai Y, Tsai CH, Lin TH, Chen CP, Tsai FJ. 2008. A/C polymorphism in the interleukin-18 coding region among Taiwanese systemic lupus erythematosus patients. Lupus 17(2):124–127 [DOI] [PubMed] [Google Scholar]
- Matsui K, Tsutsui H, Nakanishi K. 2003. Pathophysiological roles for IL-18 in inflammatory arthritis. Expert Opin Ther Targets 7(6):701–724 [DOI] [PubMed] [Google Scholar]
- Megiorni F, Pizzuti A, Mora B, Rizzuti A, Garelli V, Maxia C, Carlesimo M, Fotruna MC, Delle Chiaie R, Cavaggioni G, Rossi A. 2012. Genetic association of HLA-DQB1 and HLA-DRB1 polymorphisms with alopecia areata in the Italian population. Br J Dermatol 165(4):823–827 [DOI] [PubMed] [Google Scholar]
- Messaoudi S, Dandana M, Magdoud K, Meddeb S, Ben Slama N, Hizem S, Mahjoub T. 2012. Interleukin-18 promoter polymorphisms and risk of idiopathic recurrent pregnancy loss in a Tunisian population. J Reprod Immunol 93(2):109–113 [DOI] [PubMed] [Google Scholar]
- Muller SA, Winkelmann RK. 1963. Alopecia Areata. An Evaluation of 736 Patients. Arch Dermatol 88:290–297 [DOI] [PubMed] [Google Scholar]
- Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, Kim H, Singh P, Lee A, Chen WV, Meyer KC, Paus R, Jahoda CA, Amos CI, Gregersen PK, Christiano AM. 2010. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 466(7302):113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi KH, Muller SA, Suman VJ, Moshell AN, Melton LJ, 3rd., 1995. Incidence of alopecia areata in Olmsted County, Minnesota, 1975 through 1989. Mayo Clin Proc 70(7):628–633 [DOI] [PubMed] [Google Scholar]
- Song GG, Choi SJ, Ji JD, Lee YH. 2012. Association between interleukin-18 polymorphisms and systemic lupus erythematosus: a meta-analysis. Mol Biol Rep 40(3):2581–2587 [DOI] [PubMed] [Google Scholar]
- Thompson SR, Humphries SE. 2007. Interleukin-18 genetics and inflammatory disease susceptibility. Genes Immun 8(2):91–99 [DOI] [PubMed] [Google Scholar]
- Warchol T, Lianeri M, Wudarski M, Lacki JK, Jagodzinski PP. 2009. IL-18 105 A>C polymorphism contributes to renal manifestations in patients with SLE. Rheumatol Int 30(2):187–191 [DOI] [PubMed] [Google Scholar]
- Wong CK, Ho CY, Li EK, Tam LS, Lam CW. 2002. Elevated production of interleukin-18 is associated with renal disease in patients with systemic lupus erythematosus. Clin Exp Immunol 130(2):345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying B, Shi Y, Pan X, Song X, Huang Z, Niu Q, Cai B, Wang L. 2011. Association of polymorphisms in the human IL-10 and IL-18 genes with rheumatoid arthritis. Mol Biol Rep 38(1):379–385 [DOI] [PubMed] [Google Scholar]
- Yoshimoto T, Tsutsui H, Tominaga K, Hoshino K, Okamura H, Akira S, Paul WE, Nakanishi K. 1999. IL-18, although antiallergic when administered with IL-12, stimulates IL-4 and histamine release by basophils. Proc Natl Acad Sci U S A 96(24):13962–13966 [DOI] [PMC free article] [PubMed] [Google Scholar]


