Abstract
Adenosine-to-inosine (A-to-I) RNA editing, in which genomically encoded adenosine is changed to inosine in RNA, is catalyzed by adenosine deaminase acting on RNA (ADAR). This fine-tuning mechanism is critical during normal development and diseases, particularly in relation to brain functions. A-to-I RNA editing has also been hypothesized to be a driving force in human brain evolution. A large number of RNA editing sites have recently been identified, mostly as a result of the development of deep sequencing and bioinformatic analyses. Deciphering the functional consequences of RNA editing events is challenging, but emerging genome engineering approaches may expedite new discoveries. To understand how RNA editing is dynamically regulated, it is imperative to construct a spatiotemporal atlas at the species, tissue and cell levels. Future studies will need to identify the cis and trans regulatory factors that drive the selectivity and frequency of RNA editing. We anticipate that recent technological advancements will aid researchers in acquiring a much deeper understanding of the functions and regulation of RNA editing.
One striking observation from the Human Genome Project is that only ~20,000 protein-coding genes are present in humans, a surprisingly low number that does not scale with human developmental and cognitive complexity1. Part of the answer to this apparent paradox lies in the complexity of RNA, including alternative splicing and non-coding RNA. RNA editing, a co-transcriptional process in which the genome-encoded information is altered in RNA, provides a potentially powerful method for diversifying the transcriptome and fine-tuning biological function2.
A-to-I editing, the most common type of editing known in animals, leads to the recognition of inosine as guanosine by the translation, splicing and sequencing machineries2. A-to-I editing is carried out by the ADAR family of enzymes2, which are conserved across metazoans. All ADARs share double-stranded RNA (dsRNA)-binding domains and catalytic deaminase domains that deaminate A to I. Here we discuss recent progress and future needs for studying A-to-I RNA editing from the perspectives of identification, evolution, function, atlas and regulation (Fig. 1).
Figure 1.
Overview. We highlight the major, but not all, discussion topics.
ADAR mutants exhibit neural and behavioral phenotypes
Inactivation of ADAR family members results in mainly neuronal and behavioral phenotypes. Knockout of the single Drosophila melanogaster ADAR gene leads to brain-related phenotypes such as uncoordinated locomotion, temperature-sensitive paralysis and age-dependent neurodegeneration3. When both of its ADAR genes are deleted, Caenorhabditis elegans exhibits defective chemotaxis4. Mammalian genomes carry three members of the ADAR family, and enzymatic activity has been demonstrated for ADAR1 and ADAR2, but not ADAR3 (also known as ADAR, ADARB1 and ADARB2, respectively)5. In mice, ADAR1 is required for embryonic development, as Adar1−/− embryos die between E11.5 and E12.5 as a result of failed hematopoiesis maintenance6,7. ADAR2 is also required, as null mutants develops epileptic seizures and die several weeks after birth8. Although ADAR1 and ADAR2 are expressed in many tissues, the inactive ADAR3 is expressed exclusively in the brain5. It is intriguing to speculate on potential functions of ADAR3 in the brain.
Dysregulation of RNA editing in neurological diseases
Aberrant RNA editing has been linked with a variety of diseases, primarily neurological or psychiatric disorders. As of several years ago, only a handful of RNA editing sites had been identified and examined; however, these sites were found to be dysregulated in many diseases2,9. For example, the editing level (fraction of edited transcripts) of the glutamate receptor GluA2 (also known as GluR2, GluR-B and Gria2) Q/R site might be altered in epilepsy, amyotrophic lateral sclerosis, malignant glioma, schizophrenia and ischemia2,9. In the serotonin receptor 5-HT2CR, there are five editing sites located within 32 bases, giving rise to 24 protein isoforms10. Dysregulated 5-HT2CR (HTR2C) mRNA editing might be involved in depression and suicide, schizophrenia, Prader-Willi syndrome, and metabolic diseases such as obesity and diabetes2,9,11. Although RNA editing in GluA2 and 5-HT2CR has functional consequences under physiological conditions, the manner in which altered editing might be involved in disease pathogenesis needs to be further studied. More recently, several studies examined a larger number of sites, and the editing level of a number of these sites were found to be altered in schizophrenia, bipolar disease12, autism13 and cancers14, although replication experiments with a larger cohort may be needed to confirm these findings.
Although these results establish links between RNA editing and a number of diseases involved with the nervous system, building a functional relationship requires the following considerations. First, with the recent explosion of newly identified RNA editing sites (see below), a comprehensive screening of many, if not all, of the sites in disease contexts will provide an unbiased assessment. Second, a large number of cases and controls are needed to achieve high statistical power, particularly to pinpoint relatively subtle differences. Third, animal models of disease may be helpful because fair comparisons are possible between ‘cases’ and controls that share the same genetic background and environmental conditions. Fourth, functional in vitro or in vivo assays are needed to perturb the editing levels and correlate with disease phenotypes, as exemplified in previous work8,11,14.
Identification of RNA editing sites: a rapid expansion
The first handful of mammalian A-to-I RNA editing sites were identified serendipitously in glutamate and serotonin receptors10,15. The difficulty in finding other editing sites was primarily a result of the limitations of the sequencing technology at the time, particularly in generating the coverage necessary to delineate the single-base differences between genomic DNA and RNA. In the early 2000s, large sequencing efforts to generate expressed sequence tags (ESTs) of genomes and transcriptomes made it possible to identify RNA editing sites across the transcriptome. This is exemplified by searches using comparative genomics approaches in Drosophila16 and mammals17. However, the success of these screens was limited, mostly as a result of the relatively small amount of data available at the time and the difficulty of distinguishing RNA editing sites from genomic single nucleotide polymorphisms (SNPs). In contrast with this limited success in identifying nonrepetitive sites, several groups identified thousands of clustered editing sites in Alu repeats18–20. Primate-specific Alu repeats are heavily edited because they comprise ~1.2 million copies occupying >10% of the human genome, leading to prevalent double-stranded RNA when in inverted repeats. However, the function of Alu editing sites is largely unknown.
Next-generation sequencing is particularly helpful for comprehensively identifying RNA editing sites. In the early days of next- generation sequencing, when whole human genome and transcriptome sequencing was not feasible, we developed a targeted capture technique to sequence ~36,000 human sites that were computationally predicted to be candidates of RNA editing sites and identified hundreds of new A-to-I sites21. Recently, several groups independently developed methods for identifying RNA editing sites by comparing the genome and RNA sequencing data of the same individuals22. Unfortunately, a large number of the sites identified were noncanonical RNA editing events, which were shown to be mainly technical artifacts23–25. In our work, we were unable to identify and confirm these noncanonical RNA editing sites26,27. The false discovery of noncanonical sites is mainly derived from errors in the sequencing library preparation and the sequencing process, as well as the computational mapping of RNA sequencing (RNA-Seq) reads to the genome and transcriptome. In comparison with mapping genome sequencing reads, it is more challenging to map RNA-Seq reads to identify variants at the single-base resolution for two main reasons: the errors introduced by the random hexamer that is often used to make RNA-Seq libraries and the various RNA splicing events expressed at different levels23–25. All of the methods require both matched genome sequencing data along with the RNA-Seq data to effectively distinguish RNA editing sites from SNPs. However, many of the samples with RNA-Seq data did not have the genome sequencing data. In fact, samples with matched genome and RNA sequencing data were mostly lymphoblastoid cell lines that were used in the 1000 Genomes Project.
To fully utilize publicly available RNA-Seq data, such as those from brain tissues where RNA editing is more prevalent, we recently developed an approach for identifying RNA editing sites using only RNA-Seq data, without matched genome sequencing data. To distinguish RNA editing sites from SNPs, we developed two different approaches28. In the first approach, multiple human RNA-Seq data sets are used, RNA variants are called and known SNPs are removed. RNA editing events are enriched because rare SNPs are unlikely to be shared by different individuals. In the second approach, RNA-Seq data sets from related species are used. The RNA editing sites conserved between species are identified because SNPs independently occur in different species and are therefore not shared between species. Using these strategies allowed us to substantially expand the number of identified editing sites, as our methods permitted us to analyze a large number of samples for which RNA-Seq data, but not genome sequencing data, exists.
When RNA editing sites are identified using computational approaches described above, one often requires a high fraction (for example, >80%) among all variants to be A-to-G variants, indicative of A-to-I RNA editing. At the 80% A-to-G fraction, the false discovery rate is estimated to be ~2% (refs. 26,28). To further confirm the computational predictions, biochemical evidence will be needed. For example, using RNA-Seq data from Drosophila wild-type and ADAR knockout lines, we estimated that the false discovery rate of our computational prediction was 1.8% (ref. 28).
Over 1.4 million human RNA editing sites have been identified (this number is expected to continue growing) and the vast majority of them are located in noncoding sequences in Alu repeats28. Only about 200 sites have been identified in nonrepetitive protein coding regions, of which ~60% result in amino acid changes. The most recently identified recoding sites tend to have lower editing levels, suggesting that recoding sites that are highly or moderately edited are saturated. However, these lowly edited sites could be highly edited in specific tissues and/or cell types. Sites that are variably edited in different tissues or cell types may have specialized functionality and therefore be potentially interesting.
The recoding RNA editing sites are enriched in neuronal genes. This is not observed for the set of all genes containing RNA editing sites in Alu repeats. However, if one focuses on the subset of human genes harboring Alu repeat editing sites that are conserved between human and other primates, they are also enriched for neuronal functions28. Future work is needed to determine how the editing events in neuronal genes may affect neural functions.
RNA editing as a driving force of brain evolution?
Although many RNA editing sites, particularly recoding sites, are shared between human and mouse, the intensity of RNA editing in human is 35-fold higher than that in mouse19. This is a result of the widespread RNA editing derived from primate-specific Alu sequences. In primates, the amount of editing appears to have increased during primate evolution, based on the examination of six Alu repeats29. Human and non-human primates have highly similar genomes, but differ substantially in cognitive capacities. Given the findings of this recent study and the important roles of RNA editing in brain-related functions, it is plausible that the expansion of RNA editing in humans may have led to increased cognition and driven neural evolution.
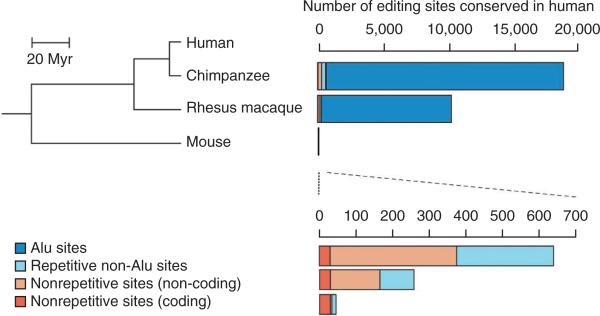
An unbiased, large-scale comparison of RNA editing events between human and non-human primates is needed to test this hypothesis. In a comparative transcriptome study, we found that the number of RNA editing sites in a given species that are shared with human editing sites decreases with increased phylogenetic distance28 (Fig. 2). Although our data indicate that many human RNA editing sites are absent in non-human primates, they cannot exclude the possibility that many of the non-human primate sites are also absent in human. Studying the evolution of RNA editing will reveal not only the functional sites that are conserved between species, but also the sites gained, lost or differentially edited across species. This provides a list of plausible candidates for future study for their potential roles in human brain cognition. Furthermore, this rich data set, when generated, will facilitate understanding of why editing events differ between human and other primates.
Figure 2.
Decreased number of RNA editing sites shared with human sites with increased phylogenetic distance. Phylogenetic relationships between human, chimpanzee, rhesus macaque and mouse are shown on the left. Myr, million years ago. Numbers of A-to-I editing sites that are conserved between human and each of the other three species are shown on the right, with the numbers in non-Alu regions magnified at the bottom. The number of editing sites conserved in human (indicated in the scales) is based on the analysis of the same number of reads from each species28. Figure is adapted from ref. 28.
In addition to studying natural variations in RNA editing between humans and other primates, a complementary approach is to alter RNA editing using genome engineering approaches. For example, given that RNA editing correlates spatiotemporally with higher cognitive functions, it would be informative to introduce specific RNA editing sites into non-human primates and determine whether they are sufficient to alter cognitive function.
Interpretation of the functions of RNA editing sites
RNA editing in the mRNA can yield multiple protein isoforms. The vast majority of the editing events occur in non-coding regions. These events could be involved in many aspects of RNA metabolism, such as splicing, trafficking, microRNA binding and ribosome binding efficiency. However, very little is currently known in the field, probably as a result of the technical challenges of deciphering the functions of an individual RNA editing event. Most of the methods widely used to study gene function (for example, RNA interference) are not suited to studying the function of an editing site. Overexpression of edited and unedited isoforms in a heterologous system provides an important first step in regards to examining the functional consequences of RNA editing events30. Until now, the gold-standard method of choice was to generate a knock-in animal with either abolished or constitutive RNA editing4,11. Because RNA editing occurs in dsRNA structures recognized by ADARs, the flanking sequence of an editing site has to be paired with an editing complementary sequence. To abolish RNA editing in a coding exon, one can remove the editing complementary sequence that may be present in the intron, thereby not directly affecting the coding regions. To produce constitutive RNA editing, one would change the editing site from adenosine to guanosine at the genomic DNA level. One classic example is the Q/R site in the GluA2 gene; when editing is abolished in mice, excess influx of Ca2+ into neurons leads to seizures and postnatal death31. Another example is the G protein–coupled serotonin receptor 5-HT2CR, which has five recoding sites located within 13 bases. In knock-in mice with constitutive RNA editing, the sympathetic nervous system is constitutively activated, the energy expenditure is enhanced and the receptor–G protein coupling is blunted, which is masked by a higher density of receptor binding sites11,32. Despite previous success, the conventional approach is very tedious, time-consuming and costly. Recent technological advancements in genome engineering, such as TALEN and CRISPR-based approaches33, have the potential to expedite studies using the knock-in mouse approach.
Given the fact that most human RNA editing sites are not present in mouse28, it is important to establish in vitro methods using human cells as the model system. In addition to using the TALEN and CRISPR-based approaches, a recently developed method using an antisense oligoribonucleotide was demonstrated to effectively inhibit RNA editing34. Alternatively, overexpression of either edited or unedited transcripts may also lead to informative phenotypes14.
Toward a spatiotemporal atlas of RNA editing
RNA editing is known to be a dynamically regulated process21,35,36. Constructing a spatiotemporal quantitative atlas of RNA editing would be a first step in understanding its dynamic regulation. The recent explosion of identified RNA editing sites in human and other species empowers a large-scale study to create an atlas of RNA editing. RNA sequencing allows for measurement of editing frequency at the transcriptome level, although the accuracy may be compromised for sites in lowly expressed genes. Alternatively, targeted approaches to sequences only those regions that harbor RNA editing sites (particularly those in functionally important regions) would enhance the sequencing accuracy in lowly expressed genes and lower the cost21,35, although current techniques need to be improved.
Previous studies have suggested that most of the sites are edited at low levels in all tissues or cell types (<20%)26,28,37. However, the lowly edited sites may be highly edited under different conditions. It appears that RNA editing is more frequent in brain tissues than in non-brain tissues, with more RNA editing sites and higher editing levels21,28. An RNA editing atlas will provide an unbiased view of RNA editing sites at the transcriptome level in brain and non-brain tissues. In addition, one can isolate different regions of the brain, which may be useful in linking RNA editing levels to specialized tasks governed by different brain regions.
At the individual cell type or single cell level, an RNA editing atlas will further reveal the dynamics of RNA editing that may not be fully appreciated at the tissue or organ levels. For example, there are hundreds of neuronal cell types with different functions in the human brain. In animal models, RNA from particular cell types can be isolated using a variety of techniques developed in mouse38, fly and worm39. Furthermore, single neurons can be isolated by laser capture or microfluidic devices and analyzed by transcriptome sequencing. RNA editing might differ radically between two cells that are similar morphologically and adjacent anatomically, or even vary subcellularly. The function of such variation may be more evident in the context of the cell types involved and/or their spatial relations. Single-cell RNA sequencing, fluorescent in situ sequencing of mRNA molecules40 and in vivo visualization of ADAR activity41 could help to illuminate such issues. These technologies may be in their infancy, but the low signal-to-noise ratio may be overcome by sampling a large number of cells. As these technologies become more mature and are adopted by more research groups, a higher resolution atlas of RNA editing will be achieved.
Understanding cis and trans regulation of RNA editing
RNA editing is a dynamically regulated process, but its regulation is poorly understood. What determines which sites are edited and at what level? At the cis regulation level, the flanking sequences affect the selectivity of editing sites and the 5′ and 3′ flanking sequences appear to be most influential42. However, these studies are often based on in vitro assays of a very limited number of substrates. In addition to the flanking sequence motifs, the local RNA structure is important, as the double-stranded RNA structure is required for ADAR binding. In addition to the stem-loop structure formed in the vicinity of an editing site, a genomically distant RNA sequence or structure can also affect ADARs editing preference43. Looking forward, new in silico, in vitro and in vivo approaches are needed to determine the RNA and ADAR protein structures to characterize ADAR selectivity or preference.
At the trans regulation level, ADARs have an important role. However, ADAR RNA and protein levels do not fully account for the spatiotemporal changes of RNA editing35, suggesting that other proteins are involved in modulating ADAR activity. It is poorly understood which other proteins beyond ADARs are involved in this regulation. The transcription factor cAMP response element–binding protein, or CREB, can induce ADAR2 expression in hippocampal CA1 neurons in rat brain44. Recently, two proteins, Pin1 and WWP2, were found to coordinately regulate ADAR2 post-translationally45. Pin1 positively regulates ADAR2 activity by binding to ADAR2 in a phosphorylation-dependent manner. WWP2 negatively regulates ADAR2 by a protein interaction that results in ubiquitination and subsequent degradation of ADAR2. In addition, two regulators have been recently revealed using Drosophila. One is the Fragile X protein, FMRP, which biochemically and genetically interacts with Drosophila ADAR and subsequently modulates RNA editing levels46. The other is Period, which is involved in circadian rhythm. The editing levels of a number of editing sites are substantially changed in the Period loss-of-function mutant47, although the relationship between ADAR and Period awaits further study.
However, barring these few examples, the number and identity of the other proteins involved in trans regulation remain mostly unknown. Thus, systematic approaches need to be developed to carry out unbiased searches of additional regulators. The fact that some sites are specifically edited in brain or other tissues hints at the existence of tissue-specific regulators that need to be identified. It is conceivable that a number of cofactors are involved in fine-tuning RNA editing levels, giving rise to its wide spectrum of dynamics. Recently, primary screens using a yeast-based RNA editing assay led to the identification of a handful of enhancers and repressors involved in ADAR RNA editing48,49. However, such screenings using yeast are suboptimal as a result of the lack of endogenous ADARs in yeast. Using metazoan systems in which ADARs are conserved, future studies may yield additional factors underlying the trans regulation of RNA editing.
Conclusions
Rapid technological developments have put us in an unprecedented position to study RNA editing. The DNA and RNA sequencing technologies make it easy to identify and measure RNA editing in different physiological and pathological scenarios. This will generate a large number of hypotheses that correlate RNA editing with biological functions. Functional studies are made possible by perturbing the status of RNA editing using genome engineering and synthetic biology. Although it is well established that A-to-I RNA editing is catalyzed by ADARs via binding to double-stranded RNA substrates, there is still much work to be done to define the code underlying the rules of cis and trans regulation. New approaches for systematically studying protein and RNA interactions will facilitate this endeavor.
The pursuit of RNA editing studies will be synergized by the recent initiative of the Brain Activity Map50. The new tools developed by the initiative will undoubtedly enable deeper understanding of RNA editing. Given the critical roles of RNA editing in brain activity, it is also important to note that understanding the function and regulation of RNA editing at single-nucleotide resolution may require the development of additional tools.
With the exciting development of technologies that are currently or will soon be available, we expect that the prime time for studying RNA editing is yet to come. The seemingly subtle differences introduced in the transcriptome, which have been largely overlooked for decades, will continue to surprise us and lead us to fully appreciate it as a fine-tuning mechanism, particularly in the complex nervous system.
ACKNOWLEDGEMENTS
We thank members of the Li laboratory for discussions and critical reading of the manuscript, and particularly R. Zhang for making Figure 2. This work was funded by US National Institutes of Health (GM102484) and the Ellison Medical Foundation (to J.B.L.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 2.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 4.Tonkin LA, et al. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CX, et al. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 7.Hartner JC, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 9.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 11.Kawahara Y, et al. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J. Neurosci. 2008;28:12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silberberg G, Lundin D, Navon R, Ohman M. Deregulation of the A-to-I RNA editing mechanism in psychiatric disorders. Hum. Mol. Genet. 2012;21:311–321. doi: 10.1093/hmg/ddr461. [DOI] [PubMed] [Google Scholar]
- 13.Eran A, et al. Comparative RNA editing in autistic and neurotypical cerebella. Mol. Psychiatry. 2012;18:1041–1048. doi: 10.1038/mp.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 16.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 17.Levanon EY, et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 19.Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 22.Li M, et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinman CL, Majewski J. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1209658. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 24.Pickrell JK, Gilad Y, Pritchard JK. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1210484. author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin W, Piskol R, Tan MH, Li JB. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1210419. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 26.Ramaswami G, et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piskol R, Peng Z, Wang J, Li JB. Lack of evidence for existence of noncanonical RNA editing. Nat. Biotechnol. 2013;31:19–20. doi: 10.1038/nbt.2472. [DOI] [PubMed] [Google Scholar]
- 28.Ramaswami G, et al. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paz-Yaacov N, et al. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc. Natl. Acad. Sci. USA. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomeli H, et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 31.Brusa R, et al. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 32.Olaghere da Silva UB, et al. Impact of RNA editing on functions of the serotonin 2C receptor in vivo. Front. Neurosci. 2010;4:26. doi: 10.3389/neuro.23.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizrahi RA, Schirle NT, Beal PA. Potent and selective inhibition of A-to-I RNA editing with 2′-O-methyl/locked nucleic acid–containing antisense oligoribonucleotides. ACS Chem. Biol. 2013;8:832–839. doi: 10.1021/cb300692k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahlstedt H, Daniel C, Enstero M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjana NE, Levanon EY, Hueske EA, Ambrose JM, Li JB. Activity-dependent A-to-I RNA editing in rat cortical neurons. Genetics. 2012;192:281–287. doi: 10.1534/genetics.112.141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng Z, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 38.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steiner FA, Talbert PB, Kasinathan S, Deal RB, Henikoff S. Cell type– specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 2012;22:766–777. doi: 10.1101/gr.131748.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ke R, et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 41.Jepson JE, Savva YA, Jay KA, Reenan RA. Visualizing adenosine-to-inosine RNA editing in the Drosophila nervous system. Nat. Methods. 2012;9:189–194. doi: 10.1038/nmeth.1827. [DOI] [PubMed] [Google Scholar]
- 42.Eggington JM, Greene T, Bass BL. Predicting sites of ADAR editing in double-stranded RNA. Nat. Commun. 2011;2:319. doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daniel C, Veno MT, Ekdahl Y, Kjems J, Ohman M. A distant cis acting intronic element induces site-selective RNA editing. Nucleic Acids Res. 2012;40:9876–9886. doi: 10.1093/nar/gks691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng PL, et al. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006;49:719–733. doi: 10.1016/j.neuron.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 45.Marcucci R, et al. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 2011;30:4211–4222. doi: 10.1038/emboj.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhogal B, et al. Modulation of dADAR-dependent RNA editing by the Drosophilafragile X mental retardation protein. Nat. Neurosci. 2011;14:1517–1524. doi: 10.1038/nn.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hughes ME, Grant GR, Paquin C, Qian J, Nitabach MN. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012;22:1266–1281. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garncarz W, Tariq A, Handl C, Pusch O, Jantsch MF. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol. 10. 2013:192–204. doi: 10.4161/rna.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tariq A, et al. RNA-interacting proteins act as site-specific repressors of ADAR2-mediated RNA editing and fluctuate upon neuronal stimulation. Nucleic Acids Res. 2013;41:2581–2593. doi: 10.1093/nar/gks1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alivisatos AP, et al. Neuroscience. The brain activity map. Science. 2013;339:1284–1285. doi: 10.1126/science.1236939. [DOI] [PMC free article] [PubMed] [Google Scholar]