The chromatin-reorganizing complex FACT functions in transcription elongation and DNA replication, yet its role in replication is not well understood. Here, Herrera-Moyano et al. find increased recombination rates and genetic instability in yeast mutants and FACT-depleted human cells. The results demonstrate a conserved function for FACT in the resolution of transcription–replication conflicts mediated by R loops. This study therefore links the roles of FACT in transcription elongation and DNA replication.
Keywords: FACT, transcription–replication collisions, R loops, genome instability, chromatin reorganization
Abstract
FACT (facilitates chromatin transcription) is a chromatin-reorganizing complex that swaps nucleosomes around the RNA polymerase during transcription elongation and has a role in replication that is not fully understood yet. Here we show that recombination factors are required for the survival of yeast FACT mutants, consistent with an accumulation of DNA breaks that we detected by Rad52 foci and transcription-dependent hyperrecombination. Breaks also accumulate in FACT-depleted human cells, as shown by γH2AX foci and single-cell electrophoresis. Furthermore, FACT-deficient yeast and human cells show replication impairment, which in yeast we demonstrate by ChIP–chip (chromatin immunoprecipitation [ChIP] coupled with microarray analysis) of Rrm3 to occur genome-wide but preferentially at highly transcribed regions. Strikingly, in yeast FACT mutants, high levels of Rad52 foci are suppressed by RNH1 overexpression; R loops accumulate at high levels, and replication becomes normal when global RNA synthesis is inhibited in FACT-depleted human cells. The results demonstrate a key function of FACT in the resolution of R-loop-mediated transcription–replication conflicts, likely associated with a specific chromatin organization.
Genome instability is a common feature of aging and cancer. Different cellular processes might become a threat to the integrity of the genomes, such as replication, DNA repair, transcription, or RNA processing (Aguilera and Gomez-Gonzalez 2008). Consistent with the fact that homologous recombination (HR) is a pathway whose main function is the repair of DNA breaks occurring preferentially during replication, increasing evidence has accumulated showing that transcription-associated recombination (TAR) is the consequence of transcription–replication collisions that can cause breakage of the replication fork (RF) (Aguilera 2002; Kim and Jinks-Robertson 2012; Gaillard et al. 2013). The relevance of transcription–replication collisions as a source of genome instability and TAR has been shown for transcription driven by different RNA polymerases (RNAPs) in different organisms (Prado and Aguilera 2005; Gottipati et al. 2008; de la Loza et al. 2009; Gaillard et al. 2013). At a genome-wide scale, it has been shown that highly transcribed RNAPII genes represent obstacles for the progression of RFs, as determined by the accumulation of the Rrm3 helicase required for RF progression through protein obstacles (Ivessa et al. 2003; Azvolinsky et al. 2009).
One element responsible for transcription–replication collisions is the cotranscriptional R loop, formed by DNA–RNA hybrids and the displaced DNA strand. Even though R loops have physiological roles in a number of processes from mitochondrial DNA replication to class-switching recombination, they are, in general, a threat to chromosome integrity (Aguilera and Garcia-Muse 2012). A major cause of genome instability associated with R loops is the stalling of RFs that can eventually lead to the recombinogenic DNA breaks (Wellinger et al. 2006; Gan et al. 2011; Gomez-Gonzalez et al. 2011a). R loops seem to occur at low levels during transcription, but defects in a number of nuclear processes can highly enhance their formation. These include defects in RNA processing and export factors such as the THO/TREX complex, as shown in yeast, Caenorhabditis elegans, and human cells (Huertas and Aguilera 2003; Dominguez-Sanchez et al. 2011; Castellano-Pozo et al. 2012); splicing factors such as ASF/SF2 in chicken and human cells (Li and Manley 2005); the Sen1/Senataxin DNA–RNA helicase involved in transcription termination (Mischo et al. 2011; Skourti-Stathaki et al. 2011); the Npl3 RNA-binding hnRNP (Santos-Pereira et al. 2013); and others identified in different genome-wide screenings from yeast to human cells that cause different forms of genome instability (Luna et al. 2005; Paulsen et al. 2009; Wahba et al. 2011; Stirling et al. 2012). Despite evidence for the role of transcription–replication collisions as a source of genome instability, whether mediated by R loops or not, we do not know the mechanisms or factors that replication uses to skip such obstacles.
The conserved FACT (facilitates chromatin transcription) complex is a factor that functions in both transcription and replication, but surprisingly little is known about its role in genome instability, in particular TAR. It is formed of Spt16/SPT16 and Pob3/SSRP1 (as named in yeast/humans) subunits. FACT promotes RNAPII transcription through chromatin templates (Mason and Struhl 2003; Saunders et al. 2003) by interacting with histones. It facilitates promoter activation by nucleosome eviction and transcription elongation by nucleosome disruption and reassembly ahead of and behind the RNAPs, respectively (Orphanides et al. 1998; Belotserkovskaya et al. 2003). FACT also has a role in replication that, even though less studied, is supported by different observations. Thus, yFACT physically interacts with DNA polymerase α (Wittmeyer and Formosa 1997), and the human and yeast complexes interact physically and functionally with other replication and checkpoint factors (Schlesinger and Formosa 2000; Gambus et al. 2006; Tan et al. 2006; VanDemark et al. 2006). Recently it has also been shown to participate in histone redeposition during replication in yeast (Foltman et al. 2013). Furthermore, replication progression is impaired in FACT-deficient Xenopus oocyte extracts in vitro and in chicken DT40 cells (Okuhara et al. 1999; Abe et al. 2011).
Using spt16 and pob3 yeast mutants and human cell lines depleted of SPT16 or SSRP1, we show that FACT solves transcription–replication conflicts to preserve genome stability. Yeast and human cells defective of FACT show DNA breaks and hyperrecombination and display different forms of instability linked to replication impairment, as determined by BrdU incorporation, two-dimensional (2D) gel electrophoresis, DNA combing, or ChIP–chip (chromatin immunoprecipitation [ChIP] combined with microarray analysis) with the Rrm3 helicase. Strikingly, replication defects are transcription-dependent, genome instability is suppressed by RNase H overexpression, and DNA–RNA hybrid immunoprecipitation (DRIP) analysis reveals a high accumulation of R loops in yeast FACT mutants and in FACT-depleted human cells. Altogether, the results demonstrate that FACT facilitates RF progression specifically through transcribed DNA regions, supporting the idea that cotranscriptional R loops are formed naturally and associate with chromatin modifications.
Results
Genome instability and recombination-dependent viability in yeast FACT mutants
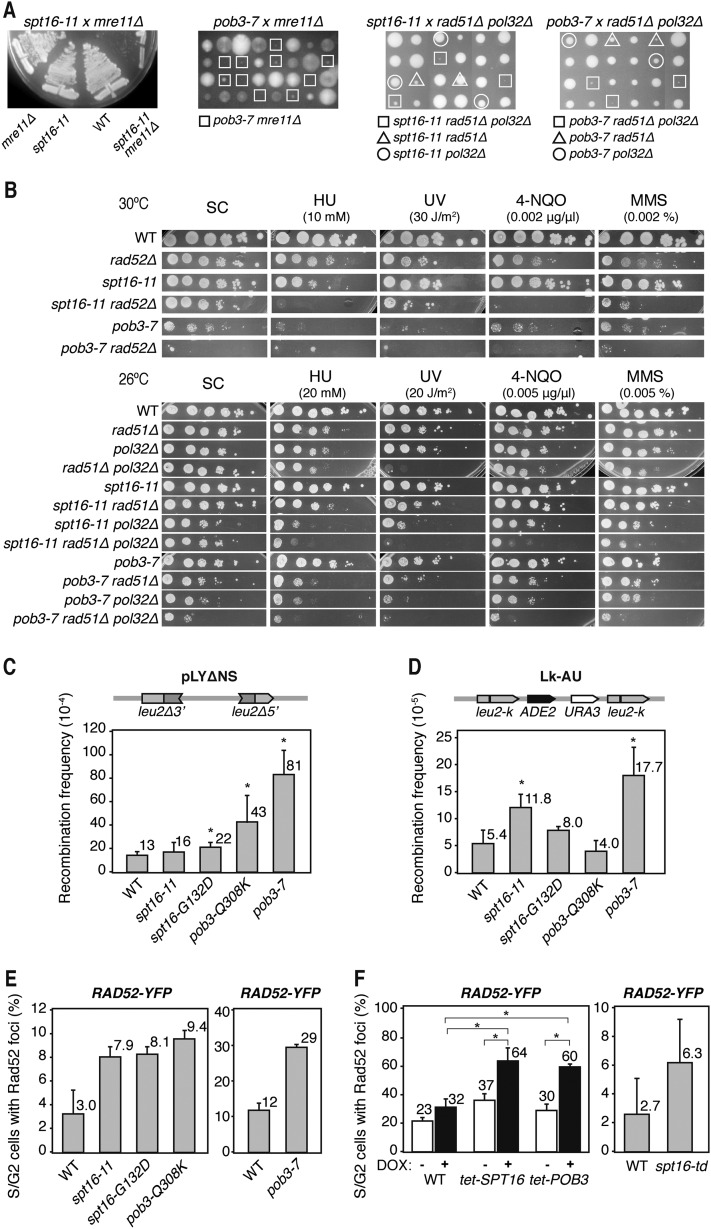
To gain insight into the molecular nature of chromatin dynamics in transcription-mediated genome instability, we selected four different thermosensitive mutants of Saccharomyces cerevisiae SPT16 and POB3 altered in different processes of DNA metabolism—the mutants spt16-11, spt16-G132D, pob3-Q308K, and pob3-7—for having the strongest phenotypes. spt16-11 and pob3-Q308K cells displayed a strong sensitivity to low doses of hydroxyurea (HU), methyl methanesulfonate (MMS), and 4-nitroquinoline N-oxide (4-NQO), and pob3-7 cells were sensitive to HU and 4-NQO (Supplemental Fig. S1A), whereas spt16-G132D was only sensitive to 4-NQO at the doses tested.
As these agents have in common their capacity to generate recombinogenic DNA breaks, we wondered whether recombination factors became essential in these mutants for cell viability. Interestingly, whereas, in the absence of Mre11, spt16-G132D and pob3-Q308K showed a mild growth defect, spt16-11 and pob3-7 cells grew poorly, indicating that the absence of HR is highly detrimental in these two mutants (Fig. 1A; Supplemental Fig. S1B). This conclusion was confirmed by assessing the importance of Rad52 for viability. spt16-11 rad52Δ cells grew poorly in synthetic complete (SC) medium and were extremely sensitive to HU, UV, 4-NQO, and MMS at doses that the single rad52Δ mutant was resistant to (Fig. 1B). pob3-7 rad52Δ cells were not viable at 30°C. These results indicate that recombinational double-strand break repair is crucial for the viability of spt16-11 and pob3-7 mutants. Interestingly, both mutations were viable in a rad51Δ background but were extremely sick if the Pol32 subunit of Pol∂ involved in break-induced replication (BIR) was also absent (Fig. 1A,B). Consistent with previous reports indicating that Rad51 and Pol32 define two repair pathways of replication-mediated breaks (Moriel-Carretero and Aguilera 2010), this result supports the idea that FACT mutations cause replication-associated DNA breaks.
Figure 1.
Genetic interaction with recombination and replication functions of yFACT-deficient cells. (A) Analysis of genetic interactions of spt16-11 (XEI-13) and pob3-7 (EIII-34) mutants with mre11Δ, rad51Δ, and pol32Δ. Single, double, and triple mutants were obtained by genetic crosses, and dissected tetrads were tested for germination on YEPD-rich medium at semipermissive temperature (26°C for pob3-7 and 30°C for spt16-11 crosses). For the spt16-11 and mre11Δ cross, several tetrads were tested for growth at 30°C. (B) Viability of single and double mutants with rad52Δ (R52-9B), rad51Δ (R51-18A), and pol32Δ (P32-1C) in synthetic complete (SC) medium and sensitivity to HU, UV-C, 4-NQO, and MMS by 10-fold serial dilutions are depicted. (C) Recombination frequency using the pLYΔNS system in wild-type (WT; W303-1AR5), spt16-11 (XEI-13), spt16-G132D (XEI-14), pob3-Q308K (XEI-16), and pob3-7 (EIII-34) cells. (D) Recombination frequency in the Lk-AU direct repeat system in wild-type (A3Y3A), spt16-11 (AYS11-1B), spt16-G132D (AYSGD-4A), pob3-Q308K (AYPQK-15B), and pob3-7 (AYP7-7A) cells. (E) Analysis of Rad52 foci formation in wild-type (W303-1A), spt16-11 (DY8107), spt16-G132D (DY5391), pob3-Q308K (DY10308), wild-type (4053-5-2), and pob3-7 (7809-7A) cells. (F) The same as in E in wild-type (R1158), tet-SPT16 (TH-5591), and tet-POB3 (TH-3699) cells in which depletion is induced with 5 μg/mL DOX and in wild-type (YKL83) and spt16-td (YKSD) cells 3 h after degron induction. Mean and standard deviation (SD) of three independent experiments are depicted. (*) P < 0.05 (Student's t-test).
Next, we determined whether the accumulation of breaks resulted in an increase in spontaneous recombination. Indeed, recombination between two leu2 direct repeats in the plasmid pLYΔNS and the chromosomal leu2-k∷ADE2-URA3∷leu2-k (Lk-AU) (Gomez-Gonzalez et al. 2011b) systems was slightly but significantly increased with respect to wild-type levels (Fig. 1C,D). Consistently, high levels of recombinogenic breaks were observed by determining the frequency of Rad52 foci in the mutants (Fig. 1E). Rad52 foci were also increased in cells harboring SPT16 or POB3 under the regulated tet promoter (Ptet) and switched off with doxycyclin (DOX) as well as in cells harboring a thermosensitive degron allele of Spt16 (spt16-td), which produces a Spt16 protein that is completely degraded after 60 min of degron induction (Fig. 1F; Supplemental Fig. S1C). Therefore, a functional FACT complex is needed to maintain genome integrity in S. cerevisiae.
Genome instability in yeast FACT mutants is transcription-dependent
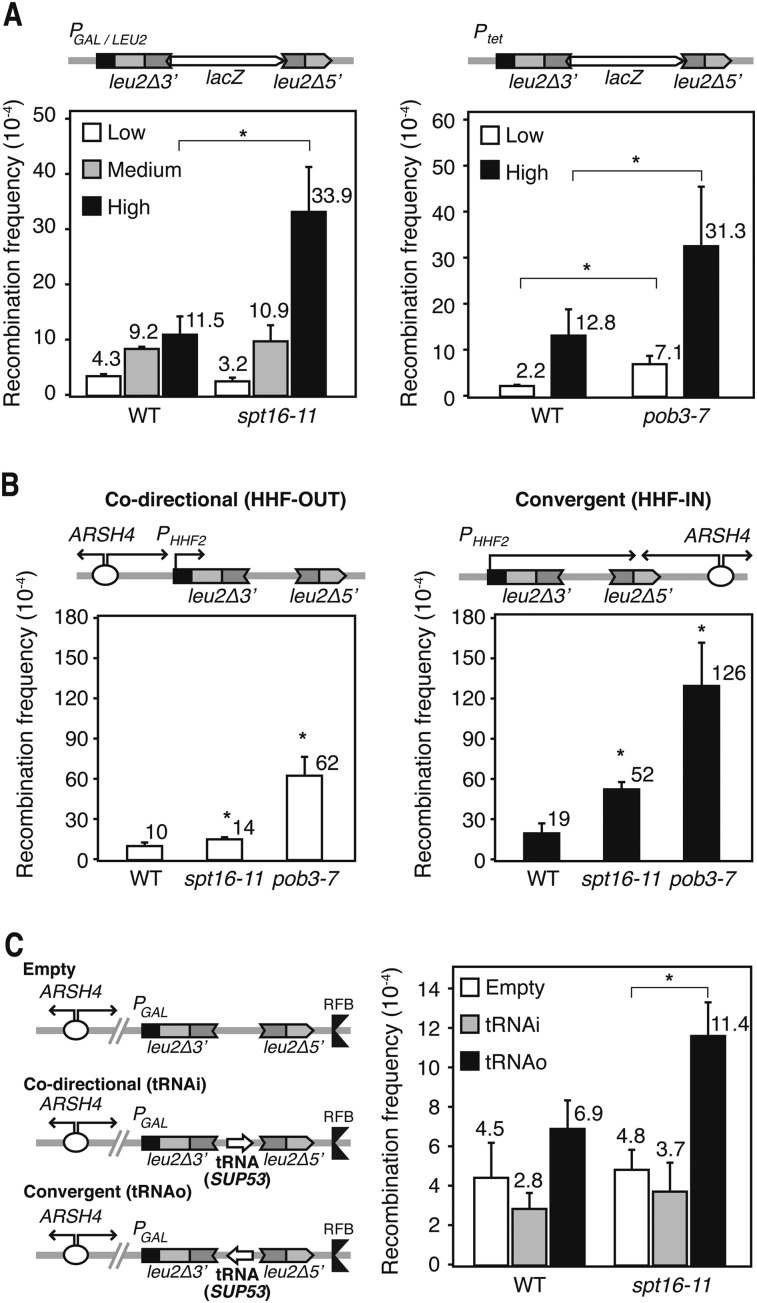
Since FACT-mediated nucleosome-reorganizing activity is necessary during transcription elongation, we wondered whether genome instability was transcription-dependent. We assayed recombination between two leu2 direct repeats separated by the GC-rich lacZ gene under the inducible GAL1,10 promoter (PGAL) or the constitutive LEU2 promoter (PLEU2) (Gomez-Gonzalez et al. 2011b). When transcription was low (PGAL in glucose), recombination levels in spt16-11, spt16-G132D, and pob3-Q308K were indistinguishable from the wild type (Fig. 2A; Supplemental Fig. S2A,B). However, when transcription was medium (PLEU2) or high (PGAL in galactose), recombination increased in all mutants, even though to different extents. The mutant with the clearest effect was spt16-11, in which recombination was stimulated 11-fold at the highest transcription levels tested. In spt16-G132D, recombination increased more at the PLEU2-driven system than at the PGAL, likely because GAL1 expression levels are lower in this mutant (Supplemental Fig. S2B). Since pob3-7 cells were Gal− and unable to activate PGAL (Supplemental Fig. S2C), they were analyzed with the TL-lacZ system, in which transcription was driven from Ptet and was even lower than in the wild type (Supplemental Fig. S2D). Recombination was significantly stimulated in pob3-7 cells under high transcription (−DOX) (Fig. 2A) and slightly even under low transcription (+DOX). Altogether, these results indicate that the genome instability phenotype of yeast FACT mutants is transcription-dependent.
Figure 2.
Transcription-associated hyperrecombination in yFACT mutants. (A) Recombination frequency in wild-type (WT; W303-1AR5), spt16-11 (XEI-13), and pob3-7 (EIII-34) cells using the plasmid-borne direct repeat L-lacZ system expressed under the control of the GAL promoter in glucose or galactose (low and high transcription levels, respectively), the LEU2 promoter (medium), or the tet promoter with or without 5 μg/mL DOX (low and high, respectively). (*) P < 0.05 (Student's t-test). (B) Recombination frequency of the direct repeat HHF-OUT and HHF-IN systems in wild-type (W303-1ARb), spt16-11 (WXEI-48), and pob3-7 (WEIII-36) cells. Replication from the ARSH4 replication origin and transcription driven by the HHF2 promoter are in codirectional (OUT) or convergent (IN) orientations. Other details are as in A. (C) Recombination frequency of the RFB-empty, RFB-tRNAi, and RFB-tRNAo systems in wild-type (W303-1ARb) and spt16-11 (WXEI-48) cells grown in glucose. Transcription of the SUP53 tRNA gene and replication initiated at ARSH4 are either codirectional (tRNAi) or convergent (tRNAo). The RF coming from downstream from the tRNA gene is paused due to the RF barrier (RFB). (*) P < 0.05 (Mann-Whitney U-test). Mean and SD of three independent experiments are depicted for each genotype.
Defective S-phase progression of yeast FACT mutants
Next, we analyzed progression of cells through the S phase. G1-synchronyzed cultures were released into the S phase at a restrictive temperature (37°C) with or without 20 mM HU, and cell cycle progression was monitored by FACS. Whereas spt16-G132D cells were arrested in G1/early S phase (Supplemental Fig. S3), pob3-Q308K cells accumulated in the S phase under replication stress (+HU). In contrast, progression through the S phase was delayed in spt16-11 and pob3-7 with and without HU, suggesting that replication is affected in all of these mutants, although to a different extent.
To confirm that the defect in S-phase progression was not allele-specific, we performed the same experiment in cells depleted of Spt16 using the degron strain. spt16-td cells were retained in the early S phase (Supplemental Fig. S3), consistent with a defect in replication. Thereafter, we decided to work only with a representative mutant from each subunit: spt16-11 and pob3-7. From these, spt16-11, the mutant most affected in TAR, was unaffected in the transcription of the systems analyzed, whereas pob3-7, the sickest mutant in the experiment, was significantly affected, as assayed by Northern of GAL10 and SPF1 and the L-lacZ construct (Supplemental Fig. S2B–D). To determine whether the genome instability phenotypes were caused by the FACT mutations directly rather than the down-regulation of specific repair genes, we analyzed the global gene expression profile of spt16-11 and pob3-7 cells. Interestingly, the microarray analysis revealed that the up-regulated and down-regulated genes (more than twofold the wild-type value) showed a high degree of coincidence between both mutants (Supplemental Fig. S4A; Supplemental Material). Neither a specific functional class of genes nor any specific replication or repair gene that to our knowledge could explain the phenotypes was down-regulated in the mutants (Supplemental Fig. S4B), supporting the conclusion that the genome instability phenotypes of the mutants are a direct consequence of a defective function of FACT.
Transcription–replication conflicts as a source of genome instability in yeast FACT mutants
We next wondered whether hyperrecombination of FACT mutants occurred at regions in which replication collided with transcription. We studied recombination in two previously described plasmid-borne recombination assays in which the RF coming from an ARSH4 replication origin encounters RNAPII at leu2 direct repeats in codirectional (HHF-OUT) or convergent (HHF-IN) orientations (Prado and Aguilera 2005). Transcription in these systems is driven from the HHF2 promoter (PHHF2), which only works during the S phase. In both the IN and OUT systems, recombination increased significantly with respect to wild type in spt16-11 and pob3-7 (Fig. 2B). These differences are not due to an enhanced transcription of the recombination systems analyzed, since their transcript levels are either similar or lower in the mutants in respect to the wild type (Supplemental Fig. S2E). A similar effect was observed in spt16-11 in a system (de la Loza et al. 2009) in which the RF encounters the RNAPIII at the SUP53 gene in convergent orientation (Fig. 2C). The results indicate that FACT maintains genome integrity at sites in which the RF encounters an elongating RNAP, the effect being more accentuated when the encounter is in the opposite orientation, and that FACT functions in RNAPIII-dependent transcribed genes in addition to those transcribed by RNAPII.
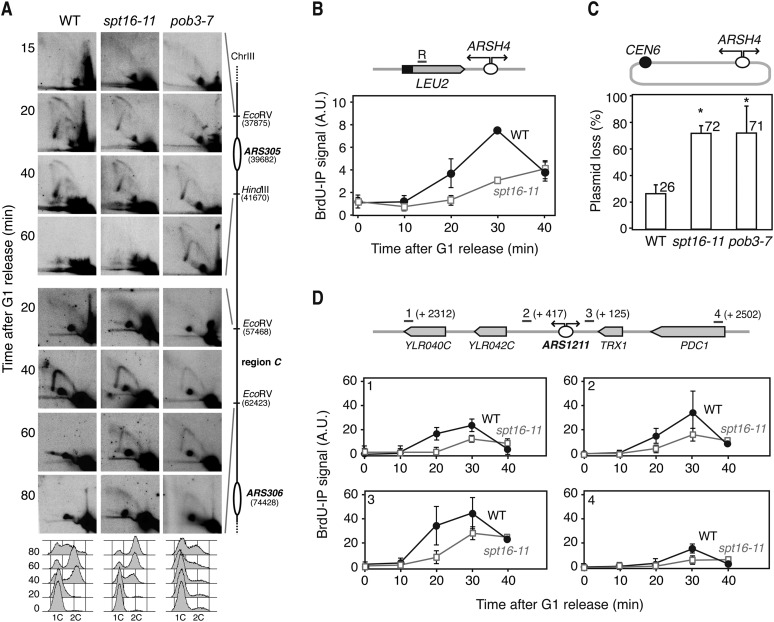
We then assayed whether replication through the transcribed regions was specifically impaired in the mutants. For this, spt16-11 and pob3-7 cells were synchronized in G1 and released into the S phase at a semipermissive temperature (30°C), and replication intermediates were visualized by 2D gel electrophoresis at the early ARS305 replication origin and in a region between ARS305 and ARS306 (region C) containing several transcribed genes at different time points. In spt16-11 cells, RF intermediates followed kinetics similar to the wild type, whereas in pob3-7, the RF showed a delay of 20 min (Fig. 3A), suggesting a defect in replication initiation. Nevertheless, the passive replication arc at region C was observed in both mutants with a considerable delay with respect to wild type (>20–40 min), indicating that the RF progresses more slowly in spt16-11 and pob3-7 than in wild-type cells (Fig. 3A).
Figure 3.
Analysis of replication in yFACT mutants. (A) Replication intermediates detected by 2D gel electrophoresis in the origin ARS305 and in the 18-kb downstream region C in wild-type (WT; W303-1ARb), spt16-11 (WXEI-48), and pob3-7 (WEIII-36) cells synchronized in G1 with α-factor and released at 30°C. FACS analysis is shown at the bottom. (B) ChIP analysis of BrdU incorporation into the DNA at the 5′ region of LEU2 of the pARSHLB-Leu2 plasmid in wild-type (WRBbL) and spt16-11 (WSRBbL) cells synchronized in G1 and released. Mean and standard error of the mean (SEM) of two independent experiments are shown. (C) Percentage of pARSHLB-Leu2 plasmid loss in wild-type (W303-1ARb), spt16-11 (WXEI-48), and pob3-7 (WEIII-36) cells growing in YEPD-rich medium. Mean and SD of three independent experiments are shown. (*) P < 0.05 (Student's t-test). (D) ChIP analysis of BrdU incorporation into the DNA at genomic regions located 2312 and 417 base pairs (bp) upstream of and 125 and 2502 bp downstream from the ARS1211 replication origin. Other details are as in B.
Next, we monitored replication by BrdU incorporation via ChIP at the 5′ region of the LEU2 gene transcribed from the PHHF2 in the plasmid-borne HHF-IN system. The kinetics of BrdU incorporation were clearly slower in spt16-11 cells with respect to the wild type (Fig. 3B), consistent with a defect in the replication of FACT mutants. In accordance, spt16-11 and pob3-7 cells showed almost threefold higher rates of plasmid loss than wild-type cells (Fig. 3C). Analysis of RF progression by BrdU incorporation at four endogenous chromosomal regions located upstream of and downstream from the ARS1211 early replication origin confirmed the replication defect (Fig. 3D). Altogether, these results demonstrate that replication is hampered in yeast FACT mutants, which can be clearly seen at transcribed DNA regions.
Genome instability in yeast FACT mutants is R-loop-dependent
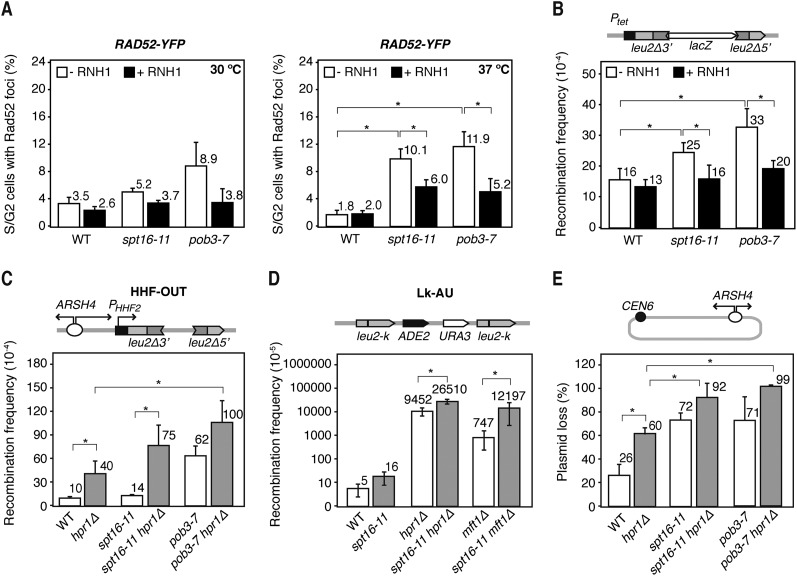
Next, we tested whether the transcription-dependent instability of the FACT mutants was dependent on R loops. For this, we assayed whether overexpression of the yeast RNH1, which degrades the RNA moiety of DNA:RNA hybrids, suppressed genome instability in spt16-11 and pob3-7 cells. Interestingly, the percentage of cells with Rad52 foci was reduced after RNH1 overexpression in both mutants at 30°C and 37°C (Fig. 4A), indicating that R loops are responsible in large part for the genetic instability of FACT-deficient cells and suggesting that FACT facilitates replication at regions where R loops accumulate. This conclusion is supported by the ability of RNH1 overexpression to suppress the increased recombination of spt16-11 and pob3-7 in the TL-lacZ direct repeat system under high transcription conditions (Fig. 4B).
Figure 4.
Effect of R-loop formation in yFACTs mutants. (A) Percentage of wild-type (WT; W303-1ARb), spt16-11 (WXEI-48), and pob3-7 (WEIII-36) S and G2 cells forming Rad52 recombination foci 3 h after a temperature shift from 26°C to 30°C or 37°C with or without RNase H1 (RNH1) overexpression from the pCM189-RNH1 plasmid. (B) Recombination frequency in wild-type (W303-1ARb), spt16-11 (WXEI-48), and pob3-7 (WEIII-36) cells in the plasmid-borne direct repeat system TL-lacZ, which is highly expressed in the absence of DOX. Other details are as in Figure 2A. (C) Recombination frequency using the direct repeats HHF-OUT in wild-type (W303-1ARb), spt16-11 (WXEI-48), and pob3-7 (WEIII-36) cells with or without the hpr1Δ mutation (26°C). (D) Recombination analysis in the Lk-AU direct repeat system in spt16-11 (AYS11-1B), wild-type (A3Y3A), hpr1Δ (AYH-2D), and mft1Δ (AYM-1D) strains plus the double mutants. (E) Percentage of pARSHLB-Leu2 plasmid loss in wild-type (W303-1ARb), spt16-11 (WXEI-48), and pob3-7 (WEIII-36) cells with or without the hpr1Δ mutation growing in YEPD-rich medium. Mean and SD of three independent experiments are shown. (*) P < 0.05 (Student's t-test).
A prediction of our conclusion is that the effect should be more accentuated in strains accumulating R loops at a high frequency. To test this, we analyzed the effect of FACT deficiency in cells lacking the THO subunit Hpr1, known to accumulate R loops at high levels (Huertas and Aguilera 2003; Gomez-Gonzalez et al. 2011a). Interestingly, spt16-11 hpr1Δ and pob3-7 hpr1Δ cells showed a synthetic growth defect even at a permissive temperature (Supplemental Fig. S5A) and were unable to grow in the presence of low HU doses. In addition, hpr1Δ increased recombination in the HHF-OUT system fourfold above wild-type levels (Fig. 4C) and twice more if SPT16 or POB3 were also mutated (Fig. 4C). We confirmed the results in the more sensitive chromosomal Lk-AU system, in which recombination was increased ∼1900-fold in hpr1Δ and threefold more in spt16-11 hpr1Δ cells (Fig. 4D). Consistently, recombination augmented 16-fold in spt16-11 mft1Δ cells compared with cells just lacking the THO subunit Mft1. Similarly, plasmid loss was significantly increased in spt16-11 hpr1Δ and pob3-7 hpr1Δ (Fig. 4E).
Altogether, the data suggest that in the absence of FACT, R loops became a major obstacle for RF progression as a putative source of recombinogenic DNA breaks. Consistently, spt16-11 hpr1Δ and pob3-7 hpr1Δ cells took longer than single mutants to progress through the S phase or accumulate in S–G2 (Supplemental Fig. S5B).
Genome-wide impairment of replication in yeast FACT mutants
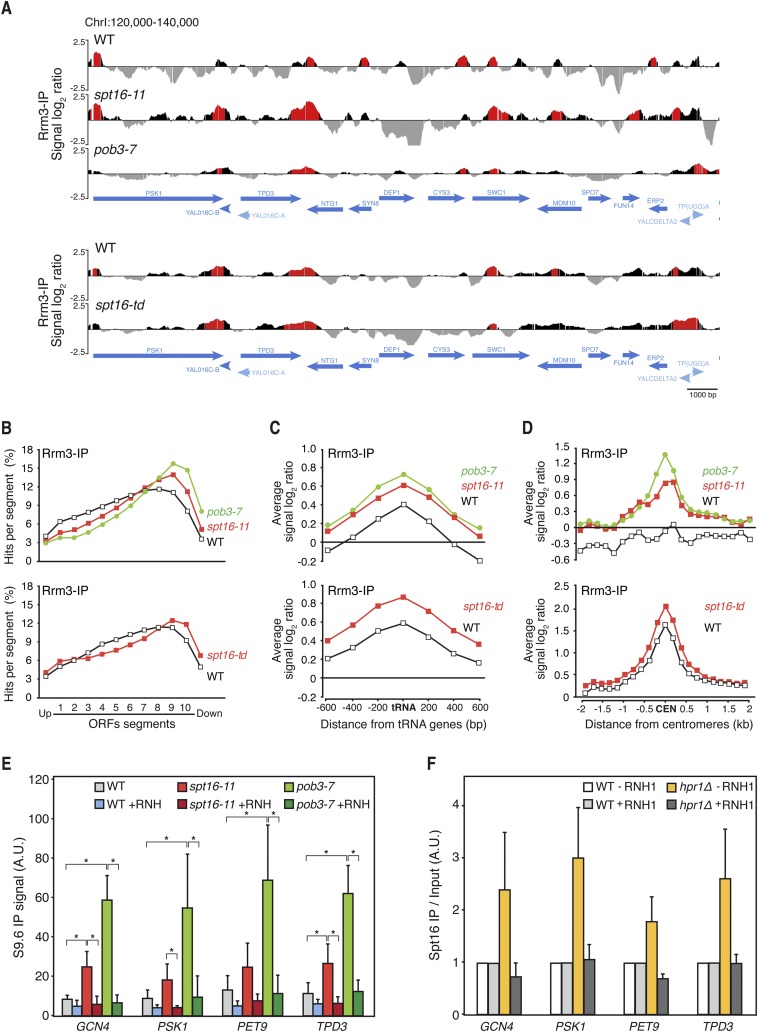
Next, we investigated the genome-wide occupancy of Rrm3 helicase, known to accumulate at specific sites of RF impairment, preferentially at highly transcribed regions (Ivessa et al. 2003; Azvolinsky et al. 2009; Gomez-Gonzalez et al. 2011a). ChIP–chip of an Rrm3-Flag fusion in asynchronous cultures showed a similar distribution of Rrm3 occupancy clusters in wild type, spt16-11, pob3-7, and spt16-td (Fig. 5A; Supplemental Figs. S6–S8). The majority of these clusters mapped in coincident ORFs, which were significantly longer and more expressed than the genome mean value (Supplemental Fig. S9A–C) and correlated positively with the RNAPII-binding clusters detected by Rpb3 ChIP to a similar extent in wild-type and mutant cells (Supplemental Fig. S9D), indicating that RF progression is impaired preferentially at long and highly expressed genes. Interestingly, the binding profile of Rrm3 to ORFs showed higher values and increased toward the 3′ ends of genes in the FACT mutants (Fig. 5B; Supplemental Fig. S9E). These regions have been shown to undergo RF progression impairment and are prone to accumulate R loops (Gomez-Gonzalez et al. 2011a). Therefore, FACT may assist RFs to pass natural transcription–replication obstacles, including those potentially mediated by R loops. Moreover, additional RF pausing/stalling sites occur at different RNAPII-driven genes in the mutant strains (Fig. 5A; Supplemental Figs. S8A, S9B). These may be potential sites for transcription–replication collisions that FACT normally resolves but that are detected in FACT-deficient cells.
Figure 5.
Genome-wide analyses of Rrm3 recruitment in yFACT mutants. (A) Genome-wide occupancy of Rrm3 in wild-type (WT; WRBb-9B), spt16-11 (WSRBb-2D), and pob3-7 (WPRBb-10D) cells 4 h after a 26°C–30°C temperature shift and in wild-type (WDRB-18A) and spt16-td (degron) (WSDR-8C) cells after 75 min of degron induction. Histogram bars in the Y-axis show the signal of loci enriched in the immunoprecipitated fraction along the indicated regions in log2 scale. Positive enrichment clusters (P-value < 0.01) are depicted in red. ORFs and other SGD genome features are indicated. (B) Composite profile of Rrm3 occupancy across the average ORF plotted as percentage of hits per segment in wild-type, spt16-11, and pob3-7 cells and in wild-type and spt16-td cells. ORFS were divided into 10 segments and studied together with the two immediately contiguous. (C) Composite profile of Rrm3-IP signal log2 ratio across the average tRNA gene and regions 600 bp around in wild-type, spt16-11, and pob3-7 cells and in wild-type and spt16-td cells. (D) Composite profile of Rrm3-IP signal log2 ratio across the average 2-kb region around centromeres in wild-type, spt16-11, and pob3-7 cells and in wild-type and spt16-td cells. (E) DRIP using the S9.6 antibody in wild-type (W303-1ARb), spt16-11 (WXEI-48), and pob3-7 (WEIII-36) asynchronously growing cells at GCN4, PSK1, PET9, and TPD3 genes. Signal values of R-loop detection are shown. (F) Spt16-Flag ChIP analysis in wild-type (WS16) and hpr1Δ (WHS16) asynchronously growing cells with or without RNase H1 (RNH1) overexpression from the pRS416-GALRNH1 plasmid in the GCN4, PSK1, PET9, and TPD3 genes. Signal ChIP values normalized in respect to the wild-type levels are shown. Mean and SD of at least three independent experiments are shown. (*) P < 0.05 (Student's t-test).
Apart from the general effect on mRNA genes, Rrm3 clusters were also observed in other regions (Supplemental Fig. S9A). Rrm3 clusters were found frequently at RNAPIII transcribed genes and in regions adjacent to tRNAs genes in FACT mutants (Fig. 5C; Supplemental Fig. S8B). The results suggest that FACT also reorganizes chromatin at regions where RF and RNAPIII encounter each other, consistent with the increased recombination observed in tRNA-dependent recombination systems in FACT mutants (Fig. 2C).
It is worth noting that the heterochromatin at pericentromeric regions also seems to hamper RF progression in FACT mutants, as detected by a strong increase in Rrm3 signal in spt16-11 and pob3-7 at these regions, as well as in the spt16-td degron mutant (Fig. 5D; Supplemental Fig. S8C). Interestingly, we showed recently that pericentromeric regions, which are also transcribed by RNAPII, accumulate high levels of R loops (Castellano-Pozo et al. 2013). Altogether, these results indicate that FACT assists RF progression at transcribed regions all over the genome. The recent finding that R loops promote chromatin compaction (Castellano-Pozo et al. 2013) raises the possibility that FACT plays a key role in assisting chromatin reorganization during R-loop-mediated transcription–replication collisions. To address this possibility, we assessed the accumulation of DNA–RNA hybrids by direct immunoprecipitation using the S9.6 monoclonal antibody at different genes where Rrm3 is accumulated in spt16-11 and pob3-7 mutants, as confirmed by ChIP analyses (Supplemental Fig. S10A,B). DNA–RNA hybrids were clearly accumulated in the mutants in all DNA regions tested, including the centromere-proximal gene GCN4 (Fig. 5E). Interestingly, the enrichment of DNA–RNA hybrids in G1-arrested cells was lower than in asynchronous cultures (Supplemental Fig. S10C), suggesting that the absence of FACT facilitates DNA–RNA hybrid accumulation differentially through the cell cycle.
Next, we asked whether FACT recruitment to chromatin was enhanced by R loops. For this, we assayed recruitment by ChIP of a Flag-tagged Spt16 in an hpr1Δ strain known to enhance R-loop accumulation. Spt16 accumulated at different genes in hpr1Δ cells, such accumulation being suppressed by overexpression of RNH1 (Fig. 5F), indicating that FACT is preferentially accumulated at R-loop-enriched DNA. Accordingly, the genome-wide Rrm3 distribution showed a profile similar to Rpb3 and Spt16 observed in the S and G1 phases (Supplemental Fig. S10D,E).
Transcription-dependent genome instability in FACT-deficient human cells
Next, we assayed whether human FACT is also involved in maintaining genome integrity and RF progression at transcribed regions. To address this question, we depleted both FACT components, SPT16 and SSRP1, via siRNA from HeLa cells and the MRC-5 fibroblast cells derived from normal lung tissue. siRNA transfected HeLa cells showed a clear reduction in SPT16 and SSRP1 mRNA and/or protein levels (Supplemental Fig. S11A,B), and SSRP1 was reduced after SPT16 depletion in both cell types, consistent with both proteins forming a protein complex.
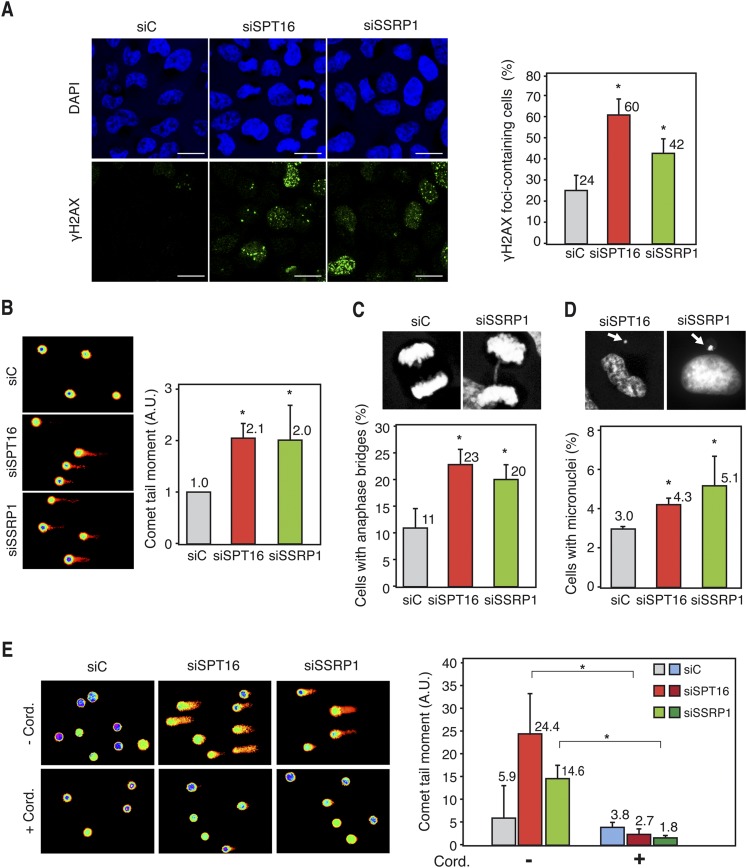
We then analyzed by immunofluorescence the formation of γH2AX foci, known to accumulate early at sites of DNA breaks. The percentage of cells with γH2AX foci was clearly increased in SPT16- and SSRP1-depleted HeLa cells with respect to the siC control (Fig. 6A). Alkaline single-cell electrophoresis revealed that FACT knockdown led to a significant increase in tail moment in both HeLa (Fig. 6E) and MRC-5 (Fig. 6B) cells, demonstrating that DNA breaks accumulate spontaneously. Finally, we depleted FACT in HeRG HeLa stable cell lines carrying the pIREC direct repeat recombination system based on two truncated copies of GFP stably integrated in the genome (Dominguez-Sanchez et al. 2011). Recombination, as determined by GFP fluorescence via FACS, was increased 2.2-fold and 1.8-fold in SPT16- and SSRP1-depleted cells with respect to the control (Supplemental Fig. S11C), consistent with an increase in DNA breaks and recombination in human FACT-deficient cells.
Figure 6.
Genome instability in FACT-depleted human cells. (A) Immunofluorescence of γH2AX in siC (control), siSPT16, and siSSRP1 transfected HeLa cells. Nuclei were stained with DAPI. Graphics show the quantification of the percentage of cells containing γH2AX foci. Bars, 25 μm. More than 300 cells were analyzed in each case. (*) P < 0.05 (Student's t-test). (B) DNA breaks measured by single-cell gel electrophoresis (comet assay) of siC, siSPT16, and siSSRP1 transfected MRC-5 cells. Graphics show the median comet tail moment. More than 300 cells were analyzed in each case. (*) P < 0.05 (Mann-Whitney U-test). (C) Anaphase bridges in siC, siSPT16, and siSSRP1 transfected HeLa cells. Pictures show DAPI staining of control and SSRP1-depleted cells in anaphase. More than 100 anaphases were analyzed in each case. Other details are as in A. (D) Percentage of HeLa cells with microuclei after siC, siSPT16, or siSSRP1 transfection. Pictures show DAPI staining of SPT16- and SSRP1-depleted cells containing micronuclei. Other details are as in B. (E) Comet assay of siC-, SPT16-, or SSRP1-depleted HeLa cells after treatment or untreated for 4 h with 50 μM cordycepin. Other details are as in B. Mean and SD of three different experiments are shown.
Next, we wondered whether these breaks could also be seen in the form of anaphase bridges and micronucleus accumulation, two well-known biomarkers of chromosomal instability resulting from unrepaired DNA breaks or of interlinked sister chromatid intermediates generated by replication stress. The percentage of anaphase cells containing bridges and the percentage of micronucleus-containing cells were significantly increased after SPT16 and SSRP1 depletion (Fig. 6C,D). Therefore, FACT is needed to maintain genome integrity in human cells, as it occurs in S. cerevisiae.
Finally, to investigate whether DNA breaks in FACT-depleted cells were dependent on transcription, we performed comet assays in HeLa cells in which transcription was inhibited with 3′ deoxyadenosine (cordycepin), a specific inhibitor of RNA chain elongation (Tuduri et al. 2009; Jones et al. 2013). Strikingly, cordycepin fully suppressed the twofold to fourfold increase in comet tail moment of SPT16- and SSRP1-depleted cells (Fig. 6E), supporting the view that, as in yeast cells, DNA breaks in the absence of human FACT are transcription-dependent.
Replication is impaired in FACT-depleted human cells in a transcription-dependent manner
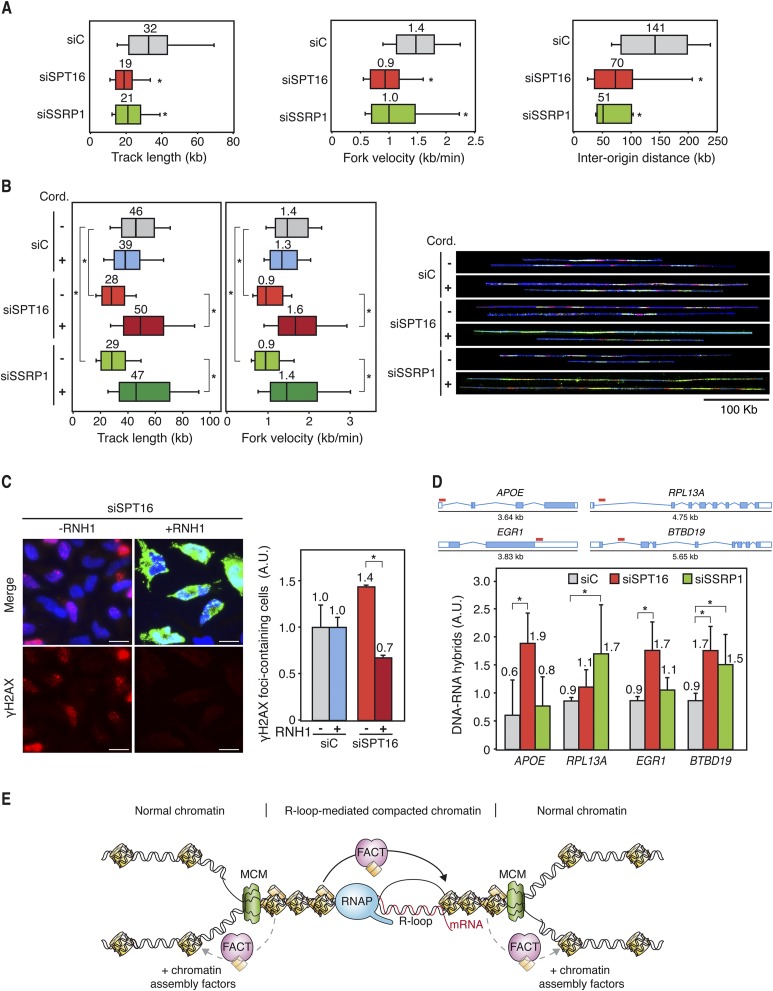
Next, we investigated how RF progression was affected by the down-regulation of SPT16 and SSRP1 in human cells. DNA combing in MRC-5 cells depleted of SPT16 or SSRP1 via siRNA for 72 h revealed shorter replicons and slower replication velocity (Fig. 7A). Similar results were observed in FACT-depleted HeLa cells (Fig. 7B), demonstrating that this phenomenon is not cell type-specific. Interestingly, the reduction in RF velocity is paralleled by an increase in the number of replication origins activated, since the distances between active origins detected by DNA combing are strongly reduced in SPT16- and SSRP1-depleted cells (Fig. 7A), consistent with similar results obtained recently in chicken DT40 cells (Abe et al. 2011).
Figure 7.
Transcription-dependent replication deficiencies in hFACT-depleted cells. (A) Single-molecule analysis of DNA replication (DNA combing) in siC, siSPT16, and siSSRP1 transfected MRC-5 cells. Cells were pulse-labeled for 10 min with IdU followed by 20 min with CldU. Distributions of CldU track lengths, RF velocity, and interorigin distance are shown. Median values are indicated. Boxes and whiskers indicate 25–75 and 10–90 percentiles, respectively. (*) P < 0.05 (median test). (B) DNA combing in siC, siSPT16, and siSSRP1 transfected HeLa cells with or without 100 min of treatment with 50 μM cordycepin. Cells were pulse-labeled for 10 min with IdU (red) followed by 30 min with CldU (green). DNA is shown in blue. Other details are as in A. (C) Immunofluorescence of γH2AX in siC (control) and siSPT16 transfected HeLa cells with or without RNase H1 (RNH1) overexpression. Cells were transfected with pcDNA3 (control) or pcDNA3-RNaseH1 (RNH1) during 24 h. Nuclei were stained with DAPI. Bars, 25 μm. Graphics show the quantification of the percentage of cells containing γH2AX foci relative to the siC (control) in each case. Mean and SEM for two independent experiments are shown. (*) P < 0.05 (Student's t-test). (D) DRIP using the S9.6 antibody, which specifically recognizes DNA:RNA hybrids in siC, siSPT16, and siSSRP1 cells at APOE, RPL13A, EGR1, and BTBD19 genes. Signal values of R-loop detection normalized in respect to the siC control are shown. Signal values from samples treated with RNase H were subtracted. Red lines indicate the regions where DRIP analyses were performed. Mean and SD for four independent experiments are shown. (*) P < 0.05 (Mann-Whitney U-test). (E) Model to explain the molecular defects occurring in FACT-deficient cells. RF progression through R-loop-altered chromatin is facilitated by the FACT complex, which would assist nucleosome swapping to facilitate nucleosome reassembly at transcription–replication collisions sites. For clarity, only the MCM helicase is depicted at the RF.
Replication impairment was not due to the impediment of the RF's progress through spontaneously damaged DNA, since treatment with MMS did not cause further reduction in track length or RF velocity in FACT-depleted cells (Supplemental Fig. S11D). Thus, we wondered whether replication failures were associated with transcription. Analysis of RF progression by DNA combing in HeLa cells in which transcription elongation was inhibited using cordycepin, which inhibits RNA synthesis, revealed that replication track length and velocity were not affected by cordycepin in the siC control. Strikingly, in cordycepin-treated SPT16- and SSRP1-depleted cells, track length and velocity values recovered to the same levels as those of the siC control (Fig. 7B), indicating that the defect in RF progression was transcription-dependent.
R loops accumulate in FACT-depleted human cells
Next, we wondered whether R loops were involved in the genome instability of human cells depleted of FACT. For this, we investigated whether DNA damage accumulation in SPT16-depleted cells was suppressed by overexpression of the human RNase H1. Indeed, the number of cells containing γH2AX foci was reduced in SPT16-depleted cells down to the siC control levels (Fig. 7C). We then assayed whether R loops accumulate in FACT-depleted HeLa cells by direct detection of R loops via DRIP using the S9.6 antibody at several genes previously reported to naturally form R loops in human Ntera2 cells (Ginno et al. 2012, 2013). As can be seen in Figure 7D and Supplemental Figure S12A, R loops accumulate in siC control cells and at higher levels in SPT16- and SSRP1-depleted cells in the analyzed genes (APOE, RPL13A, EGR1, and BTBD19). In addition, ChIP experiments showed that both FACT subunits bind to these regions (Supplemental Fig. S12B). The results suggest that FACT facilitates RF progression at transcribed chromatin, which, when it accumulates R loops, likely associates with specific chromatin modifications.
Discussion
We show that the FACT chromatin-reorganizing complex facilitates RF progression through transcribed DNA in both yeast and human cells. As a consequence, FACT-deficient cells accumulate DNA breaks and show different kinds of genome instability that are in a significant part mediated by R loops. Our study supports the idea that cotranscriptional R loops are formed naturally as a major obstacle of replication and that a key function of FACT during replication is to facilitate the resolution of R-loop-mediated transcription–replication conflicts likely associated with a particular chromatin structure that demands the action of FACT for its reorganization.
The role of FACT as a histone chaperone during transcription is relatively well defined. In vivo and in vitro experiments support the view that FACT destabilizes nucleosomes by disrupting interactions between the DNA and H2A/H2B dimers, thereby facilitating the progression of RNAP through a less repressive chromatin. In addition, FACT activity is known to be important to prevent histone eviction during this process and promote nucleosome recovery afterward (Reinberg and Sims 2006; Formosa 2012). During replication, chromatin is disrupted to allow RF progression, but assembly of new nucleosomes behind the fork requires factors such as ASF1 and CAF1 (Alabert and Groth 2012). FACT's physical interaction with DNA polymerase α (Wittmeyer and Formosa 1997) and its isolation as a RF component in yeast (Gambus et al. 2006) suggest that it participates in nucleosome dynamics during replication.
Here, we demonstrate that replication elongation and initiation are altered in yeast and human FACT-deficient cells by 2D gel electrophoresis, ChIP of BrdU incorporation, and DNA combing assays at transcribed regions (Figs. 3, 7). Accordingly, it has been shown that yeast H2Bub1, the modification that stabilizes Spt16 association with transcribed DNA, is essential to guarantee RF progression and histone deposition during DNA replication, suggesting that H2Bub1 also stabilizes Spt16 on replicating chromatin (Trujillo and Osley 2012). Thus, FACT could also help nucleosome reassembly factors to build new nucleosomes on replicated DNA strands, as proposed in yeast (VanDemark et al. 2006), and has been recently shown to facilitate the reincorporation of histones onto the DNA via its collaboration with Mcm2 replication helicase subunit (Foltman et al. 2013). This model is consistent with our results supporting that FACT could travel with the RF, somehow ensuring its stability. However, despite the fact that RF progression is perturbed in the absence of a functional FACT in Xenopus oocytes in vitro and in chicken DT40 cells (Okuhara et al. 1999; Abe et al. 2011), the function of FACT in preventing RF instability during DNA replication elongation is not well established.
Importantly, FACT assists RFs at transcribed regions, since the replication defect of FACT mutants is not observed all over the genome but mainly at transcribed regions, as determined by the genome-wide ChIP–chip analyses of Rrm3 (Fig. 5; Supplemental Figs. S8, S9). The RF is not only slower but also instable in FACT-defective cells, since genome integrity is highly compromised in the absence of FACT (Figs. 1, 6), which could explain the severe growth defect reported in pob3 mutants in the absence of Rad53 and Mec1 DNA damage checkpoint factors (Schlesinger and Formosa 2000; VanDemark et al. 2006).
One major obstacle for RF progression is the transcription machinery, which has a stronger impact on replication if R loops are also present (Wellinger et al. 2006; Gan et al. 2011). Strikingly, our study reveals that FACT's key function during replication is to assist the RF to overcome transcription conflicts, many of which are R-loop-dependent. Hyperrecombination in yeast FACT mutants is higher at sites where transcription–recombination collisions occur (Fig. 2), and RF progression is impaired in the absence of FACT (Fig. 3). Importantly, replication impairment and DNA break accumulation in human FACT-deficient cells are suppressed when transcription is shut off (Figs. 6E, 7B). Therefore, beyond the idea that FACT's putative role in replication is general, our results indicate that the main function of FACT during RF progression is critical at transcribed chromatin.
R loops may form at a low level at transcribed chromatin but increase by defects in RNA processing, export, and splicing factors (Aguilera and Garcia-Muse 2012). Importantly, the accumulation of recombinogenic DNA breaks in yeast FACT mutants is suppressed by RNH1 overexpression, which eliminates R loops. Furthermore, replication and genome stability are highly compromised in yeast FACT mutants in the absence of THO components, which lead to an increase of R loops (Fig. 4; Supplemental Fig. S5), and the spt16-197 is lethal in combination with the RNA export mutant yra1-1 (Burckin et al. 2005). Furthermore, yeast FACT mutants and FACT-depleted human cells accumulate R loops at high levels, as detected by DRIP (Figs. 5E, 7D). All of these data are consistent with the view that R loops are a major natural obstacle for RF progression. In yeast and human FACT-deficient cells, R loops could trigger genome instability. FACT could thus help the DNA polymerase solve collisions with the transcription machinery in DNA regions prone to form R loops.
It is possible that the inefficient chromatin reorganization during transcription elongation in the absence of FACT could facilitate the formation of R loops that could impair RF progression. However, the fact that R loops are clearly observed in normal cells, that FACT is required during replication when transcription is active, and that FACT is preferentially accumulated in R-loop-rich regions supports the conclusion that a key function of FACT is to reorganize chromatin at transcription–replication collisions, with a major role at regions enriched in R loops. Consistently, a specific enrichment of FACT has been observed in the hypermutable regions of Immunoglobulin genes prone to accumulate R loops (Aida et al. 2013).
Given the function of FACT as a chromatin reorganizer, our study raises the possibility that R loops promote specific chromatin modifications that may demand the obligatory action of FACT for replication to reorganize chromatin. It is worth noting that the thermosensitivity and HU sensitivity of spt16-11 mutant are suppressed by histone mutations that weaken the interface between histones within nucleosomes (McCullough et al. 2011), consistent with the idea that the spt16-11 FACT mutant indeed may have difficulties in reorganizing nucleosomes and thereby may hamper the progression of RF through R-loop-enriched chromatin. Interestingly, R loops have recently been detected at centromeres (Nakama et al. 2012), in which we clearly observed that FACT is required to prevent RF stalling (Fig. 5D). Importantly, we found recently that R loops promote compaction and heterochromatinization of DNA, as detected by histone H3 Ser10 phosphorylation and/or Lys9 dimethylation in yeast, C. elegans, and human cells (Castellano-Pozo et al. 2013). It would be interesting to see whether the FACT relationship to R-loop accumulation might be mediated by histone modifications.
Therefore, we conclude that transcription–replication collisions may be associated with R loops, which cause chromatin compaction as a major obstacle to RF progression. For an efficient replication through these regions, a specific chromatin-reorganizing activity would be required to help reorganize and reassemble chromatin after the RF has passed. We propose a model in which FACT helps the replisome to overcome transcription-mediated obstacles whether they are mediated by R loops or not (Fig. 7E). The need for FACT at the sites of transcription–replication conflicts could be explained by the need to swap nucleosomes from ahead of the RNA and DNA polymerases to behind to allow the progression of both machineries and nucleosome reassembly. In addition, a more compacted chromatin structure formed at the R-loop-containing regions (Castellano-Pozo et al. 2013) would make FACT function critical at those sites to allow replication to resume (Fig. 7E) and, consequently, prevent genome instability.
Materials and methods
Replication analysis in yeast
2D gel electrophoresis of CTAB (cetyltrimethylammonium bromide)-extracted DNA of cultures growing in YEPD was performed as described (Moriel-Carretero and Aguilera 2010). BrdU ChIPs were performed in cells released from G1 in the presence of 200 µg/mL BrdU using monoclonal anti-BrdU (MBL) antibody. For details, see the Supplemental Material.
ChIP–chip experiments
ChIP–chip experiments of yeast asynchronous mid-log cultures were performed after a 26°C–30°C temperature shift (4 h) in the spt16-11 and pob3-7 mutants or 75 min after Spt16 degron induction in spt16-td cells as described (see the Supplemental Material; Gomez-Gonzalez et al. 2011a). High-density oligonucleotide tiling arrays (Affymetrix) were used for the analysis of yeast chromosomes at a 300-base-pair (bp) resolution, each 300-bp region being covered by at least 60 probes. Data were analyzed using the Affymetrix Tiling Array Suite software (TAS). Protein chromosomal distribution was analyzed using binding clusters defined as ranges that respected an estimated signal (IP/SUP-binding ratio) positive in the whole range, a P-value < 0.01, minimum run of 100 bp, and maximum gap of 250 bp. ChIP–chip data can be accessed at the Gene Expression Omnibus database (GSE51653).
Genome instability and replication analysis in human cell lines
Immunofluorescence, comet assay, and DNA combing were performed as described previously (Dominguez-Sanchez et al. 2011). For micronucleus and anaphase bridge analyses, DNA was stained with DAPI. Representative images of DNA fibers were assembled from different microscopic fields of view.
DRIP assays
DNA:RNA hybrids were immunoprecipitated using the S9.6 antibody from gently extracted and enzymatically digested DNA, treated or not with RNase H as described (Ginno et al. 2012). Quantitative PCR was performed at the indicated regions. The relative abundance of DNA:RNA hybrid immunoprecipitated in each region was normalized to the signal at the negative control region SNRPN gene in human cell lines. For details, see the Supplemental Material.
Miscellanea
Strains, plasmids, and primers are shown in the Supplemental Material. Recombination and plasmid loss assays; Rad52-YPF foci formation of mid-log S–G2 cells; FACS of yeast cells; Northern, Western, and ChIP assays; microarray analyses (Gene Expression Omnibus database, GSE54340); cell transfection; image acquisition; data analysis; and quantitative PCR were performed using standard procedures, as indicated in the Supplemental Material.
Acknowledgments
We thank D. Stillman, T. Formosa, and M. Amor-Guéret for reagents; F. Chédin for the DRIP protocol; U. Galindo for technical assistance; E. Andújar and M. Pérez-Alegre (Genomic Unit, CABIMER) for assistance with ChIP–chip and microarray assays; A. Marín for assistance with ChIP–chip analyses; R. Nakato and K. Shirahige for providing Spt16 ChIP sequencing (ChIP-seq) data,; R. Luna for critical reading of the manuscript; and D. Haun for style supervision. Research was funded by grants from the Spanish Ministry of Economy and Competitiveness (Consolider CSD2007-00015 and BFU2010-16372), the Junta de Andalucía (CVI4567), and the European Union (FEDER). E.H.-M. was recipient of a Predoctoral Formación en Investigación en Salud training grant from the Spanish Ministry of Health.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.234070.113.
References
- Abe T, Sugimura K, Hosono Y, Takami Y, Akita M, Yoshimura A, Tada S, Nakayama T, Murofushi H, Okumura K, et al. 2011. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem 286: 30504–30512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A 2002. The connection between transcription and genomic instability. EMBO J 21: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T 2012. R loops: from transcription byproducts to threats to genome stability. Mol Cell 46: 115–124 [DOI] [PubMed] [Google Scholar]
- Aguilera A, Gomez-Gonzalez B 2008. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Aida M, Hamad N, Stanlie A, Begum NA, Honjo T 2013. Accumulation of the FACT complex, as well as histone H3.3, serves as a target marker for somatic hypermutation. Proc Natl Acad Sci 110: 7784–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Groth A 2012. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13: 153–167 [DOI] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 [DOI] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M Jr 2005. Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol 12: 175–182 [DOI] [PubMed] [Google Scholar]
- Castellano-Pozo M, Garcia-Muse T, Aguilera A 2012. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep 13: 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Santos-Pereira JM, Rondon AG, Barroso S, Andujar E, Perez-Alegre M, Garcia-Muse T, Aguilera A 2013. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell 52: 583–590 [DOI] [PubMed] [Google Scholar]
- de la Loza MC, Wellinger RE, Aguilera A 2009. Stimulation of direct-repeat recombination by RNA polymerase III transcription. DNA Repair (Amst) 8: 620–626 [DOI] [PubMed] [Google Scholar]
- Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A 2011. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet 7: e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltman M, Evrin C, De Piccoli G, Jones RC, Edmondson RD, Katou Y, Nakato R, Shirahige K, Labib K 2013. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep 3: 892–904 [DOI] [PubMed] [Google Scholar]
- Formosa T 2012. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta 1819: 247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, Herrera-Moyano E, Aguilera A 2013. Transcription-associated genome instability. Chem Rev 113: 8638–8661 [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X 2011. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes & Dev 25: 2041–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F 2012. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45: 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lim YW, Lott PL, Korf I, Chedin F 2013. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res 23: 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Garcia-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marin A, Foiani M, Aguilera A 2011a. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J 30: 3106–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Ruiz JF, Aguilera A 2011b. Genetic and molecular analysis of mitotic recombination in Saccharomyces cerevisiae. Methods Mol Biol 745: 151–172 [DOI] [PubMed] [Google Scholar]
- Gottipati P, Cassel TN, Savolainen L, Helleday T 2008. Transcription-associated recombination is dependent on replication in mammalian cells. Mol Cell Biol 28: 154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, Petermann E 2013. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene 32: 3744–3753 [DOI] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S 2012. Transcription as a source of genome instability. Nat Rev Genet 13: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378 [DOI] [PubMed] [Google Scholar]
- Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A 2005. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol Cell 18: 711–722 [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol 23: 8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough L, Rawlins R, Olsen A, Xin H, Stillman DJ, Formosa T 2011. Insight into the mechanism of nucleosome reorganization from histone mutants that suppress defects in the FACT histone chaperone. Genetics 188: 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriel-Carretero M, Aguilera A 2010. A postincision-deficient TFIIH causes replication fork breakage and uncovers alternative Rad51- or Pol32-mediated restart mechanisms. Mol Cell 37: 690–701 [DOI] [PubMed] [Google Scholar]
- Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y 2012. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 17: 218–233 [DOI] [PubMed] [Google Scholar]
- Okuhara K, Ohta K, Seo H, Shioda M, Yamada T, Tanaka Y, Dohmae N, Seyama Y, Shibata T, Murofushi H 1999. A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr Biol 9: 341–350 [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116 [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A 2005. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J 24: 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D, Sims RJ 3rd 2006. De FACTo nucleosome dynamics. J Biol Chem 281: 23297–23301 [DOI] [PubMed] [Google Scholar]
- Santos-Pereira JM, Herrero AB, Garcia-Rubio ML, Marin A, Moreno S, Aguilera A 2013. The Npl3 hnRNP prevents R-loop-mediated transcription–replication conflicts and genome instability. Genes Dev 27: 2445–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, Reinberg D, Lis JT 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301: 1094–1096 [DOI] [PubMed] [Google Scholar]
- Schlesinger MB, Formosa T 2000. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 155: 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42: 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, Hieter P 2012. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev 26: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Chien CT, Hirose S, Lee SC 2006. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J 25: 3975–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Osley MA 2012. A role for H2B ubiquitylation in DNA replication. Mol Cell 48: 734–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11: 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP, Formosa T 2006. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell 22: 363–374 [DOI] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M 2011. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell 44: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A 2006. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol 26: 3327–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J, Formosa T 1997. The Saccharomyces cerevisiae DNA polymerase α catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol 17: 4178–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]